Abstract
Phosphatidylinositol 4,5-bisphosphate (PI4,5P2) modulates a plethora of cytoskeletal interactions that control the dynamics of actin assembly and, ultimately, cell migration. We show that the type Iγ phosphatidylinositol phosphate kinase 661 (PIPKIγ661), an enzyme that generates PI4,5P2, is required for growth factor but not G protein–coupled receptor–stimulated directional migration. By generating PI4,5P2 and regulating talin assembly, PIPKIγ661 modulates nascent adhesion formation at the leading edge to facilitate cell migration. The epidermal growth factor (EGF) receptor directly phosphorylates PIPKIγ661 at tyrosine 634, and this event is required for EGF-induced migration. This phosphorylation regulates the interaction between PIPKIγ661 and phospholipase Cγ1 (PLCγ1, an enzyme previously shown to be involved in the regulation of EGF-stimulated migration). Our results suggest that phosphorylation events regulating specific PIPKIγ661 interactions are required for growth factor–induced migration. These interactions in turn define the spatial and temporal generation of PI4,5P2 and derived messengers required for directional migration.
Introduction
Phosphatidylinositol 4,5-bisphosphate (PI4,5P2) has been implicated in many biological processes, including vesicular trafficking (Downes et al., 2005), secretion (Martin, 2001), focal adhesion and cytoskeleton assembly (Ling et al., 2006), regulation of ion channels (Delmas et al., 2005), and nuclear signaling pathways (Gonzales and Anderson, 2006). PI4,5P2 has a role not only as a substrate of PLC and phosphoinositol 3-kinase–mediated second messenger production, but also as a direct effector that binds to and regulates the function of many PI4,5P2-interacting proteins (Anderson and Marchesi, 1985; Heck et al., 2007). The generation of PI4,5P2 in cells primarily occurs through the phosphorylation of phosphatidylinositol(4) phosphate (PI[4]P) by type I phosphatidylinositol phosphate kinases (PIPKI; Doughman et al., 2003a). Three isoforms of PIPKI (Iα, Iβ, and Iγ) have been characterized along with several splice variants. By associating with their unique binding partners, different PIPKI isoforms produce PI4,5P2 with distinct subcellular distributions, from which they perform individual biological functions (Anderson et al., 1999; Coppolino et al., 2002; Doughman et al., 2003b; Heck et al., 2007).
Cell migration requires the coordination of many biochemical events, including organized adhesion formation and turnover as well as dynamic cytoskeletal rearrangements (Horwitz and Parsons, 1999; Webb et al., 2002). PI4,5P2 binds to and regulates many proteins that are crucial for the assembly of the migratory machinery. For example, PI4,5P2 regulates reorganization of the actin cytoskeleton by associating with α-actinin, WASP/N-WASP, gelsolin, cofilin, profilin, and villin (Niggli, 2005; Ling et al., 2006). PI4,5P2 has also been proposed to regulate adhesions by binding to and modulating talin, vinculin, ezrin/radixin/moesin, calpain, and other proteins involved in adhesion dynamics (Niggli, 2005; Ling et al., 2006). PI4,5P2 is therefore positioned to play key roles in migration by modulating adhesion dynamics and cytoskeleton rearrangement.
Many observations indicate that PI4,5P2 is a key signaling molecule in the regulation of cell migration, yet the role of specific PIP kinases in the regulation of cell migration remains to be clarified. PIPKIγ is alternatively spliced in cells, resulting in at least two major variants, PIPKIγ635 and PIPKIγ661, which differ by a 26-amino-acid C-terminal extension (Ishihara et al., 1998). Most interesting, the 26-amino-acid C-terminal extension binds to talin and targets PIPKIγ661, but not PIPKIγ635, to adhesions (Di Paolo et al., 2002; Ling et al., 2002). This specific targeting of PIPKIγ661 allows for the generation of PI4,5P2 at adhesions, which is known to enhance the association between integrin and talin (Martel et al., 2001).
The binding of talin to β-integrin enhances the affinity of integrin for its ligands and activates the integrin heterodimer (Tadokoro et al., 2003). As a result, PIPKIγ661 may manipulate the inside-out activation of integrin signaling and the adhesion formation through its association with talin. Other than facilitating the assembly of talin into adhesions, PIPKIγ661 may also influence the recruitment and activation of other adhesion components through the local generation of PI4,5P2. These combined data indicate that PIPKIγ661 may play key roles in regulating adhesion dynamics that are critical for cell migration.
Unlike PI3,4,5P3, total levels of cellular PI4,5P2 are relatively high and undergo only modest changes upon stimulation of cell migration (Ling et al., 2006). This suggests that PI4,5P2 generation is tightly controlled, both spatially and temporally, to fulfill the requirements of rapid adhesion turnover and cytoskeleton rearrangement that are critical to the process of cell migration. Here, we demonstrate that PIPKIγ661 is required specifically for growth factor–stimulated directional migration, supporting a role for PIPKIγ661 in generation of the PI4,5P2 required for cell migration toward an EGF concentration gradient.
Results
Knockdown of PIPKIγ661 attenuates EGF but not lysophosphatidic acid (LPA)– or stromal cell–derived factor (SDF) 1α–stimulated migration
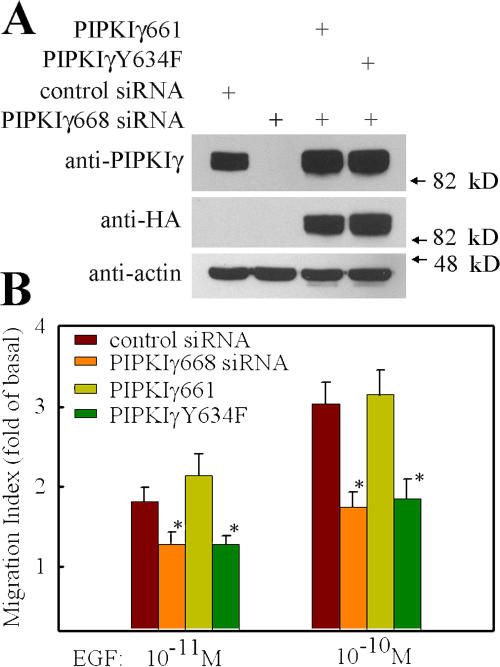
Previous studies have demonstrated that PIPKIγ661 is targeted very specifically to adhesions (Di Paolo et al., 2002; Ling et al., 2002; Calderwood et al., 2004). The localized generation of PI4,5P2 at adhesions has been proposed to have roles in both integrin activation and adhesion formation (Ling et al., 2006), and these events are key for cell migration. To explore the possible role of PIPKIγ661 in cell migration, siRNA specifically targeting PIPKIγ668 (human homologue of mouse PIPKIγ661) was designed. As shown in Fig. S1 A (available at http://www.jcb.org/cgi/content/full/jcb.200701078/DC1), PIPKIγ668-specific siRNA could specifically knock down expression of PIPKIγ668 but had no effect on the expression of PIPKIγ640 (human homologue of PIPKIγ635).
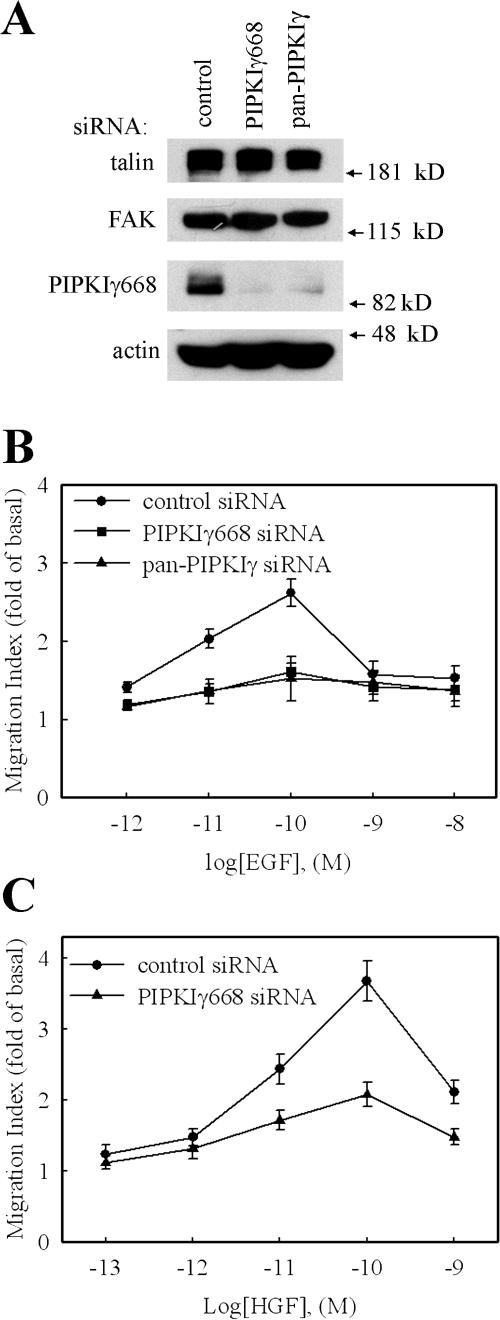
The effect of PIPKIγ knockdown on EGF-stimulated cell migration was quantified using a modified Boyden chamber transwell assay (Neptune and Bourne, 1997; Sun et al., 2002). As shown in Fig. 1 B, the knockdown of global PIPKIγ by pan-PIPKIγ siRNA blocked EGF-stimulated migration of HeLa cells. To determine if this effect is specifically due to the knockdown of the PIPKIγ661 splice variant, PIPKIγ668-specific siRNA was used in the same assay. Specific PIPKIγ668 knockdown had the same effect as global PIPKIγ knockdown on attenuation of EGF-stimulated migration of HeLa cells (Fig. 1 B). The efficacy of these two siRNAs on PIPKIγ668 knockdown is shown in Fig. 1 A; the expression of PIPKIγ668 in HeLa cells was efficiently knocked down by both of these two siRNAs without affecting the expression level of other proteins, such as talin, FAK, and actin. To distinguish the role of PIPKIγ668 in directed migration or random migration, a checkerboard assay was used. As shown in Fig. S1 B, knockdown of PIPKIγ668 attenuated EGF-induced directional migration but not random migration. These results provide the first evidence that PIPKIγ661 plays a role in directional migration. Equivalent results were observed using MtLn3 and A431 cell lines (unpublished data), confirming the observation that PIPKIγ661 is required for EGF-stimulated cell migration in these cells as well.
Figure 1.
Knockdown of PIPKIγ attenuated EGF-stimulated cell migration. HeLa cells were transfected with control siRNA, pan-PIPKIγ siRNA, or PIPKIγ668-specific siRNA separately as indicated. (A) Expression of PIPKIγ668, talin, FAK, and actin were detected by their specific antibodies. (B) EGF (10−12 to 10−8 M) was used to stimulate cell migration. (C) HGF (10−13 to 10−9 M) was used to stimulate cell migration. Quantifications are means ± SEM of three separate experiments.
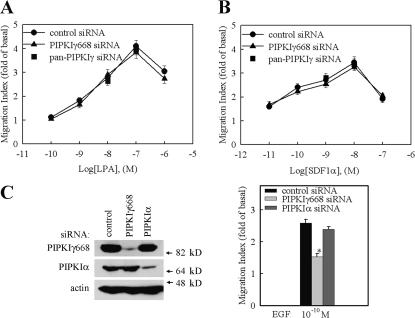
To determine if the function of PIPKIγ661 in regulating migration is specific for EGF, the effect of PIPKIγ661 knockdown on migration induced by other chemoattractants, such as hepatocyte growth factor (HGF), LPA, and SDF1α, was investigated. HGF-induced cell migration was blocked by PIPKIγ661 knockdown (Fig. 1 C). This result further confirmed the role of PIPKIγ661 in growth factor–induced migration. Remarkably, the results from these migration assays showed that the knockdown of PIPKIγ661 did not affect LPA- or SDF1α-stimulated cell migration (Fig. 2, A and B; and Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200701078/DC1). SDF1α is the ligand of CXCR4 chemokine receptor. Both CXCR4 and LPA receptor belong to the G protein–coupled receptor family. These data indicate that PIPKIγ661 plays a specific role in growth factor receptor–stimulated cell migration.
Figure 2.
The specificity of PIPKIγ in regulating cell migration. HeLa cells were transfected with control siRNA or PIPKIγ668-specific siRNA separately as indicated, and LPA (A) or SDF1α (B) was used to stimulate cell migration. (C) HeLa cells were transfected with control siRNA or PIPKIγ668- or PIPKIα-specific siRNA separately as indicated, and EGF was used to stimulate cell migration. Expression of PIPKIγ668, PIPKIα, and actin was detected by their specific antibodies. Quantifications are expressed as means ± SEM of three separate experiments (*, P < 0.01 compared with control siRNA–transfected HeLa cells).
In addition, the effect of PIPKIα knockdown on EGF-stimulated migration was also investigated. siRNAs specifically targeting PIPKIα were used to knock down the expression of PIPKIα in HeLa cells. As shown in Fig. 2 C, ∼85% of endogenous PIPKIα was knocked down by PIPKIα-specific siRNA in HeLa cells. However, the knockdown of PIPKIα did not affect EGF-stimulated migration (Fig. 2 C), demonstrating specificity for the PIPKIγ661 isoform.
EGF-stimulated cell migration requires PIPKIγ661 kinase activity and its talin binding ability
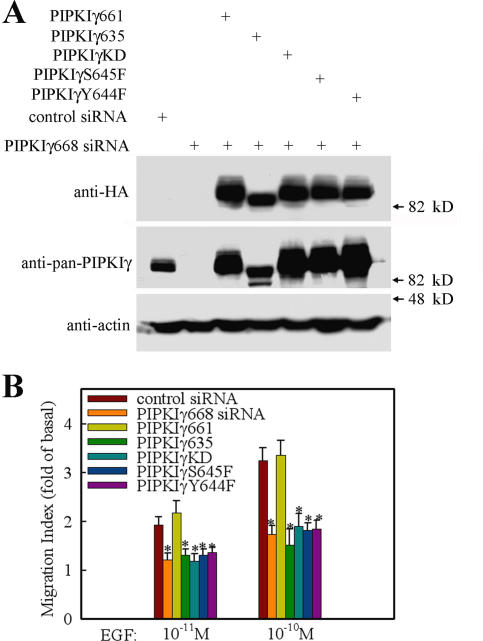
To further investigate the mechanism of how PIPKIγ661 regulates EGF-stimulated cell migration, HeLa stable cell lines expressing wild-type and mutant mouse PIPKIγ661 using a tet-off expression approach were established with the goal of rescuing endogenous PIPKIγ668 knockdown phenotype through the expression of specific mouse PIPKIγ661 mutants. siRNA specific for PIPKIγ668 was used to knock down PIPKIγ668 in these HeLa cell lines, and the rescuing effects of wild-type and mutant mouse PIPKIγ661 expression on EGF-stimulated cell migration were quantified.
To determine if PI4,5P2 generation was required for PIPKIγ661 control of EGF-stimulated migration, the kinase-dead PIPKIγ661 (PIPKIγKD)–expressing cell line was used in the rescue experiment. As shown in Fig. 3, expression of PIPKIγKD could not rescue EGF-stimulated cell migration in PIPKIγ668 knockdown cells. However, the expression of the wild-type mouse PIPKIγ661 fully rescued EGF-stimulated cell migration. These findings demonstrate that the ability of PIPKIγ661 to produce PI4,5P2 is required. PIPKIγ661 binds to talin, and binding is regulated by tyrosine phosphorylation on Y644 (Ling et al., 2002, 2003). Talin is a key protein in regulating adhesion turnover (Cram and Schwarzbauer, 2004; Nayal et al., 2004). To determine if talin binding is necessary for PIPKIγ661 to mediate EGF-stimulated migration, the Y644 to phenylalanine mutant of PIPKIγ661 (PIPKIγY644F) was assayed to rescue the endogenous knockdown. PIPKIγY644F has in vivo defects in the association with talin (Ling et al., 2003). As shown in Fig. 3, the mutant could not rescue the effect of PIPKIγ668 knockdown on EGF-stimulated cell migration. Furthermore, expression of PIPKIγ635, the short splice variant of PIPKIγ, which does not bind talin, was also unable to rescue the effect of PIPKIγ668 knockdown on EGF-stimulated migration.
Figure 3.
EGF-induced cell migration requires PIPKIγ661 kinase activity and its talin binding ability. Parental HeLa cells or HeLa tet-off stable cell lines expressing HA-tagged wild-type PIPKIγ661, PIPKIγ635, PIPKIγKD, PIPKIγY644F, or PIPKIγS645F were transfected with control siRNA or PIPKIγ668-specific siRNA separately as indicated. (A) The expression of PIPKIγ in these cell lines was detected by anti–pan-PIPKIγ antibody or anti-HA antibody separately. As the loading control, the amount of actin was detected by anti-actin antibody. (B) EGF was used to stimulate the migration of these cell lines. Quantifications are expressed as means ± SEM of three separate experiments (*, P < 0.01 compared with control siRNA–transfected HeLa cells).
The same region of PIPKIγ661 that interacts with talin also associates with the μ subunits of the adaptor protein (AP) 1B and AP2 complexes. Both PIPKIγY644F and PIPKIγ635 also have in vivo defects in the association with μ subunits (Bairstow et al., 2006; Ling et al., 2007). To exclude the possibility that PIPKIγ661 effects on EGF-induced migration is due to its binding with μ subunits of the AP1B and AP2 complexes but not with talin, an S645 to phenylalanine mutant of PIPKIγ661 (PIPKIγS645F) was used. PIPKIγS645F has in vivo defects in the association with talin but maintains the ability to associate with μ subunits of the AP1B and AP2 complexes (Bairstow et al., 2006; Ling et al., 2007). As shown in Fig. 3, the mutant could not rescue the effect of PIPKIγ668 knockdown on EGF-stimulated cell migration. It is important to note that the PIPKIγY644F also loses the ability to associate with the AP1B and AP2 complexes (Bairstow et al., 2006; Ling et al., 2007). Together, these findings indicate that PIP kinase activity, talin binding, and possibly AP complex binding, are required for PIPKIγ661 regulation of EGF-stimulated migration.
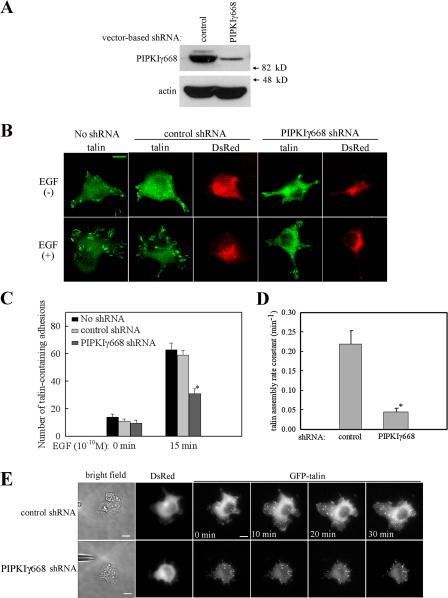
PIPKIγ661 is necessary for EGF-induced talin assembly into adhesions
Because talin binding is required for PIPKIγ661 effects on EGF- induced migration, regulating talin assembly into adhesions is a likely mechanism by which PIPKIγ661 regulates EGF-induced migration. To determine if talin assembly into adhesions is altered in PIPKIγ661-deficient cells, the impact of PIPKIγ661 knockdown on EGF-induced talin assembly of adhesions was assessed. A vector-based short hairpin RNA (shRNA) was used to knock down PIPKIγ668 expression in HeLa cells. This vector also expresses a red fluorescent protein, DsRed, alongside the expression of the PIPKIγ668-specific shRNA. In this way, the PIPKIγ668 knockdown cells can be identified as the red fluorescence positive cells. Transfection of this vector-based shRNA into HeLa cells resulted in efficient knockdown of PIPKIγ668 (Fig. 4 A). As shown in Fig. 4 B, in the absence of EGF, there are relatively few talin-containing adhesions found in either the control shRNA–transfected HeLa cells or in the PIPKIγ668 shRNA–transfected HeLa cells. Stimulation with EGF leads to talin recruitment to adhesions in DsRed-negative or control shRNA–expressing HeLa cells. However, in the PIPKIγ668 knockdown HeLa cells, the EGF-induced talin recruitment to adhesions was significantly decreased (Fig. 4, B and C). This result indicates that PIPKIγ668 is required for EGF-induced talin assembly into adhesions. Furthermore, in the PIPKIγ668 knockdown HeLa cells, the EGF-induced vinculin recruitment to adhesions was also decreased (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200701078/DC1). This result further confirmed that PIPKIγ668 is required for EGF-induced adhesion formation.
Figure 4.
PIPKIγ is required for EGF-induced talin assembly to adhesions. (A) The expression of PIPKIγ668 in control shRNA– or PIPKIγ668-specific shRNA–transfected HeLa cells was detected. (B) Control shRNA or PIPKIγ668-specific shRNA–transfected HeLa cells were stimulated with 10−10 M EGF for 15 min. Cells were fixed and stained with anti-talin antibody. (C) EGF-induced talin assembly to adhesions was quantified. (D) Control shRNA or PIPKIγ668-specific shRNA were cotransfected with GFP-talin into HeLa cells. A micropipette filled with EGF was put near the cell to stimulate GFP-talin recruitment. GFP-talin assembly kinetics was quantified. Rate constants for assembly of individual adhesions were calculated as described in Materials and methods. Quantifications are expressed as means ± SEM (*, P < 0.01 compared with control siRNA–transfected HeLa cells). (E) GFP-talin assembly to adhesions at different time point of micropipette stimulation was shown. See Videos 1 and 2 (available at http://www.jcb.org/cgi/content/full/jcb.200701078/DC1). Bars, 10 μm.
PIPKIγ661 is required for EGF-induced talin assembly into nascent adhesions at the leading edge
At the onset of cell migration, cells extend protrusions of the plasma membrane at their leading edge and then assemble nascent adhesions serving as points of traction and help to establish cell polarity (Smilenov et al., 1999; Webb et al., 2002). As PIPKIγ661 is required for EGF-induced talin assembly into adhesions, it is possible that PIPKIγ661 is required for EGF-induced talin recruitment to the leading edge and to regulate adhesion assembly.
To determine if PIPKIγ661 is required for the polarized recruitment of talin to the leading edge in a gradient of chemoattractant, a micropipette-stimulation assay (Mouneimne et al., 2006) that can selectively stimulate cells with locally released EGF was used. As shown in Fig. 4 E, a micropipette filled with EGF was placed near cells, and constant pressure was added to the micropipette to create a concentration gradient of EGF. Vector-based shRNA was used to knock down the expression of PIPKIγ668, and knockdown cells were visualized with DsRed. To assess talin turnover at the leading edge in real time, a GFP-talin construct (Franco et al., 2004) was transfected into HeLa cells to detect its dynamic assembly at adhesions. GFP-talin assembly rate was quantified in Fig. 4 D. Rate constants for talin assembly of individual adhesions were calculated as described in Materials and methods. GFP-talin assembly into adhesions at the leading edge at different time points of micropipette stimulation is shown in Fig. 4 E. In the cells expressing control shRNA, local stimulation with EGF by the pipette led to rapid recruitment of GFP-talin to the leading edge and assembly into nascent adhesions. In the cells expressing PIPKIγ668 shRNA, however, GFP-talin recruitment to nascent adhesions was significantly decreased (Fig. 4, D and E; and Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200701078/DC1). These findings indicate that EGF induces new adhesions in the direction of the growth factor concentration gradient, and this requires PIPKIγ661.
Phosphorylation of PIPKIγ661 at Y634 is required for EGF-induced talin assembly and migration
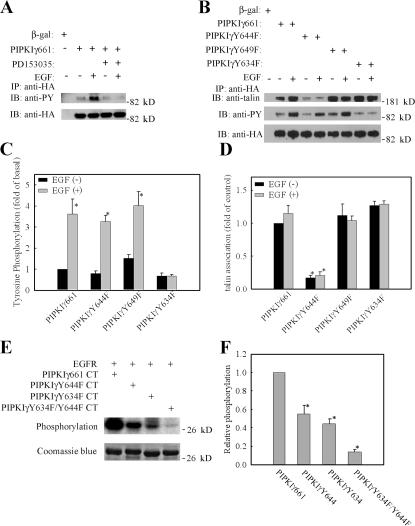
The function of PIPKIγ661 is regulated by its tyrosine phosphorylation. Src-mediated phosphorylation of Y644 increases the PIPKIγ661-talin binding affinity (Ling et al., 2003) and blocks the PIPKIγ661 interaction with AP complexes (Bairstow et al., 2006). To determine if EGF can stimulate PIPKIγ661 tyrosine phosphorylation, the tyrosine phosphorylation of PIPKIγ661 was quantified. As shown in Fig. 5 A, EGF stimulation noticeably increased tyrosine phosphorylation of PIPKIγ661. To determine if this effect is specifically due to the activation of EGF receptor (EGFR), cells were preincubated with EGFR-specific inhibitor PD153035, and this blocked EGF-induced tyrosine phosphorylation of PIPKIγ661. Interestingly, when Y644 of PIPKIγ661, the known Src phosphorylation site, was mutated to phenylalanine (PIPKIγY644F), the EGF-stimulated tyrosine phosphorylation was not affected (Fig. 5, B and C), suggesting that Y644 is not required for EGF-induced tyrosine phosphorylation of PIPKIγ661 in vivo. To identify the key tyrosine residues for the EGF-induced tyrosine phosphorylation, a series of tyrosine residues were mutated, and the Y634 to phenylalanine mutant of PIPKIγ661 (PIPKIγY634F) was found to ablate EGF-induced tyrosine phosphorylation (Fig. 5, B and C). These findings demonstrate that Y634 is the key tyrosine residue for EGF-induced tyrosine phosphorylation.
Figure 5.
PIPKIγ661 is tyrosine phosphorylated by EGF stimulation. (A) HeLa cells were transfected with β-gal or HA-PIPKIγ661 separately, pretreated with or without EGFR-specific inhibitor PD153035, and stimulated with 10−9 M EGF for 5 min. Cells were used in immunoprecipitation with anti-HA antibody, and the tyrosine phosphorylation was detected by anti-phosphotyrosine antibody. (B) HA-tagged PIPKIγ661, PIPKIγY644F, PIPKIγY649F, or PIPKIγY634F were transfected into HeLa cells separately and stimulated with 10−9 M EGF for 5 min. Cells were subjected into immunoprecipitation with anti-HA antibody, and the tyrosine phosphorylation was detected. The amount of talin in the immunoprecipitation complex was detected by anti-talin antibody. (C) Tyrosine phosphorylation of HA-tagged PIPKIγ661, PIPKIγY644F, PIPKIγY649F, or PIPKIγY634F with or without EGF stimulation was quantified. (D) Talin association with HA-tagged PIPKIγ661, PIPKIγY644F, PIPKIγY649F, or PIPKIγY634F with or without EGF stimulation was quantified. (E) Reconstituted c-tail of wild-type PIPKIγ661, PIPKIγY644F, PIPKIγY634F, or PIPKIγY634F/Y644F was used as substrate of purified EGFR in the in vitro kinase assay. (F) The relative phosphorylation of wild-type PIPKIγ661, PIPKIγY644F, PIPKIγY634F, or PIPKIγY634F/Y644F was quantified. The phosphorylation of wild-type PIPKIγ661 was set as 100% (*, P < 0.01 compared with wild-type PIPKIγ661). Quantifications are expressed as means ± SEM of three separate experiments.
EGFR is a tyrosine kinase, and ligand binding to the extracellular portion of the EGFR leads to autophosphorylation of specific tyrosine residues in its cytoplasmic region, which stimulate the intrinsic kinase activity of the receptor (Carpenter and Cohen, 1990). To determine if EGFR could directly phosphorylate PIPKIγ661, an in vitro EGFR kinase assay was used. The reconstructed wild-type PIPKIγ661 C terminus or the mutant PIPKIγY634F C terminus was purified from Escherichia coli and subjected to in vitro EGFR kinase assay as substrates of purified EGFR. As shown in Fig. 5 (E and F), wild-type PIPKIγ661 C terminus can be phosphorylated directly by purified EGFR. In this assay, mutant PIPKIγY634F C terminus lost EGFR-induced phosphorylation compared with wild-type PIPKIγ661. Interestingly, in the Y644 to F mutant of PIPKIγ661 (PIPKIγY644F), EGFR-induced phosphorylation was also reduced compared with wild-type PIPKIγ661. Mutation of both Y634 and Y644 in PIPKIγ661 to phenylalanine resulted in a loss of the EGFR-induced phosphorylation compared with the wild type. In vivo, the mutation of Y634, but not Y644, resulted in the loss of EGF-induced tyrosine phosphorylation of PIPKIγ661. These data demonstrate that EGFR phosphorylates Y634 in vivo and in vitro and are also consistent with results showing that Y644 is preferentially phosphorylated by Src (Ling et al., 2003).
The results demonstrate that Y634 of PIPKIγ661 is phosphorylated upon EGF stimulation. To determine if Y634 phosphorylation is required for PIPKIγ661 effects on EGF-induced migration, the PIPKIγY634F-expressing stable HeLa cell line was used in a cell migration assay to demonstrate its effect on EGF-induced migration. As shown in Fig. 6, the expression of PIPKIγY634F could not rescue the effect of PIPKIγ668 knockdown on EGF-induced directional migration. The results indicate that Y634 phosphorylation is required for the role of PIPKIγ661 in EGF-induced migration.
Figure 6.
Expression of PIPKIγY634F could not rescue EGF-stimulated cell migration in PIPKIγ668 knockdown HeLa cells. Parental HeLa cells or HeLa tet-off stable cell lines expressing HA-tagged wild-type PIPKIγ661 or PIPKIγY634F were transfected with control or PIPKIγ668 siRNA separately as indicated. (A) The expression of PIPKIγ in these cell lines was detected by anti-PIPKIγ antibody or anti-HA antibody. As the loading control, the amount of actin was detected by anti-actin antibody. (B) EGF was used to stimulate the migration of these cell lines. Quantifications are expressed as means ± SEM of three separate experiments (*, P < 0.01 compared with control siRNA–transfected HeLa cells).
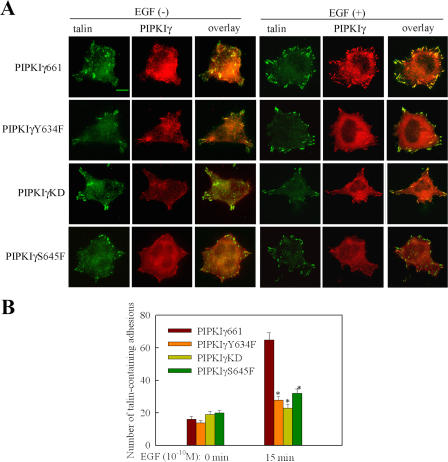
As shown in Fig. 5 (B and D), PIPKIγY634F still retains the ability to bind talin. Quantification of PIP kinase activity demonstrated that PIPKIγY634F retains kinase activity on a level that is comparable to wild-type PIPKIγ661 (unpublished data). It is interesting to determine if Y634 phosphorylation is required for EGF-induced talin assembly to facilitate migration. As shown in Fig. 7, both wild-type PIPKIγ661 and talin are recruited to adhesions after EGF stimulation. However, in comparison, PIPKIγY634F was less efficiently recruited to adhesions by EGF stimulation. Also, talin recruitment to adhesions was decreased in these same cells. The expression of PIPKIγKD (kinase dead) also decreased talin recruitment to adhesions after EGF stimulation, similar to the PIPKIγY634F mutant. These data demonstrate that both Y634 phosphorylation and the kinase activity of the PIPKIγ661 are required for efficient talin assembly into adhesions induced by EGF stimulation. In addition, PIPKIγS645F, the mutant that has in vivo defects in association with talin and does not rescue directional migration, also was not recruited to adhesions after EGF stimulation. Correspondingly, cells expressing PIPKIγS645F showed decreased talin recruitment to adhesions after EGF stimulation compared with cells expressing wild-type PIPKIγ661. This result indicates that the collaboration of PIPKIγ661 and talin is required for their recruitment to adhesions induced by EGF. Furthermore, Y634 phosphorylation and the kinase activity of the PIPKIγ661 are also required for efficient vinculin assembly into adhesions induced by EGF stimulation (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200701078/DC1). This result further confirms that Y634 phosphorylation and the kinase activity of the PIPKIγ661 are required for EGF-induced adhesion formation.
Figure 7.
The effect of PIPKIγ661, PIPKIγY634F, PIPKIγKD, or PIPKIγS645F on EGF-induced talin assembly into adhesions. (A) HA-tagged PIPKIγ661, PIPKIγY634F, PIPKIγKD, or PIPKIγS645F stably expressing HeLa cells were stimulated with 10−10 M EGF for 15 min. Cells were fixed and stained with anti-HA and anti-talin antibodies. (B) EGF-induced talin assembly to adhesions in these cells was quantified. Quantifications are expressed as means ± SEM (*, P < 0.01 compared with wild-type PIPKIγ661). Bar, 10 μm.
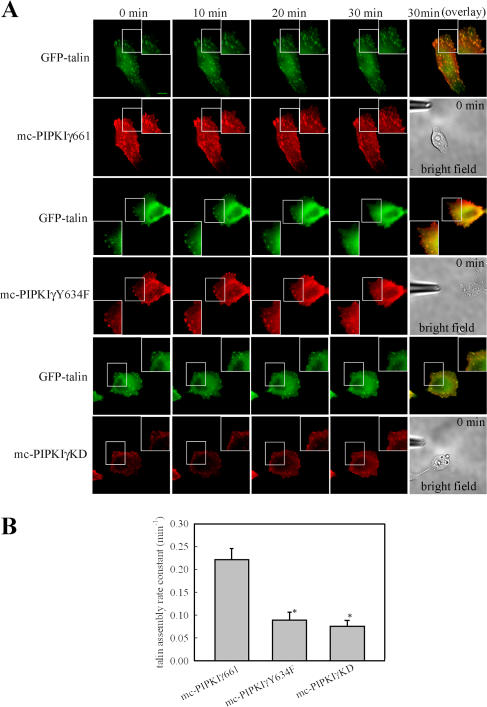
To determine if Y634 phosphorylation of PIPKIγ661 is required for the polarized recruitment of talin to the leading edge, HeLa cells expressing mCherry-tagged wild-type PIPKIγ661, PIPKIγY634F, or PIPKIγKD were stimulated with a gradient of EGF using the micropipette-stimulation assay for directional migration. A micropipette filled with 10 nM EGF was placed near the cell as in Fig. 8 A. GFP-talin was used to allow for the detection of talin dynamics at adhesions in real time. As shown in Fig. 8 A, local stimulation by EGF induced rapid co-translocation of both mCherry-tagged wild- type PIPKIγ661 (mc-PIPKIγ661) and GFP-talin to the leading edge and assembly into nascent adhesions. However, mCherry-tagged PIPKIγY634F and PIPKIγKD (mc-PIPKIγY634F and mc-PIPKIγKD) were less efficiently recruited to adhesions by EGF stimulation. Correspondingly, EGF-induced GFP-talin assembly to nascent adhesions at the leading edge was also decreased both in mc-PIPKIγY634F– and mc-PIPKIγKD–expressing HeLa cells (Fig. 8, A and B; and Videos 3–8, available at http://www.jcb.org/cgi/content/full/jcb.200701078/DC1). These findings further demonstrate that both Y634 phosphorylation and the kinase activity of PIPKIγ661 are required for the polarized recruitment of talin to the leading edge in a gradient of EGF.
Figure 8.
Expression of PIPKIγY634F decreased EGF-induced talin assembly to nascent adhesions at the leading edge. (A) GFP-talin was cotransfected with mc-PIPKIγ661, mc-PIPKIγY634F, or mc-PIPKIγKD into HeLa cells. A micropipette filled with EGF was put near the cell to stimulate talin recruitment. See Videos 3–8 (available at http://www.jcb.org/cgi/content/full/jcb.200701078/DC1). Bar, 10 μm. (B) GFP- talin assembly kinetics in mc-PIPKIγ661–, mc-PIPKIγY634F–, or mc-PIPKIγKD–expressing HeLa cells were quantified. Quantifications are expressed as means ± SEM (*, P < 0.01 compared with mc-PIPKIγ661–expressing HeLa cells).
These combined data support a requirement for EGF-induced phosphorylation of PIPKIγ661 at Y634 for the polarized assembly of adhesions at the leading edge, and this is crucial for EGF-stimulated cell migration. As the PIPKIγ661 is assembled into multiple protein complexes regulated by phosphorylation, these results suggested that the phosphorylation of Y634 may also modulate protein–protein interactions.
Phosphorylation of PIPKIγ661 at Y634 regulates the association of PLCγ1 with PIPKIγ661
Although phosphorylation of PIPKIγ661 at Y634 does not alter the ability of PIPKIγ661 to bind talin (Fig. 5, B and D), Y634 phosphorylation may affect the association of PIPKIγ661 with other proteins involved in regulating migration. To explore this possibility, the binding ability of wild-type PIPKIγ661 and PIPKIγY634F with PIPKIγ-associating proteins other than talin was compared. AP1B and AP2 complexes bind to PIPKIγ661 with the same sequence that binds talin (Bairstow et al., 2006). When PIPKIγY634F was assayed for binding to AP1B and AP2 complexes, this mutant associated with both AP complexes in a manner that was indistinguishable from the wild-type PIPKIγ661 (unpublished data).
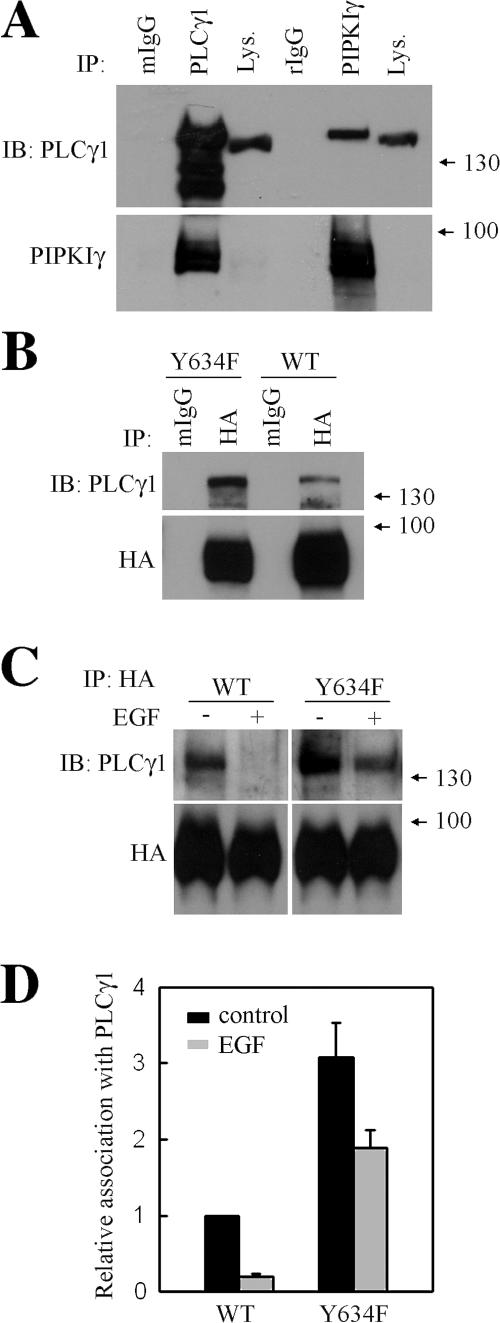
Interestingly, we have shown that PLCγ1, an enzyme also required for EGF-stimulated directional migration (Piccolo et al., 2002), associates with the PIPKIγ661 complex. Further, PLCγ1 was found to be differentially associated with wild-type PIPKIγ661 and PIPKIγY634F. As shown in Fig. 9 A, PLCγ1 was coimmunoprecipitated with endogenous PIPKIγ and is the first evidence that PLCγ1 could associate with the PIPKIγ661 complex. Both wild-type PIPKIγ661 and PIPKIγY634F associate with PLCγ1, whereas PIPKIγY634F had a stronger interaction with PLCγ1 than did wild-type PIPKIγ661 (Fig. 9 B). Intriguingly, association of PIPKIγ661 with PLCγ1 was lost after EGF stimulation, whereas the PLCγ1 association with PIPKIγY634F could not be efficiently disrupted by EGF treatment (Fig. 9, C and D). These results indicate that phosphorylation of PIPKIγ661 at Y634 regulates PLCγ1 association with PIPKIγ661.
Figure 9.
PLCγ differentially associates with PIPKIγ661 and PIPKIγY634F. (A) HeLa cells were put into immunoprecipitation assay with the antibodies indicated. The amount of endogenous PIPKIγ or PLCγ1 in the immunoprecipitation complex was detected by anti-PIPKIγ antibody or anti-PLCγ1 antibody separately. (B) HeLa cells were transfected with HA-PIPKIγ661 or HA-PIPKIγY634F. Cells were put into immunoprecipitation assay with anti-HA antibody, and the amount of PLCγ1 in the immunoprecipitation complex was detected. (C) HeLa cells transfected with HA-PIPKIγ661 or HA-PIPKIγY634F were stimulated with 10−9 M EGF for 5 min. Cells were subjected into immunoprecipitation with anti-HA antibody, and the amount of PLCγ1 in the immunoprecipitation complex was detected. (D) The effect of EGF stimulation on PLCγ1 association with wild-type PIPKIγ661 or PIPKIγY634F was quantified. Quantifications are expressed as means ± SEM of three separate experiments.
Discussion
Directional cell migration is critical to many biological and pathological processes, including embryogenesis, the inflammatory response, atherosclerosis, tissue repair and regeneration, and cancer metastasis (Lauffenburger and Horwitz, 1996; Webb et al., 2002). PI4,5P2 modulates many key components of the cell migration machinery and is proposed to be synthesized in a highly spatial and temporal fashion to regulate this process (Doughman et al., 2003a; Ling et al., 2006; Santarius et al., 2006). However, the underlying mechanism for the restricted generation of PI4,5P2 that regulates migration is poorly understood. PIPKIγ was proposed to be the major enzyme responsible for PI4,5P2 synthesis at synapses (Wenk et al., 2001), and much of the work on the biological functions of PIPKIγ since then have been focused on this system. Recently, compelling observations revealing the critical biological roles of PIPKIγ in nonneuronal systems have emerged. PIPKIγ635 has been reported to have important functions in G protein–coupled receptor–mediated IP3 generation (Wang et al., 2004). More interesting, PIPKIγ661 is implicated in focal adhesion assembly (Ling et al., 2002), AP2-mediated endocytosis (Bairstow et al., 2006), and the endocytosis and basolateral sorting of E-cadherin (Ling et al., 2007). Our current findings demonstrate that PIPKIγ661 is required for EGF-stimulated directional migration.
The role of PIPKIγ661 in regulating EGF-stimulated directional migration is unique, as neither PIPKIγ635 nor PIPKIα can compensate for the loss of PIPKIγ661. In coordination with Rac signaling, PIPKIα plays roles in dorsal membrane ruffling stimulated by PDGF (Doughman et al., 2003b; Kisseleva et al., 2005), and PIPKIα interacts with the LIM domain protein Ajuba and appears to coordinate the targeting to membrane ruffles (Kisseleva et al., 2005). Membrane ruffles are often found on the cell surface and at the advancing front of a lamellipodium and serve as sites of actin polymerization (Cheresh et al., 1999). Although membrane ruffling is thought to be important for migration, it is not necessary for migration of all cells. For example, Rac1-deficient macrophages exhibit defects in membrane ruffling but show normal directional migration (Wells et al., 2004). Investigation in epidermal keratinocytes demonstrated that high membrane ruffling rates correlated with low lamellipodia persistence and inefficient migration (Borm et al., 2005). PIPKIα-induced membrane ruffling may be required for certain types of migration but may not be essential for the EGF-stimulated migration of epithelial cells like HeLa, MtLn3, and A431.
Cell migration is an integrated process that requires the continuous, coordinated formation and disassembly of adhesions (Smilenov et al., 1999; Webb et al., 2002). Our results demonstrate that PIPKIγ661 is required for talin assembly into nascent adhesions forming at the leading edge toward the direction of the growth factor concentration gradient. This supports the hypothesis that PIPKIγ661 regulates growth factor–mediated migration ultimately through modulation of adhesion dynamics. These data indicate that PIPKIγ661 works as an effector of growth factor signaling to provide a link to the corresponding intracellular adhesion dynamics required for migration. Talin plays key roles in adhesion turnover and cell migration by providing a link between integrin and the cytoskeleton (Priddle et al., 1998; Critchley, 2000; Cram and Schwarzbauer, 2004). Reduction of talin expression leads to defects in normal cell migration in Caenorhabditis elegans (Cram et al., 2003). Despite the critical roles in cell migration, how talin is recruited to and regulated at adhesions is poorly understood. The results presented here support a role for PIPKIγ661 in talin recruitment and regulation downstream of EGF-induced directional migration.
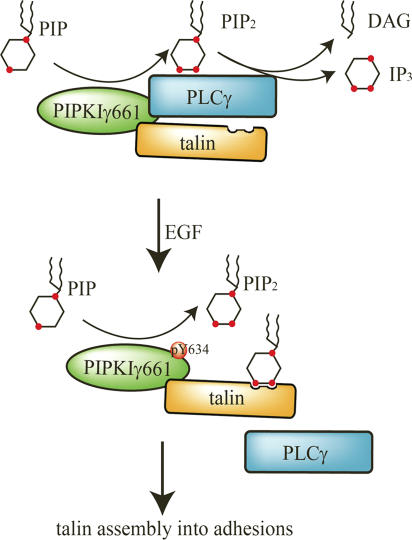
Y634 phosphorylation of PIPKIγ661 is required for talin recruitment in response to EGF. It is interesting that the phosphorylation of Y634 does not obviously affect the kinase activity of PIPKIγ661 or its talin binding affinity. The phosphorylation of Y634 regulates the interaction between PIPKIγ661 and PLCγ. Therefore, current results provide a possible model of how Y634 phosphorylation of PIPKIγ661 is involved in EGF-induced migration (Fig. 10). By associating with PIPKIγ661, PLCγ1 could hydrolyze the PI4,5P2 produced by PIPKIγ661; the hydrolysis would diminish the PI4,5P2 required to regulate talin assembly to adhesions. EGF-induced phosphorylation of PIPKIγ661 at Y634 causes a disassembly of the PLCγ1–PIPKIγ661 complex, and this could enhance PI4,5P2 accumulation and thus enhance talin assembly into adhesions.
Figure 10.
Model of how PIPKIγ661 is involved in EGF-induced migration. By associating with PIPKIγ661, PLCγ1 could hydrolyze the PI4,5P2 produced by PIPKIγ661; the hydrolysis would diminish the PI4,5P2 required to regulate talin assembly into adhesions. EGF-induced phosphorylation of PIPKIγ661 at Y634 causes a disassembly of the PLCγ1–PIPKIγ661 complex, and this could enhance PI4,5P2 accumulation and thus enhance talin assembly into adhesions. By modulating talin assembly, PIPKIγ661 regulates adhesion formation at the leading edge and facilitates EGF-induced migration.
It is established that PLCγ1 regulates EGF-induced migration (Piccolo et al., 2002). Activation of PLCγ is required for protrusion formation at the leading edge. PLCγ1 cleaves PI4,5P2 and releases actin binding proteins gelsolin and cofilin to initiate protrusion and define the direction of cell migration (Chen et al., 1996; Chou et al., 2002; Mouneimne et al., 2004). Furthermore, PLCγ1 also modulates the polarized localization of m-calpain and regulates the adhesion detachment (Shao et al., 2006). In this context, PIPKIγ661 and PLCγ likely work together to regulate local PI4,5P2 level at the leading edge to facilitate the protrusion formation and stabilization of adhesions. As the involvement of PIPKIγ661 seems specific for growth factor stimulation and not LPA and SDF1α stimulation, it is likely that G protein–coupled receptors use distinct downstream elements to modulate the formation of protrusions. Together, this unexpectedly indicates that PIPKIγ661 is a unique and key signaling component specific for growth factor–induced directional cell migration.
Tyrosine 644 is another residue site on PIPKIγ661 that is phosphorylated by Src. This phosphorylation enhances the PIPKIγ661-talin binding affinity (Ling et al., 2003). Src can be activated by EGFR via a Ral-GTPase–dependent mechanism (Goi et al., 2000). It is possible that EGF treatment could also lead to PIPKIγ661 phosphorylation at Y644 by activating Src. Although our results show that EGF stimulation under these conditions did not lead to obvious change of cellular PIPKIγ661-talin binding affinity, EGF stimulation may modulate PIPKIγ661-talin association at specific sites, such as adhesions via Src-mediated phosphorylation of Y644. EGF stimulation may lead to an ordered set of phosphorylation events that regulate PIPKIγ661 interactions with its partners, such as PLCγ1 and talin, and, via this mechanism, regulate directional migration.
By regulating adhesion dynamics at the leading edge, PIPKIγ661 may play a role in the regulation of cell protrusion toward the direction of stimulation. Cell adhesion and protrusion are highly interrelated during migration. Protrusion results primarily from actin polymerization at the leading edge of migrating cells. Nascent adhesions forming at leading edge could stabilize the new protrusion and help establish cell polarity during migration. Talin binds actin and provides a molecular linkage between adhesion and actin that inhibits retrograde flow and thus regulates the rate of protrusion by counterbalancing the forward movement of actin polymerization (Mitchison and Kirschner, 1988; Schwartz and Horwitz, 2006). It is plausible that PIPKIγ661 participates in stabilization of protrusions by enhancing the recruitment of talin to the leading edge; via this mechanism, PIPKIγ661 would determine the direction of migration.
The same region of PIPKIγ661 that interacts with talin also associates with the μ subunit of the AP1B and AP2 complexes (Bairstow et al., 2006; Ling et al., 2007). Indeed, membrane trafficking also regulates the directional migration of cells (Bershadsky and Futerman, 1994; Etienne-Manneville, 2004; Tayeb et al., 2005; Caswell and Norman, 2006; Rosse et al., 2006). Thus, our results are equally consistent with a role for the PIPKIγ661 in membrane trafficking in controlling EGF-stimulated directional migration. Endocytosis of receptor tyrosine kinases plays a key role in growth factor–stimulated chemotaxis (Jekely et al., 2005; Le Roy and Wrana, 2005). In Drosophila, endocytic trafficking is required for directional migration stimulated by EGF and EGFR homologues in vivo (Jekely et al., 2005). We have explored the possibility that PIPKIγ661 modulates EGFR endocytosis. Knockdown of PIPKIγ661 or expression of the PIPKIγ661 kinase-dead mutants did not effect EGFR internalization upon agonist binding (unpublished data). Nevertheless, this result does not eliminate a potential role for PIPKIγ661 in modulation of membrane trafficking within the EGF-stimulated migration pathway. There are other trafficking events that may be crucial for PIPKIγ661 to regulate EGF-stimulated migration, such as trafficking of integrins (Martel et al., 2000; Caswell and Norman, 2006).
PIPKIγ661 also regulates the ability of epithelial cells to assemble E-cadherin based cell–cell contacts (Ling et al., 2007), and here we show that it also regulates the ability of cells to migrate toward an EGF gradient. Therefore, PIPKIγ661 has been implicated to be required for two key physiological processes: cell–cell adhesion and directional cell migration. This is very significant, as these two processes are fundamental in early stages of the metastasis of cancers of epithelial origin. In breast cancer metastasis, the sequential loss of E-cadherin and cell–cell contacts allows tumor cells to migrate (Yang et al., 2004). The migration toward blood vessels is stimulated in some cases by a gradient of EGF, and this facilitates a key step called intravasation, where the tumor cells migrate through the vessel wall to be transported throughout the body (Wang et al., 2005; Xue et al., 2006). Further exploration of the underlying signals and mechanisms that regulate PIPKIγ661 will be crucial to understanding the complete role of this enzyme in cell migration and tumor metastasis.
Materials and methods
Constructs
Site-directed mutagenesis for the PIPKIγ661 mutants was performed using PCR-primer overlap extension with mutagenic primers. The mutations were confirmed by DNA sequence analysis. The siRNA sequence targeting pan-PIPKIγ is 5′-GGACCUGGACUUCAUGCAG-3′. The siRNA sequence targeting human PIPKIγ668 is 5′-GAGCGACACAUAAUUUCUA-3′. The sequence of control scrambled siRNA is 5′-GUACCUGUACUUCAUGCAG-3′. The mCherry vector was provided by R.Y. Tsien (University of California, San Diego, La Jolla, CA).
Antibodies
Anti-talin and anti-actin antibody were purchased from Sigma-Aldrich. Monoclonal mouse anti-PY (4G10), anti-FAK (4.47), and anti-PLCγ1 antibodies were obtained from Upstate Biotechnology. Anti-HA antibody was purchased from Covance and Roche. Polyclonal PIPKIγ anti-serum was generated as described previously (Ling et al., 2002). Anti-PIPKIγ661 specific antibody was purified on an affinity column generated by coupling the 26-amino-acid C-terminal peptide of PIPKIγ661 to cyanogen bromide–activated Sepharose 4B (Sigma-Aldrich) as described previously (Ling et al., 2002). Secondary antibodies were obtained from Jackson ImmunoResearch Laboratories.
Cell cultures and transfection
HeLa cells and A431 cells were cultured using DME supplemented with 10% FBS. MTLn3-EGFR cells, provided by J. Condeelis (Yeshiva University, New York, NY), were maintained in MEM α supplemented with 5% FBS. For plasmid transfection, HeLa cells were transfected by using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. For siRNA transfection, MTLn3-EGFR and A431 cells were transfected with Oligofectamine, and HeLa cells were transfected with Lipofectamine 2000 following the manufacturer's instructions.
HeLa tet-off stable cell lines
Tet-off HeLa cells (CLONTECH Laboratories, Inc.) were stably transfected with various PIPKIγ constructs, which are under the control of the tetracycline responsive promoter. The transfected HeLa cells were maintained in DME containing 10% FBS, 200 μg/ml G418, and 100 μg/ml hygromycin B to select for stable transfection. The medium was supplemented with 2 μg/ml doxycycline to suppress transgene expression, as doxycycline withdrawal results in expression of transfected PIPKIγ.
Cell migration assay
The assays were performed in modified Boyden chamber transwell (Neuroprobe) as described previously (Neptune and Bourne, 1997; Sun et al., 2002). The membrane was precoated with 10 μg/ml type I collagen. 50,000 cells were applied per well. Most chemotaxis assays were done at 37°C in humidified air with 5% CO2 for 4 h. For a different time course, cells were allowed to migrate for 2, 4, or 8 h. For each agonist concentration tested, cells migrated through to the underside of the membrane were counted in five high-power fields, in a blinded fashion. The migration index for each experiment was calculated as the mean number of cells that migrated toward medium-containing agonist divided by the mean number of cells that migrated toward medium-containing bovine serum albumin only.
Immunoprecipitation and immunoblotting
Immunoprecipitation was performed as described previously (Ling et al., 2003). In brief, 48 h after transfection, HeLa cells were starved with serum-free DME overnight and then stimulated with 10−9 M EGF for 5 min. Then cells were harvested and lysed in 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% NP-40, 5.0 mM NaF, 2 mM Na3VO4, 4 mM Na2P2O7, 1 mM EDTA, 0.1 mM EGTA, 10% glycerol, and proteinase inhibitor cocktail, centrifuged, and incubated with protein A–Sepharose and 2 μg anti-HA antibody as indicated at 4°C overnight. The immunocomplexes were separated by SDS-PAGE and analyzed as indicated.
In vitro EGFR protein kinase assay
The in vitro kinase assays were performed with EGFR (Promega) and recombinant purified PIPKIγ661 C terminus, encompassing residues 439–661 in the pET28 vector, as previously described (Ling et al., 2003). The reaction was performed with a half unit of EGFR (as defined by Promega) in 5 mM Hepes, pH 7.4, 50 μM Na3VO4, 5 mM MgCl2, 2 mM MnCl2, and 250 mM (NH4)2SO4 with 5 μg protein substrate. The reaction was initiated with the addition of 20 μM ATP with 5 μCi γ-[32P]adenosine triphosphate (GE Healthcare) and terminated by the addition of sample buffer after 15 min. The substrates were resolved by SDS-PAGE and fixed and stained using GelCode Blue (Pierce Chemical Co.). Image analysis was performed using NIH ImageJ.
Immunofluorescence
Immunofluorescence was performed as described previously (Ling et al., 2002). Glass coverslips were acid washed and coated with 10 μg/ml type I collagen. Cells were resuspended and plated on the coverslips in serum-free DME and allowed to adhere for 3 h. Then, cells were stimulated with 10−10 M EGF for different time course (from 0 to 15 min) and fixed by methanol at −20°C for 10 min. Cells were then blocked by 3% BSA in PBS at RT for 30 min, incubated with the primary antibody overnight at 4°C, washed with 0.1% Triton X-100 in PBS, incubated with fluorescence-labeled secondary antibody at RT for 30 min, and washed with 0.1% Triton X-100 in PBS. Cells were maintained and examined using a 60× Plan oil-immersion lens on an inverted microscope (Eclipse TE200-U; Nikon). Images were processed as described previously (Ling et al., 2002) using Photoshop 7.0 (Adobe).
The micropipette assay
The micropipette assay was performed as described previously (Mouneimne et al., 2004, 2006). A Femtojet Micromanipulator 5171 (Eppendorf) and a pump (Femtojet; Eppendorf) were used to control the position of the micropipette and the pressure required for the chemoattractant flow. To induce the formation of nascent adhesions at the leading edge, a micropipette was filled with 10 nM EGF and was placed ∼10 μm from the edge of a cell, and a constant pressure was exerted to induce flow.
Live fluorescence microscopy
Fluorescence imaging of live cells was performed using a 60× objective on the Eclipse TE200-U inverted microscope housed in a closed system to maintain the temperature at 37°C. Glass-bottomed dishes were acid washed and coated with 10 μg/ml type I collagen. Cells were plated in DME media and allowed to adhere for 1 h, after which time the media was replaced with serum-free L15 media supplemented with 0.2% fatty acid–free BSA. Fluorescent images then captured every 1 min for 30 min using MetaMorph Imaging software (Universal Imaging Corp.).
Quantification of adhesion dynamics
The dynamics of fluorescently tagged talin was quantified according to the described protocols (Franco et al., 2004; Webb et al., 2004). Fluorescence intensities of individual adhesions from background-subtracted images were measured over time using MetaMorph Imaging software. For rate constant measurements, periods of assembly (increasing fluorescence intensity) of adhesions containing GFP-talin were plotted on separate semilogarithmic graphs representing fluorescence intensity ratios over time. Semilogarithmic plots of fluorescence intensities as a function of time were generated using the following formula: Ln([I]/[I0]) for assembly, where I0 is the initial fluorescence intensity and I is the fluorescence intensity at various time points. The slopes of linear regression trend lines fitted to the semilogarithmic plots were then calculated to determine apparent rate constants of assembly. For each rate constant, measurements were made on at least 10 individual adhesions of the cell, for a total of >50 adhesions in six separate cells. All measurements shown are the mean ± SEM. P values were calculated using t test.
Online supplemental material
Fig. S1 shows that knockdown of PIPKIγ668 attenuated EGF-stimulated directional migration. Fig. S2 shows that knockdown of PIPKIγ does not affect LPA- or SDF1α-stimulated cell migration in a different time course. Fig. S3 shows that PIPKIγ is required for EGF-induced vinculin assembly into adhesions. Fig. S4 shows the different effects of PIPKIγ661, PIPKIγY634F, PIPKIγKD, or PIPKIγS645F on EGF-induced vinculin assembly into adhesions. Videos 1 and 2 show the polarized recruitment of GFP-talin to the leading edge in a gradient of EGF in control shRNA– or PIPKIγ668 shRNA–transfected HeLa cells. Videos 3–8 show the polarized recruitment of GFP-talin to the leading edge in a gradient of EGF in mc-PIPKIγ661–, mc-PIPKIγY634F–, or mc-PIPKIγKD–expressing HeLa cells.
Supplementary Material
Acknowledgments
We thank Dr. John S. Condeelis at Albert Einstein College of Medicine of Yeshiva University for the MtLn3 cell lines. We thank Santos J. Franco and Anna Huttenlocher for the initial GFP-talin constructs, early discussions, and comments on the manuscript. We thank Dr. Roger Y. Tsien at University of California San Diego for providing the mCherry vector.
We acknowledge the following grant support: National Institutes of Health grants CA104708, GM057549, and P30-CA-014520 to R.A. Anderson, American Heart Association fellowship 0520121Z to Y. Sun, and American Heart Association fellowship 0425731Z and American Heart Association National Scientist Development grant 0535552N to K. Ling.
Abbreviations used in this paper: AP, adaptor protein; EGFR, EGF receptor; HGF, hepatocyte growth factor; LPA, lysophosphatidic acid; PI4,5P2, phosphatidylinositol 4,5-bisphosphate; PIPKI, type I phosphatidylinositol phosphate kinase; SDF, stromal cell–derived factor; shRNA, short hairpin RNA.
References
- Anderson, R.A., and V.T. Marchesi. 1985. Regulation of the association of membrane skeletal protein 4.1 with glycophorin by a polyphosphoinositide. Nature. 318:295–298. [DOI] [PubMed] [Google Scholar]
- Anderson, R.A., I.V. Boronenkov, S.D. Doughman, J. Kunz, and J.C. Loijens. 1999. Phosphatidylinositol phosphate kinases, a multifaceted family of signaling enzymes. J. Biol. Chem. 274:9907–9910. [DOI] [PubMed] [Google Scholar]
- Bairstow, S.F., K. Ling, X. Su, A.J. Firestone, C. Carbonara, and R.A. Anderson. 2006. Type Igamma661 phosphatidylinositol phosphate kinase directly interacts with AP2 and regulates endocytosis. J. Biol. Chem. 281:20632–20642. [DOI] [PubMed] [Google Scholar]
- Bershadsky, A.D., and A.H. Futerman. 1994. Disruption of the Golgi apparatus by brefeldin A blocks cell polarization and inhibits directed cell migration. Proc. Natl. Acad. Sci. USA. 91:5686–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borm, B., R.P. Requardt, V. Herzog, and G. Kirfel. 2005. Membrane ruffles in cell migration: indicators of inefficient lamellipodia adhesion and compartments of actin filament reorganization. Exp. Cell Res. 302:83–95. [DOI] [PubMed] [Google Scholar]
- Calderwood, D.A., V. Tai, G. Di Paolo, P. De Camilli, and M.H. Ginsberg. 2004. Competition for talin results in trans-dominant inhibition of integrin activation. J. Biol. Chem. 279:28889–28895. [DOI] [PubMed] [Google Scholar]
- Carpenter, G., and S. Cohen. 1990. Epidermal growth factor. J. Biol. Chem. 265:7709–7712. [PubMed] [Google Scholar]
- Caswell, P.T., and J.C. Norman. 2006. Integrin trafficking and the control of cell migration. Traffic. 7:14–21. [DOI] [PubMed] [Google Scholar]
- Chen, P., J.E. Murphy-Ullrich, and A. Wells. 1996. A role for gelsolin in actuating epidermal growth factor receptor-mediated cell motility. J. Cell Biol. 134:689–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheresh, D.A., J. Leng, and R.L. Klemke. 1999. Regulation of cell contraction and membrane ruffling by distinct signals in migratory cells. J. Cell Biol. 146:1107–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, J., D.B. Stolz, N.A. Burke, S.C. Watkins, and A. Wells. 2002. Distribution of gelsolin and phosphoinositol 4,5-bisphosphate in lamellipodia during EGF-induced motility. Int. J. Biochem. Cell Biol. 34:776–790. [DOI] [PubMed] [Google Scholar]
- Coppolino, M.G., R. Dierckman, J. Loijens, R.F. Collins, M. Pouladi, J. Jongstra-Bilen, A.D. Schreiber, W.S. Trimble, R. Anderson, and S. Grinstein. 2002. Inhibition of phosphatidylinositol-4-phosphate 5-kinase Ialpha impairs localized actin remodeling and suppresses phagocytosis. J. Biol. Chem. 277:43849–43857. [DOI] [PubMed] [Google Scholar]
- Cram, E.J., and J.E. Schwarzbauer. 2004. The talin wags the dog: new insights into integrin activation. Trends Cell Biol. 14:55–57. [DOI] [PubMed] [Google Scholar]
- Cram, E.J., S.G. Clark, and J.E. Schwarzbauer. 2003. Talin loss-of-function uncovers roles in cell contractility and migration in C. elegans. J. Cell Sci. 116:3871–3878. [DOI] [PubMed] [Google Scholar]
- Critchley, D.R. 2000. Focal adhesions—the cytoskeletal connection. Curr. Opin. Cell Biol. 12:133–139. [DOI] [PubMed] [Google Scholar]
- Delmas, P., B. Coste, N. Gamper, and M.S. Shapiro. 2005. Phosphoinositide lipid second messengers: new paradigms for calcium channel modulation. Neuron. 47:179–182. [DOI] [PubMed] [Google Scholar]
- Di Paolo, G., L. Pellegrini, K. Letinic, G. Cestra, R. Zoncu, S. Voronov, S. Chang, J. Guo, M.R. Wenk, and P. De Camilli. 2002. Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature. 420:85–89. [DOI] [PubMed] [Google Scholar]
- Doughman, R.L., A.J. Firestone, and R.A. Anderson. 2003. a. Phosphatidylinositol phosphate kinases put PI4,5P(2) in its place. J. Membr. Biol. 194:77–89. [DOI] [PubMed] [Google Scholar]
- Doughman, R.L., A.J. Firestone, M.L. Wojtasiak, M.W. Bunce, and R.A. Anderson. 2003. b. Membrane ruffling requires coordination between type Ialpha phosphatidylinositol phosphate kinase and Rac signaling. J. Biol. Chem. 278:23036–23045. [DOI] [PubMed] [Google Scholar]
- Downes, C.P., A. Gray, and J.M. Lucocq. 2005. Probing phosphoinositide functions in signaling and membrane trafficking. Trends Cell Biol. 15:259–268. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville, S. 2004. Actin and microtubules in cell motility: which one is in control? Traffic. 5:470–477. [DOI] [PubMed] [Google Scholar]
- Franco, S.J., M.A. Rodgers, B.J. Perrin, J. Han, D.A. Bennin, D.R. Critchley, and A. Huttenlocher. 2004. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat. Cell Biol. 6:977–983. [DOI] [PubMed] [Google Scholar]
- Goi, T., M. Shipitsin, Z. Lu, D.A. Foster, S.G. Klinz, and L.A. Feig. 2000. An EGF receptor/Ral-GTPase signaling cascade regulates c-Src activity and substrate specificity. EMBO J. 19:623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales, M.L., and R.A. Anderson. 2006. Nuclear phosphoinositide kinases and inositol phospholipids. J. Cell. Biochem. 97:252–260. [DOI] [PubMed] [Google Scholar]
- Heck, J.N., D.L. Mellman, K. Ling, Y. Sun, M.P. Wagoner, N.J. Schill, and R.A. Anderson. 2007. A conspicuous connection: structure defines function for the phosphatidylinositol-phosphate kinase family. Crit. Rev. Biochem. Mol. Biol. 42:15–39. [DOI] [PubMed] [Google Scholar]
- Horwitz, A.R., and J.T. Parsons. 1999. Cell migration—movin' on. Science. 286:1102–1103. [DOI] [PubMed] [Google Scholar]
- Ishihara, H., Y. Shibasaki, N. Kizuki, T. Wada, Y. Yazaki, T. Asano, and Y. Oka. 1998. Type I phosphatidylinositol-4-phosphate 5-kinases. Cloning of the third isoform and deletion/substitution analysis of members of this novel lipid kinase family. J. Biol. Chem. 273:8741–8748. [DOI] [PubMed] [Google Scholar]
- Jekely, G., H.H. Sung, C.M. Luque, and P. Rorth. 2005. Regulators of endocytosis maintain localized receptor tyrosine kinase signaling in guided migration. Dev. Cell. 9:197–207. [DOI] [PubMed] [Google Scholar]
- Kisseleva, M., Y. Feng, M. Ward, C. Song, R.A. Anderson, and G.D. Longmore. 2005. The LIM protein Ajuba regulates phosphatidylinositol 4,5-bisphosphate levels in migrating cells through an interaction with and activation of PIPKI alpha. Mol. Cell. Biol. 25:3956–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger, D.A., and A.F. Horwitz. 1996. Cell migration: a physically integrated molecular process. Cell. 84:359–369. [DOI] [PubMed] [Google Scholar]
- Le Roy, C., and J.L. Wrana. 2005. Signaling and endocytosis: a team effort for cell migration. Dev. Cell. 9:167–168. [DOI] [PubMed] [Google Scholar]
- Ling, K., R.L. Doughman, A.J. Firestone, M.W. Bunce, and R.A. Anderson. 2002. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 420:89–93. [DOI] [PubMed] [Google Scholar]
- Ling, K., R.L. Doughman, V.V. Iyer, A.J. Firestone, S.F. Bairstow, D.F. Mosher, M.D. Schaller, and R.A. Anderson. 2003. Tyrosine phosphorylation of type Iγ phosphatidylinositol phosphate kinase by Src regulates an integrin-talin switch. J. Cell Biol. 163:1339–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, K., N.J. Schill, M.P. Wagoner, Y. Sun, and R.A. Anderson. 2006. Movin' on up: the role of PtdIns(4,5)P(2) in cell migration. Trends Cell Biol. 16:276–284. [DOI] [PubMed] [Google Scholar]
- Ling, K., S.F. Bairstow, C. Carbonara, D.A. Turbin, D.G. Huntsman, and R.A. Anderson. 2007. Type Iγ phosphatidylinositol phosphate kinase modulates adherens junction and E-cadherin trafficking via a direct interaction with μ1B adaptin. J. Cell Biol. 176:343–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel, V., L. Vignoud, S. Dupe, P. Frachet, M.R. Block, and C. Albiges-Rizo. 2000. Talin controls the exit of the integrin alpha 5 beta 1 from an early compartment of the secretory pathway. J. Cell Sci. 113:1951–1961. [DOI] [PubMed] [Google Scholar]
- Martel, V., C. Racaud-Sultan, S. Dupe, C. Marie, F. Paulhe, A. Galmiche, M.R. Block, and C. Albiges-Rizo. 2001. Conformation, localization, and integrin binding of talin depend on its interaction with phosphoinositides. J. Biol. Chem. 276:21217–21227. [DOI] [PubMed] [Google Scholar]
- Martin, T.F. 2001. PI(4,5)P(2) regulation of surface membrane traffic. Curr. Opin. Cell Biol. 13:493–499. [DOI] [PubMed] [Google Scholar]
- Mitchison, T., and M. Kirschner. 1988. Cytoskeletal dynamics and nerve growth. Neuron. 1:761–772. [DOI] [PubMed] [Google Scholar]
- Mouneimne, G., L. Soon, V. DesMarais, M. Sidani, X. Song, S.C. Yip, M. Ghosh, R. Eddy, J.M. Backer, and J. Condeelis. 2004. Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation. J. Cell Biol. 166:697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimne, G., V. DesMarais, M. Sidani, E. Scemes, W. Wang, X. Song, R. Eddy, and J. Condeelis. 2006. Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr. Biol. 16:2193–2205. [DOI] [PubMed] [Google Scholar]
- Nayal, A., D.J. Webb, and A.F. Horwitz. 2004. Talin: an emerging focal point of adhesion dynamics. Curr. Opin. Cell Biol. 16:94–98. [DOI] [PubMed] [Google Scholar]
- Neptune, E.R., and H.R. Bourne. 1997. Receptors induce chemotaxis by releasing the betagamma subunit of Gi, not by activating Gq or Gs. Proc. Natl. Acad. Sci. USA. 94:14489–14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggli, V. 2005. Regulation of protein activities by phosphoinositide phosphates. Annu. Rev. Cell Dev. Biol. 21:57–79. [DOI] [PubMed] [Google Scholar]
- Piccolo, E., P.F. Innominato, M.A. Mariggio, T. Maffucci, S. Iacobelli, and M. Falasca. 2002. The mechanism involved in the regulation of phospholipase Cgamma1 activity in cell migration. Oncogene. 21:6520–6529. [DOI] [PubMed] [Google Scholar]
- Priddle, H., L. Hemmings, S. Monkley, A. Woods, B. Patel, D. Sutton, G.A. Dunn, D. Zicha, and D.R. Critchley. 1998. Disruption of the talin gene compromises focal adhesion assembly in undifferentiated but not differentiated embryonic stem cells. J. Cell Biol. 142:1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosse, C., A. Hatzoglou, M.C. Parrini, M.A. White, P. Chavrier, and J. Camonis. 2006. RalB mobilizes the exocyst to drive cell migration. Mol. Cell. Biol. 26:727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarius, M., C.H. Lee, and R.A. Anderson. 2006. Supervised membrane swimming: small G-protein lifeguards regulate PIPK signalling and monitor intracellular PtdIns(4,5)P2 pools. Biochem. J. 398:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, M.A., and A.R. Horwitz. 2006. Integrating adhesion, protrusion, and contraction during cell migration. Cell. 125:1223–1225. [DOI] [PubMed] [Google Scholar]
- Shao, H., J. Chou, C.J. Baty, N.A. Burke, S.C. Watkins, D.B. Stolz, and A. Wells. 2006. Spatial localization of m-calpain to the plasma membrane by phosphoinositide biphosphate binding during epidermal growth factor receptor-mediated activation. Mol. Cell. Biol. 26:5481–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilenov, L.B., A. Mikhailov, R.J. Pelham, E.E. Marcantonio, and G.G. Gundersen. 1999. Focal adhesion motility revealed in stationary fibroblasts. Science. 286:1172–1174. [DOI] [PubMed] [Google Scholar]
- Sun, Y., Z. Cheng, L. Ma, and G. Pei. 2002. Beta-arrestin2 is critically involved in CXCR4-mediated chemotaxis, and this is mediated by its enhancement of p38 MAPK activation. J. Biol. Chem. 277:49212–49219. [DOI] [PubMed] [Google Scholar]
- Tadokoro, S., S.J. Shattil, K. Eto, V. Tai, R.C. Liddington, J.M. de Pereda, M.H. Ginsberg, and D.A. Calderwood. 2003. Talin binding to integrin beta tails: a final common step in integrin activation. Science. 302:103–106. [DOI] [PubMed] [Google Scholar]
- Tayeb, M.A., M. Skalski, M.C. Cha, M.J. Kean, M. Scaife, and M.G. Coppolino. 2005. Inhibition of SNARE-mediated membrane traffic impairs cell migration. Exp. Cell Res. 305:63–73. [DOI] [PubMed] [Google Scholar]
- Wang, W., S. Goswami, E. Sahai, J.B. Wyckoff, J.E. Segall, and J.S. Condeelis. 2005. Tumor cells caught in the act of invading: their strategy for enhanced cell motility. Trends Cell Biol. 15:138–145. [DOI] [PubMed] [Google Scholar]
- Wang, Y.J., W.H. Li, J. Wang, K. Xu, P. Dong, X. Luo, and H.L. Yin. 2004. Critical role of PIP5KIγ87 in InsP3-mediated Ca2+ signaling. J. Cell Biol. 167:1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, D.J., J.T. Parsons, and A.F. Horwitz. 2002. Adhesion assembly, disassembly and turnover in migrating cells—over and over and over again. Nat. Cell Biol. 4:E97–100. [DOI] [PubMed] [Google Scholar]
- Webb, D.J., K. Donais, L.A. Whitmore, S.M. Thomas, C.E. Turner, J.T. Parsons, and A.F. Horwitz. 2004. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 6:154–161. [DOI] [PubMed] [Google Scholar]
- Wells, C.M., M. Walmsley, S. Ooi, V. Tybulewicz, and A.J. Ridley. 2004. Rac1-deficient macrophages exhibit defects in cell spreading and membrane ruffling but not migration. J. Cell Sci. 117:1259–1268. [DOI] [PubMed] [Google Scholar]
- Wenk, M.R., L. Pellegrini, V.A. Klenchin, G. Di Paolo, S. Chang, L. Daniell, M. Arioka, T.F. Martin, and P. De Camilli. 2001. PIP kinase Igamma is the major PI(4,5)P(2) synthesizing enzyme at the synapse. Neuron. 32:79–88. [DOI] [PubMed] [Google Scholar]
- Xue, C., J. Wyckoff, F. Liang, M. Sidani, S. Violini, K.L. Tsai, Z.Y. Zhang, E. Sahai, J. Condeelis, and J.E. Segall. 2006. Epidermal growth factor receptor overexpression results in increased tumor cell motility in vivo coordinately with enhanced intravasation and metastasis. Cancer Res. 66:192–197. [DOI] [PubMed] [Google Scholar]
- Yang, J., S.A. Mani, J.L. Donaher, S. Ramaswamy, R.A. Itzykson, C. Come, P. Savagner, I. Gitelman, A. Richardson, and R.A. Weinberg. 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 117:927–939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.