Abstract
Although the structure and function of components of the nuclear pore complex (NPC) have been the focus of many studies, relatively little is known about NPC biogenesis. In this study, we report that Apq12 is required for efficient NPC biogenesis in Saccharomyces cerevisiae. Apq12 is an integral membrane protein of the nuclear envelope (NE) and endoplasmic reticulum. Cells lacking Apq12 are cold sensitive for growth, and a subset of their nucleoporins (Nups), those that are primarily components of the cytoplasmic fibrils of the NPC, mislocalize to the cytoplasm. APQ12 deletion also causes defects in NE morphology. In the absence of Apq12, most NPCs appear to be associated with the inner but not the outer nuclear membrane. Low levels of benzyl alcohol, which increases membrane fluidity, prevented Nup mislocalization and restored the proper localization of Nups that had accumulated in cytoplasmic foci upon a shift to lower temperature. Thus, Apq12p connects nuclear pore biogenesis to the dynamics of the NE.
Introduction
In eukaryotic cells, all nucleocytoplasmic transport occurs through nuclear pore complexes (NPCs), which are large macromolecular assemblies (∼44 MD in yeast) that span the nuclear envelope (NE; for review see Tran and Wente, 2006). NPCs show eightfold rotational symmetry in a plane perpendicular to the NE and are constructed using multiple copies of ∼30 proteins, which are termed nucleoporins (Nups). Remarkably, more than half of yeast Nups are individually dispensable for growth, although strains lacking some are temperature sensitive for growth and nucleocytoplasmic transport. The nuclear pore itself can be divided roughly into three domains: the nuclear basket, the central core, and the cytoplasmic filaments. The basket and cytoplasmic filaments are composed of Nups that are found solely in those structures, whereas most other Nups are localized symmetrically on both the nuclear and cytoplasmic sides of the plane of the NE. Three integral membrane proteins are components of NPCs and have been implicated in both the organization and proper assembly of NPCs. Although genetic and biochemical analyses have advanced the identification of the Nups as well as their localizations and interactions within the NPC, the mechanism of NPC biogenesis is poorly understood.
Most nucleocytoplasmic transport is mediated by members of the karyopherin family of receptors. These receptors recognize localization signals in their cargoes and move with their cargoes through the central channel of the NPC. mRNA export is not mediated by karyopherins, and the actual complex exported consists of the mRNA in a complex with proteins, forming a messenger RNP complex. The NPC plays a mechanistic role in transport of molecules between the nucleus and the cytoplasm by providing docking sites for these complexes. FG repeat domains are found in approximately one third of yeast Nups and contain FG repeat domains that have multiple copies of GLFG, XFXFG, or XXFG separated by spacers rich in polar amino acids. Structural studies indicate that FG domains are natively unfolded and are able to bind karyopherins and karyopherin–cargo complexes. It is not known how these complexes selectively penetrate the FG repeat milieu of the NPC channel (Denning et al., 2003).
Screening the collection of ∼4,500 yeast strains each disrupted for one nonessential gene led to the observation that cells lacking Apq12p have defects in both nuclear 3′ pre-mRNA processing (Baker et al., 2004) and mRNA export (Hieronymus et al., 2004). Apq12-GFP localizes to the nuclear periphery and the ER, but it is not a Nup because its distribution is unaffected by mutations that cause NPCs to cluster in one or a few regions of the NE (Baker et al., 2004). More recently, Apq12 was postulated to have a role in cell division, as loss of APQ12 led to synthetic growth defects when combined with mutations affecting genes coding for spindle pole body (SPB) proteins and other proteins involved in cell division. When Apq12p was not present, anaphase was delayed, and re-replication of DNA before completion of cytokinesis was also observed (Montpetit et al., 2005).
Because of our interest in mRNA biogenesis and export, we examined how the absence of Apq12p affected various aspects of nucleocytoplasmic transport. We report that Apq12p is an integral membrane protein found within the NE and in the ER (Baker et al., 2004). In addition to a partial block in mRNA export, the absence of Apq12p led to cold-sensitive defects in the growth and localization of a subset of Nups, particularly those asymmetrically localized to the cytoplasmic fibrils. In addition, cells lacking Apq12 displayed defects both in NPC biogenesis and in the morphology of the NE. We suggest that these defects are caused by alterations in the dynamics and properties of the NE because the proper localization of Nups in apq12Δ cells was restored upon addition to the medium of benzyl alcohol (BA), which is thought to increase the fluidity and flexibility of membranes (Colley and Metcalfe, 1972; Gordon et al., 1980). Thus, it is likely that the reported defects in mRNA export, pre-mRNA processing, and cell division of apq12Δ cells are indirect consequences of altered membrane dynamics. Collectively, our results demonstrate the dependence of NPC biogenesis and function on the physical properties of the nuclear membranes.
Results
Apq12p is an integral membrane protein
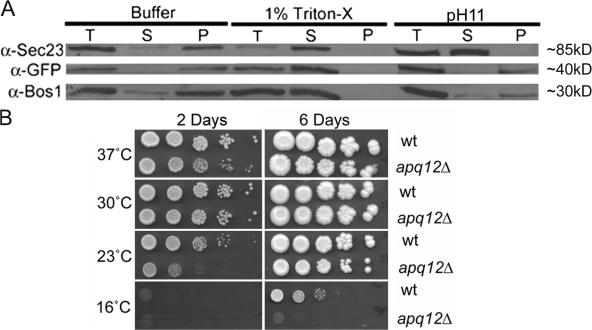
A previous study demonstrated that Apq12 localizes to the NE/ER and that the protein is not associated exclusively with NPCs (Baker et al., 2004). Hydropathy analyses using the program TMHMM (Krogh et al., 2001) revealed two predicted transmembrane domains (amino acids 40–62 and 69–91). To determine biochemically whether Apq12 is a transmembrane protein, a lysate was prepared from a strain that produces a C-terminally tagged Apq12p-GFP from the genomic APQ12 locus. Lysates were treated with either Triton X-100, high pH, or buffer alone and subsequently separated into supernatant and pellet fractions by centrifugation. Transmembrane proteins will be found predominantly in the pellet fraction after treatment with high pH or buffer alone. In contrast, high pH treatment causes the release from the membrane of peripherally associated proteins, and they will be found in the supernatant. Immunoblotting of the different fractions with α-GFP antibodies revealed that Apq12-GFP remained in the pellet fraction during high pH treatment and only shifted to the supernatant when lysates were treated with detergent (Fig. 1 A). As controls, Sec23, a peripherally associated ER protein, was found in the supernatant fraction after high pH treatment, whereas the integral membrane ER protein Bos1, like Apq12-GFP, remained in the pellet fraction after high pH treatment. These results prove that Apq12 is an integral membrane protein.
Figure 1.
APQ12 encodes a transmembrane protein that, when absent, results in cold sensitivity. (A) Differential centrifugation to determine membrane association. In addition to antibodies against GFP, the blot was also probed using antibodies directed against a control integral membrane protein (Bos1) as well as a peripherally associated membrane protein, Sec23. (B) Growth assays comparing WT and apq12Δ cells at various temperatures after 2 and 6 d. T, total; S, soluble; P, pellet.
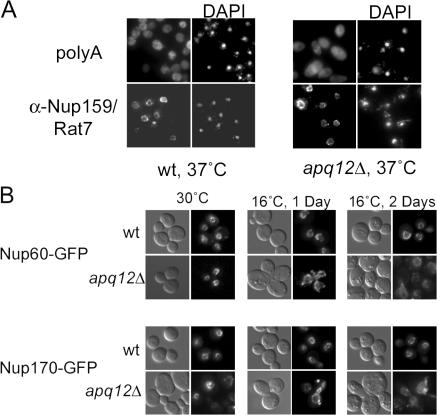
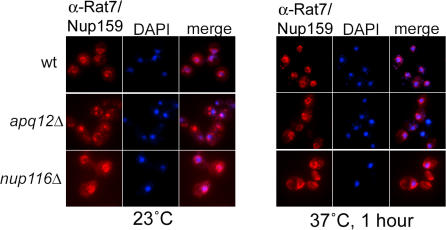
Although APQ12 is not essential, cells lacking it grow more slowly at 23°C than do wild-type (WT) cells (Fig. 1 B). We compared the growth behavior of apq12Δ and WT at 16, 23, 30, and 37°C. apq12Δ cells grew as well as WT at both 30 and 37°C but were cold sensitive and barely able to grow at 16°C. Notably, the previously described apical cell morphology (Baker et al., 2004) and defect in mRNA export (Baker et al., 2004; Hieronymus et al., 2004) were observed in apq12Δ cells grown at 23°C but were not seen in cells maintained at 37°C (see Fig. 3 A).
Figure 3.
apq12Δ cell growth rates correlate with Nup localization and function. (A) Images of WT and apq12Δ cells processed for either FISH assays or indirect IF using α-Nup159/Rat7 antibodies after overnight growth at 37°C. (B) WT or apq12Δ cells expressing either NUP60- or NUP170-GFP from the proper chromosomal locus were grown overnight at 30°C, shifted to 16°C, and viewed after either 1 or 2 d.
APQ12 interacts genetically with genes coding for Nups
Because the deletion of APQ12 led to defects in mRNA export, we investigated whether there were genetic interactions between apq12Δ and mutations affecting genes required for nucleocytoplasmic transport. We crossed the apq12Δ strain with haploid strains harboring deletions of genes encoding nonessential Nups or ts alleles of genes encoding essential Nups and mRNA export factors, including rat8-2, rat7ΔN/nup159ΔN, rss1-37/gle1-37, nup120Δ, and mex67-5. Heterozygous diploids were sporulated, tetrads were dissected, and haploid progeny were scored for the presence of both mutations. Growth of double mutant haploid strains was analyzed at temperatures ranging from 23 to 37°C. Of the five mutants listed above, nup120Δ had the most severe effect on the growth of apq12Δ (Table I). Nup120 is a nonessential structural component of the NPC (Heath et al., 1995). Strong synthetic growth defects were seen with rat8-2 and rat7ΔN/nup159ΔN, but no enhanced growth defect was seen when apq12Δ was combined with mex67-5. Mex67 is the mRNA export receptor and mediates interactions between the messenger RNP and NPCs during mRNA export (Segref, et al., 1997).
Table I.
Synthetic interactions of apq12Δ
| Relevant genotype | 23°C | 30°C | 34°C | 37°C |
|---|---|---|---|---|
| apq12Δ | ++ | +++ | +++ | +++ |
| rat8-2 (dbp5) apq12Δ/rat8-2 | ++ + | +++ + | + − | − − |
| mex67-5 apq12Δ/mex67-5 | ++ ++ | +++ +++ | + + | − − |
| rat7ΔN(nup159ΔN apq12Δ/rat7ΔN | ++ + | +++ + | ++ + | − − |
| gle1-37(rss1-37) apq12Δ/gle1-37 | ++ + | ++ ++ | − − | − − |
| nup42Δ (rip1Δ) apq12Δ/nup42Δ | +++ +++ | +++ +++ | +++ +++ | +++ +++ |
| nup116Δ apq12Δ/nup116Δ | ++ + | ++ + | + + | − − |
| nup120Δ apq12Δ/nup120Δ | ++ + | +++ + | + − | − − |
| nup85Δ apq12Δ/nup85Δ | ++ + | +++ + | + − | − − |
| nup188Δ apq12Δ/nup188Δ | +++ + | +++ + | +++ + | +++ − |
| nup170Δ apq12Δ/nup170Δ | ++ + | +++ + | +++ + | +++ − |
| nup100Δ apq12Δ/nup100Δ | +++ ++ | +++ +++ | +++ +++ | +++ +++ |
| nup59Δ apq12Δ/nup59Δ | +++ +++ | +++ +++ | +++ +++ | +++ +++ |
| nup2Δ apq12Δ/nup2Δ | +++ ++ | +++ ++ | +++ ++ | +++ ++ |
| nup60Δ apq12Δ/nup60Δ | +++ ++ | +++ ++ | +++ ++ | +++ ++ |
| pom34Δ apq12Δ/pom34Δ | +++ + | +++ ++ | +++ ++ | +++ ++ |
| pom152Δ apq12Δ/pom152Δ | +++ + | +++ ++ | +++ + | +++ + |
| ndc1-39 apq12Δ/ndc1-39 | +++ + | ++ + | + − | − − |
Synthetic interaction chart for strains that contain the apq12Δ disruption in combination with mutations in genes coding for Nups and Nup-associated proteins. +++, WT growth; ++ and +, synthetically sick; −, synthetically lethal.
We expanded our genetic analyses to include additional strains in which a gene encoding a nonessential Nup was disrupted. The data are summarized in Table I. Of the others tested, the most severe growth defects were seen when apq12Δ was combined with disruptions of NUP170, NUP188, NUP85, NUP116, and POM152. A strong growth defect was also seen in an apq12Δ/ndc1-39 strain. Ndc1 is an essential integral membrane protein and is the only protein found in both NPCs and the SPB. Note that little or no defect was seen when apq12Δ was combined with either nup60Δ or nup2Δ. Nup2 and Nup60 are components of the nuclear basket of the NPC. Together, these results suggest that Apq12 is important for NPC function or biogenesis.
Loss of Apq12 affects the localization of several Nups
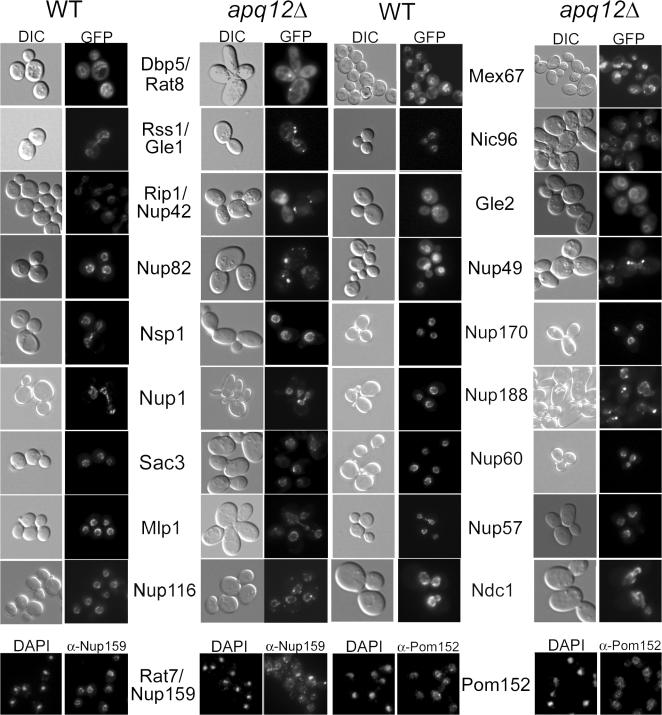
Because of these genetic interactions, we examined the localization of several Nup-GFP fusion proteins in apq12Δ cells at 23°C (Fig. 2). We also examined the localization of Nup159/Rat7 and Pom152 by indirect immunofluorescence (IF) using antibodies directed against each protein. Nuclear basket components Nup1 and Nup60 were not mislocalized in apq12Δ cells nor were Sac3 or Mlp1, two proteins that associate with nuclear basket Nups (Strambio-de-Castillia et al., 1999; Fischer et al., 2002). Similarly, we observed little or no mislocalization of Nup170, Gle2, Nsp1, or Nup57 in apq12Δ cells, all of which are thought to be components of the central structural framework of the NPC. Normal localization was also seen for two integral membrane Nups, Pom152 and Ndc1. Although Nup188, Nup49, and Nic96, which are also core components of the NPC, were not entirely lost from the nuclear periphery, there were subtle differences in their localization in apq12Δ cells compared with WT. For example, in cells lacking Apq12, there were studs or bright foci of Nup188- and Nup49-GFP distributed around the nuclear periphery in contrast to their relatively uniform punctate distribution around the nuclear periphery in WT cells.
Figure 2.
Loss of APQ12 causes the severe mislocalization of cytoplasmic filament Nups. Localizations of various Nups at 23°C in a WT versus apq12Δ background. Images for Nup159/Rat7 and Pom152 were from indirect IF experiments using antibodies directed against the respective proteins; all others were viewed via live microscopy using Nup-GFP fusions. DIC, differential interference contrast.
In contrast to the majority of nuclear basket and core Nups, we observed dramatic defects in the localization of all Nups that are components of the cytoplasmic filaments of the NPC. In the absence of Apq12, Nup42/Rip1, Gle1/Rss1, Nup82, and Nup159/Rat7 mislocalized to foci. Some foci were adjacent to the nuclear periphery, and others were cytoplasmic and appeared to be completely detached from the NE/NPC. We used Western blotting to compare the levels of several Nups in apq12Δ and WT cells and saw no differences in those analyzed (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200702120/DC1). In addition to Nup mislocalization, we observed the dramatic mislocalization of Dbp5/Rat8-GFP, an essential mRNA export factor that shuttles between the nucleus and cytoplasm and performs key functions during mRNA export when bound to the cytoplasmic filaments of the NPC (Hodge et al., 1999, Weirich et al., 2004).
To ensure that the mislocalization observed did not result from a synthetic growth defect caused by the presence of a Nup-GFP fusion and the absence of Apq12, we compared the growth of apq12Δ cells with several apq12Δ strains expressing Nup-GFP fusions. All grew at similar rates (unpublished data). Furthermore, using antibodies directed against the GLFG repeats, we also saw the mislocalization of Nups recognized by this antibody as well as an overall decrease in staining at the nuclear periphery (unpublished data).
Because apq12Δ cells grow as well as WT at 37°C (Fig. 1 B), we analyzed at 37°C both mRNA export and the localization of Nups that were mislocalized at 23°C. At this optimal growth temperature, the localization of the cytoplasmic filament Nups (Nup159/Rat7 and Nup82) was normal, and no defect in mRNA export was seen (Fig. 3 A; also see Fig. 6 A).
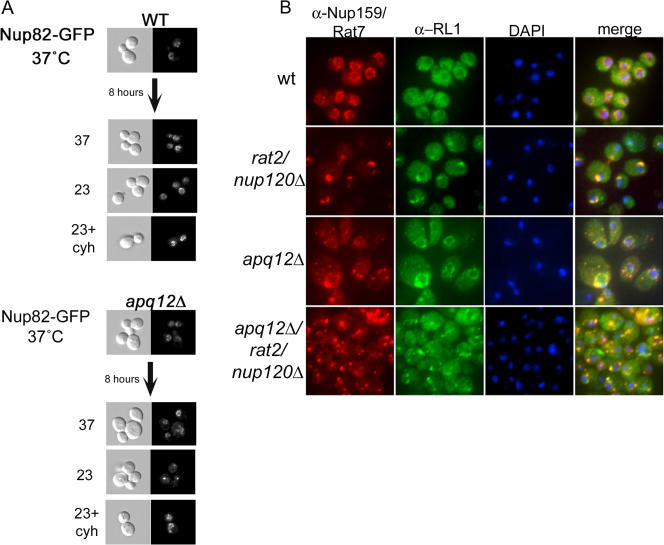
Figure 6.
Apq12 is required for optimal NPC biogenesis. (A) WT or apq12Δ cells expressing Nup82-GFP were grown overnight at 37°C and either maintained at 37°C or shifted to 23°C with or without the translational inhibitor cycloheximide. Cells were then imaged after 8 h. (B) WT and apq12Δ cells lacking NUP120 were grown overnight at 23°C and processed for indirect IF using both α-Nup159/Rat7 and α-RL1 antibodies.
Both Nup60- and Nup170-GFP were properly localized in apq12Δ cells at both 23 (Fig. 2) and 30°C (Fig. 3 B). We shifted WT and apq12Δ cells expressing Nup60- or Nup170-GFP from 30 to 16°C and examined their localization after 1 and 2 d. Both Nup170- and Nup60-GFP became abnormally distributed in apq12Δ cells while retaining normal distribution in WT cells (Fig. 3 B). Punctate Nup-GFP foci were seen, and the fluorescent signal became more diffuse, with a notable increase in the intranuclear signal. Thus, even Nups whose distribution was normal in slow growing apq12Δ cells at 23°C became mislocalized in cells shifted to a more restrictive temperature (16°C). In some apq12Δ cells, the nucleus itself became misshapen at 16°C (Fig. 3 B), which is consistent with cold-sensitive defects in the NE. Collectively, the data indicate that Nup localization defects increased in severity and extent as the temperature was reduced.
In the absence of Apq12, the NE is abnormal, and defects in the distribution of NPCs occur
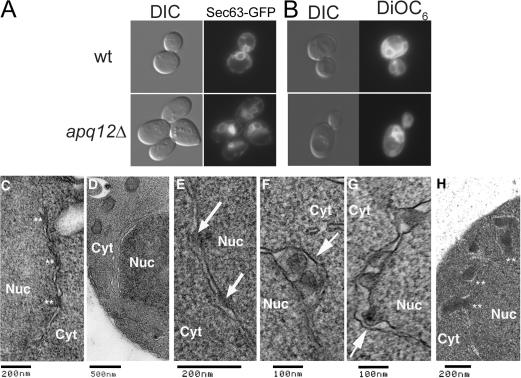
Because Apq12 localizes to the NE and cells lacking Apq12 have NPC defects, we examined the NE by light and electron microscopy. Many ER proteins are present in the outer nuclear membrane (ONM) because the two are continuous. Therefore, we assayed for nuclear membrane deformities by examining the distribution of a GFP fusion to Sec63, a resident ER protein (Prinz et al., 2000), and also by staining live cells with 3,3′- dihexyloxacarbocyanine iodide (DiOC6), a fluorescent lipophylic dye that permits easy visualization of ER and nuclear membranes in yeast (Koning et al., 1993). In WT cells maintained at 23°C, both Sec63-GFP (Fig. 4 A) and DiOC6 staining (Fig. 4 B) formed continuous rings surrounding the nucleus. In apq12Δ cells, there were several abnormalities, including membranous divisions within the nucleus, extra protrusions of ER membrane not normally seen in WT cells, and studs of fluorescent signal adjacent to the NE.
Figure 4.
Loss of APQ12 causes NE abnormalities. (A and B) WT and apq12Δ cells either harboring a SEC63-GFP plasmid (A) or stained with DiOC6 (B) were viewed via live microscopy. (C–G) Electron microscopy images of either WT (C) or apq12Δ cells (D–G) at 23°C. (H) apq12Δ cells processed for and imaged via electron microscopy after overnight growth at 37°C. Asterisks in C indicate normal NPCs. Arrows in E point to NPCs contacting only the INM. The arrow in F points to an electron-dense inclusion within the lumen of the NE. The arrow in G points to an electron-dense inclusion extending into the lumen of the NE. Asterisks in H indicate protrusions of the NE containing electron-dense material. DIC, differential interference contrast.
To gain further insight into the apq12Δ defect, we performed electron microscopy to examine NE ultrastructure (Fig. 4, C–H). In WT cells, NPCs appear as electron-dense material extending from the inner nuclear membrane (INM) to the ONM (Fig. 4 C, asterisks). We observed a range of defects in both NPCs and the NE in apq12Δ cells. In apq12Δ cells grown overnight at 23°C, >90% of NPCs examined contacted only the INM (Fig. 4 E, arrows). Groups of NPCs associated with the NE were seen in some apq12Δ cells (Fig. 4 F, arrow), and these probably correspond to the bright fluorescent foci seen in Fig. 3 A and with some Nup-GFPs. Invaginations and/or extensions of the NE (Fig. 4 D) were seen in 30–40% of the nuclei examined. However, because apq12Δ cells are able to grow at 23°C, apq12Δ cells must have some normal nuclear pores, and these were seen, although rarely (unpublished data). We also observed many cases in which large electron-dense inclusions extended into the lumen of the NE (Fig. 4 G, arrow). Sometimes, these were located entirely within the lumen (Fig. 4 F, arrow).
Because growth at 37°C prevented the Nup82 localization defect seen at 23°C, we also performed electron microscope on cells grown continuously at 37°C. Although NPCs that were unable to associate with the ONM were still seen (unpublished data), >85% of NPCs appeared normal. Protrusions of the NE that contained electron-dense material (Fig. 4 H, asterisks) were still seen in ∼50% of nuclei examined, and only rarely did these contain multiple inclusions, which is in contrast to what was seen at 23°C (Fig. 4 G). More than 200 NPCs were examined in determining these percentages.
Nuclear membrane herniations and membrane seals covering some NPCs are phenotypes associated with the deletion of nonessential NUP116 (Wente and Blobel, 1993). Because apq12Δ cells also showed membrane seals covering some NPCs, we compared the localization of Nup159/Rat7 in apq12Δ and nup116Δ cells. The data in Fig. 5 show that there was a much more severe defect in the localization of Nup159/Rat7 in apq12Δ than in nup116Δ. We also constructed an apq12Δ/nup116Δ double mutant strain, and none of the morphological defects seen in either single mutant were exacerbated by combining the mutations (unpublished data). However, the double mutant strain grew considerably less well than either single mutant at 23 or 30°C and did not grow at 37°C (Table I).
Figure 5.
nup116Δ cells display normal localization of Nup159/Rat7. WT, apq12Δ, and nup116Δ cells were grown overnight at 23°C, either maintained at 23°C or shifted to 37°C for 1 h, and processed for indirect IF using α-Nup159/Rat7 antibodies.
Normal ER function in cells lacking apq12Δ
Although there were no readily observed morphological defects in the peripheral ER in apq12Δ cells (Fig. 4, A and B), we analyzed apq12Δ cells for defects in ER function because Apq12 is present throughout the ER. No defect was observed in the maturation of carboxy-peptidase Y, a recognized cargo of ER protein transport that is modified within the Golgi and is ultimately delivered to the vacuole (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200702120/DC1). The accumulation of unfolded proteins in the ER activates the unfolded protein response (UPR; Sidrauski et al., 1998). In yeast strains defective in ER protein trafficking and secretion, cells typically display a constitutively active UPR. The UPR leads to the activation of several genes that encode proteins needed for addressing the increased level of unfolded proteins in the ER. A conserved DNA sequence element (the UPR element [UPRE]) is a feature of promoters activated by the UPR (Mori et al., 1992; Cox et al., 1993). Thus, the expression of GFP from a promoter containing a UPRE has been used as a measure of the extent to which the UPR has been activated in different genetic backgrounds (Pollard et al., 1998; Travers et al., 2000). We saw no induction of a UPRE-GFP reporter in apq12Δ cells, although the reporter was activated in the control erv25Δ strain (Fig. S2 B).
Apq12 is required for proper NPC biogenesis
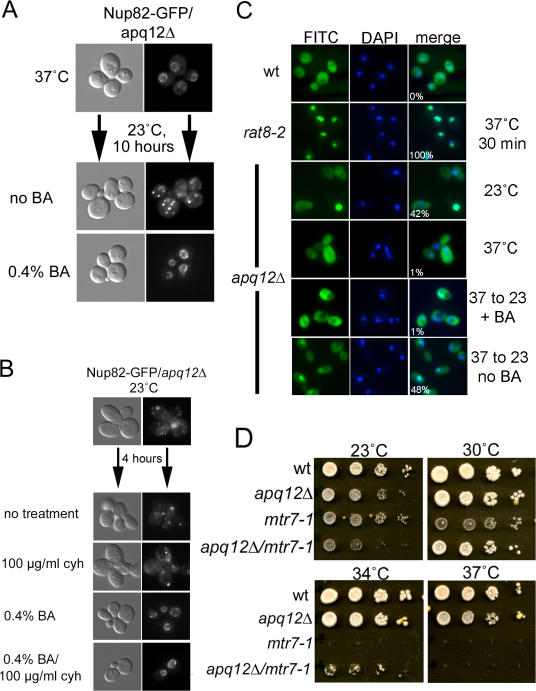
Mislocalization of Nups could reflect defects in NPC biogenesis, NPC stability, or both. We took advantage of the cold sensitivity of apq12Δ cells to ask whether Apq12 is required for proper NPC biogenesis. First, we determined how the distribution of Nup82-GFP changed over time in cells shifted from 37 to 23°C. We diluted cells grown overnight at 37°C to restore exponential growth and shifted them to 23°C. Mislocalization of Nup82-GFP was detectable but minimal 2 h after the shift (unpublished data) and was complete by 8 h (Fig. 6 A). No mislocalization was seen in apq12Δ cells maintained at 37°C.
To distinguish between defects in NPC assembly and stability, we treated cells with the translation inhibitor cycloheximide to prevent the synthesis of new Nups. We reasoned that if mislocalization did not occur in cycloheximide-treated cells, this would indicate that NPCs produced at 37°C were stable at 23°C and that the cytoplasmic foci of GFP-tagged Nups resulted from a defect in NPC assembly. This approach was used earlier by Ryan et al. (2003, 2006) to investigate the genetic requirements for yeast NPC biogenesis. Cells were grown overnight at 37°C, diluted to restore exponential growth, and shifted to 23°C with or without the addition of cycloheximide. As shown in Fig. 6 A, the normal localization of Nup82-GFP was retained in cycloheximide-treated apq12Δ cells shifted to 23°C but not in the untreated control. We conclude that apq12Δ cells are defective in NPC biogenesis.
As another approach to examine whether Apq12 plays a role in NPC biogenesis, we examined the localization of Nup159/Rat7 (using anti-Nup159/Rat7 antisera) and the various Nups (including Nsp1) recognized by the RL1 monoclonal antibody (Copeland and Synder, 1993) in an apq12Δ/nup120Δ double mutant strain (Fig. 6 B). In nup120Δ cells, NPCs cluster to one area within the NE (Heath et al., 1995), and both antibodies recognized these clusters (Fig. 6 B; see merge in the nup120Δ row). In contrast, in apq12Δ cells, RL1 antibody stained the nuclear periphery as well as cytoplasmic foci, presumably as a result of the antibody recognizing some cytoplasmic filament Nups. In the double mutant, very few cells had NPC clusters, and the mislocalization defects seen in apq12Δ were enhanced: cytoplasmic foci (stained using RL1) were much brighter in apq12Δ/nup120Δ than in apq12Δ cells, indicating that additional Nups became mislocalized in double mutant cells, and some accumulated in cytoplasmic foci.
Although both Nup159/Rat7 and Gle1/Rss1 are located asymmetrically on the cytoplasmic side of NPCs, a direct interaction between them has not been detected. However, indirect IF indicated that these two Nups colocalized to cytoplasmic foci in apq12Δ cells (unpublished data). Further investigation of these foci using indirect IF also demonstrated the colocalization of Nup82-GFP with Nup159/Rat7 and with a fraction of Nup170-GFP (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200702120/DC1). We suggest that the foci contain Nup subcomplexes that are unable to be assembled into NPCs.
Alteration of membrane dynamics suppresses Nup mislocalization in apq12Δ cells
The aforementioned results suggest that the observed mislocalization of some Nups and the defect in NPC biogenesis might result from altered physical properties of the NE. Cold sensitivity is a phenotype sometimes observed with defective membrane proteins (Baba et al., 1990; Lau et al., 2004), and apq12Δ cells are cold sensitive for the growth and proper organization of NPCs within the NE. One mechanism cells use to cope with a reduction in growth temperature is to modify the composition of membranes by increasing the abundance of phospholipids that contain shorter and unsaturated acyl chains (Nishida and Murata, 1996). This allows the maintenance of membrane fluidity and flexibility at lower temperatures. Alterations in the protein components of membranes may also contribute to the maintenance of normal membrane properties after temperature shifts. Recently, it was demonstrated that ethanol-induced changes in fluidity of the NE in yeast cells caused defects in both NPC organization and nuclear transport (Izawa et al., 2004). This suggests that the apq12Δ-associated phenotypes might be caused by the mutant strain's inability to modulate membrane composition at lower temperatures. To test this hypothesis, we added low levels of BA to the media to increase membrane fluidity (Colley and Metcalfe, 1972; Gordon et al., 1980). apq12Δ cells expressing Nup82-GFP were incubated overnight at 37°C and shifted to 23°C for 10 h with or without 0.4% BA. In cells cultured at 23°C with BA, the mislocalization of Nup82-GFP was prevented (Fig. 7 A). This suggests that apq12Δ cells are defective in adjusting the composition of the NE so as to maintain proper flexibility or fluidity at lower growth temperatures.
Figure 7.
In apq12Δ cells, the membrane fluidizer BA suppresses defects in NPC biogenesis and mRNA export. (A) apq12Δ cells expressing Nup82-GFP were grown overnight at 37°C and shifted to 23°C in the presence (0.4% BA) or absence (no BA) of BA. These cells were then imaged after 10 h at 23°C. (B) apq12Δ cells expressing Nup82-GFP were grown overnight at 23°C, treated with either BA, cycloheximide, or both, and imaged after 4 h of treatment. (C) analysis of mRNA export in apq12Δ cells treated with BA before the shift to 23°C. (D) Growth assay comparing WT, apq12Δ, mtr7 –1(acc1), and the double mutant apq12Δ/mtr7-1 strains. Plates were incubated at the temperatures shown for 3 d.
We extended this experiment by adding BA to cells that had been grown overnight at 23°C so that Nup82-GFP was already mislocalized when BA was added (Fig. 7 B). Within 4 h of BA addition, cells had regained the normal localization for Nup82-GFP at the nuclear periphery, suggesting that modifying membrane properties could correct Nup mislocalization. This normal pattern was also restored if cycloheximide was added simultaneously with BA. This indicates that restoration of the normal pattern reflects assembly into NPCs of Nups or Nup complexes already present and mislocalized in cells grown overnight at 23°C. Cycloheximide added without BA had no effect on previously mislocalized Nup82-GFP. We repeated this assay with an apq12Δ strain expressing both a Nup82-GFP fusion and a Sec63-RFP (fusion from their respective chromosomal loci) so that NPC localization and ER morphology could be monitored simultaneously. As shown in Fig. S4 A (available at http://www.jcb.org/cgi/content/full/jcb.200702120/DC1), the morphology of the NE was restored to near normal, but some defects in Sec63-RFP localization were still observed.
We next wanted to determine whether the properly localized NPCs in apq12Δ cells treated with BA were functional. To do this, we examined mRNA export in BA-treated apq12Δ cells. Only a low percentage of these cells exhibited a defect, suggesting that BA treatment had enhanced the formation of functional NPCs in apq12 cells (Fig. 7 C). Likewise, BA corrected the mislocalization of Nup188-GFP in apq12Δ cells (Fig. S4 B). Because extended exposure to 0.4% BA had deleterious effects on the growth of WT cells, we were unable to analyze the ability of BA to suppress the cold-sensitive growth defect of apq12Δ cells.
Nup mislocalization has been observed in other mutant yeast strains. Both brr6-1 and prp20-G282S cells accumulate Nups in cytoplasmic foci, similar to what was seen with apq12Δ. BRR6 was identified in a screen for cold-sensitive mutants defective in mRNA export and, like APQ12, encodes an integral membrane protein of the NE and ER (de Bruyn Kops and Guthrie, 2001). prp20-G282S is a novel allele of PRP20, encoding the guanine nucleotide exchange factor for Gsp1 (Ran), and, unlike other prp20 alleles, this mutation is unique in that it causes defects in NPC biogenesis. In contrast to other mutant alleles, prp20-G282S does not affect nuclear transport in general (Ryan et al., 2003). The addition of BA was able to partially correct for the mislocalization of Nic96-GFP and Nup170-GFP in the prp20-G282-S strain (Fig. S4 C). Little change was seen in the location of Nup159 in brr6-1 cells shifted to 16°C (Fig. S4 D).
Genetic interactions occur between the mtr7-1 mutation and apq12Δ
The observation that the addition of BA suppresses the NPC biogenesis and mRNA export defects of apq12Δ cells suggests that the properties and composition of the NE are affected directly by the absence of Apq12. The mtr7-1 allele of ACC1 (encoding acetyl-CoA carboxylase) is known to have defects in the lipid composition of its membranes. Acc1/Mtr7 catalyzes the rate-limiting step in synthesis of fatty acids, and the mtr7-1 mutant was particularly defective in the synthesis of very long chain fatty acids. We constructed a strain carrying both apq12Δ and mtr7-1 and examined its growth at several temperatures (Fig. 7 D). At 23°C, the double mutant strain grew less well than either single mutant, but at both 30 and 34°C, the double mutant strain showed enhanced growth in comparison with the mtr7-1 single mutant. At 37°C, both the double mutant and mtr7-1 cells could not grow. The enhanced defects at lower temperature suggest that double mutant cells are less able to respond to a reduction in temperature and support the hypotheses that both Apq12 and Acc1/Mtr7 affect the composition and, therefore, the properties of the NE. Interestingly, at 23°C, the apq12Δ/mtr7-1 double mutant had a more severe mRNA export defect than did apq12Δ cells (unpublished data).
The cell division defects resulting from the absence of Apq12 are independent of mRNA export defects of apq12Δ
During interphase, the nucleus maintains a relatively stable morphology, which is in contrast to the dynamic changes it undergoes in location and structure during cell division. These alterations can be observed easily by staining live cells with DiOC6 during various stages of the cell cycle (Koning et al., 1993). apq12Δ cells are known to have cell shape and cell division defects (Montpetit et al., 2005). We wondered whether these phenotypes might be related to and were a consequence of defects in the NE and NPCs.
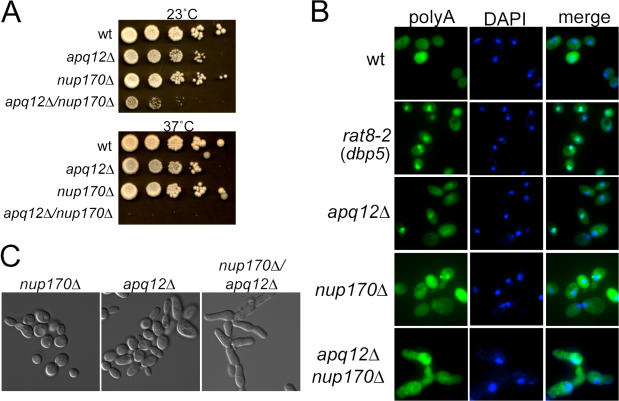
Nup170 is a structural component of NPCs (Aitchison et al., 1995). Much like Apq12, Nup170 has also been implicated in playing a role in cytokinesis, as the mutation of NUP170 leads to defects in chromosome segregation and kinetochore integrity (Kerscher et al., 2001). Cells carrying deletions of both apq12 and nup170 grew considerably less well at 23°C than either single deletion mutant, and synthetic lethality between apq12Δ and nup170Δ was seen at 37°C (Fig. 8 A). Because the absence of either Apq12 or Nup170 leads to defects in cell division and nuclear transport/structure, we saw this dramatic synthetic growth defect as an opportunity to determine whether or not there was interdependence between the two defects. We assayed both single mutants and the apq12Δ/nup170Δ double mutant for defects in mRNA export and Nup159/Rat7 localization. Surprisingly, double mutant cells were no more defective for mRNA export at 23°C than apq12Δ cells (Fig. 8 B), and the mislocalization of Nup159/Rat7 seen in apq12Δ cells was also reduced in double mutant cells at 23°C (unpublished data). We next assessed whether or not the deletion of NUP170 enhanced the cell division defects of apq12Δ. Fig. 8 C shows that double mutant cells were morphologically more defective than either of the parental mutants, and acquired a pseudohyphal appearance. DiOC6 staining of double mutant cells showed that the deletion of NUP170 enhanced the apq12Δ nuclear dynamic defects during division, and many cells appeared to have multiple nuclei (unpublished data). Collectively, the data indicate that the deletion of NUP170 enhanced the nuclear membrane and cell division defects of apq12Δ cells, had relatively little effect on the mRNA export defect of apq12Δ cells, and partially suppressed the Nup mislocalization defect of apq12Δ cells. These results suggest that the apq12Δ defects in cell division and NPC function do not depend on each other and are both consequences of NE abnormalities.
Figure 8.
Defects in NEs/NPCs may also be the cause of apq12Δ-associated defects in cell division. (A) Growth assays comparing the growth of WT, apq12Δ, nup170Δ, and apq12Δ/nup170Δ at 23 and 37°C. (B) FISH of various strains after growth overnight at 23°C. (C) Differential interference contrast images of nup170Δ, apq12Δ, and nup170Δ/apq12Δ cells after growth overnight at 23°C.
Discussion
Because of our long-standing interest in mRNA export, the reported defect in mRNA export in apq12Δ cells (Baker et al., 2004; Hieronymus et al., 2004), and the synthetic lethality we observed between apq12Δ and rat8-2, we decided to investigate the function of Apq12. Apq12 is an integral membrane protein (Fig. 1) present in both the ER and NE (Huh et al., 2003; Baker et al., 2004). apq12Δ showed synthetic growth defects when combined with the deletion of any one of several nonessential Nups and with ts alleles affecting several essential Nups (Table I).
The absence of APQ12 led to defects in NPC biogenesis and the mislocalization of several Nups, including those that comprise the cytoplasmic filaments of the NPC (Nup159/Rat7, Nup82, Gle1/Rss1, and Nup42/Rip1; Fig. 2). Dbp5/Rat8, which binds to these filaments, was also mislocalized in apq12Δ cells. Nups that were not NPC associated in apq12Δ cells could be detected in foci that contained multiple Nups and Dbp5/Rat8 (Fig. 6 B and not depicted). Strikingly, the Nups in these aggregates retained the ability to be incorporated into NPCs, as the addition of BA restored their localization to NPCs under conditions in which no new Nups could be synthesized (Fig. 7 B).
NPC biogenesis and nuclear membrane dynamics
It is not surprising that some Nups are essential for early steps in NPC biogenesis. Pom152, Pom34, and Ndc1 are transmembrane Nups enriched in NPC-containing subcellular fractions (Rout et al., 2000). Of these, only Ndc1 is essential, and it is also found in SPBs. It is thought that transmembrane Nups initiate NPC construction within the NE and coordinate the early steps of NPC biogenesis, but little is known about how these Nups find one another or the extent to which Nups associate into subcomplexes before their assembly into NPCs. The deletion of POM152 or mutation of NDC1 causes genetic defects when combined with the mutation of NIC96 (Aitchison et al., 1995; Lau et al., 2004), a Nup with a demonstrated role in NPC assembly (Zabel et al., 1996). Similarly, perturbations of NPC structure and function were seen in pom34Δ/nup188Δ double mutant cells (Miao et al., 2006). Electron microscopy studies revealed that the pores of ndc1/pom152 double mutant strains had larger diameters than WT cells, and, as a consequence, NPC transport was perturbed (Madrid et al., 2006). This suggests that in addition to a role in biogenesis, these proteins contribute to the overall structural integrity of the pore.
NPC biogenesis also depends on factors that are not components of NPCs. Recent work suggests that the Ran cycle is required for targeting NPC components such as integral membrane Nups to the NE via a vesicular intermediate (Ryan et al., 2003). Mutations affecting Brr6, another NE protein, result in the altered localization of some Nups (de Bruyn Kops and Guthrie, 2001). Like Apq12, Brr6 was not found in NPC-containing subcellular fractions (Rout et al., 2000) and does not cluster in yeast mutant strains where NPCs cluster. Defects in lipid metabolism have effects on the NE that lead to defects in nuclear transport, presumably by affecting NPCs. An earlier screen for mRNA export mutants identified mtr7-1, a temperature-sensitive allele of acetyl coA carboxylase (Acc1/Mtr7; Schneiter et al., 1996). Some of the gross abnormalities of the NE observed in mtr7-1 cells (Kadowaki et al., 1994; Schneiter et al., 1996) were similar to what we observed in apq12Δ cells (Fig. 4). It is likely that the mRNA export defect of acc1/mtr7 mutants is an indirect consequence of defects in the nuclear membrane that impact NPC biogenesis and function. We hypothesize that this is also the case for apq12Δ.
Biological membranes respond to changes in temperature. Membranes contain a diverse mixture of lipids, and phospholipid acyl chains vary in length and in degree of saturation. By an unknown signaling mechanism, cells perceive changes in temperature, and one response is the alteration of membrane lipid composition. A downward shift in growth temperature generally leads to an increase in the proportion of shorter acyl chains and unsaturated acyl chains in membrane phospholipids, resulting in the maintenance of normal fluidity and membrane functions at lower temperatures (Nishida and Murata, 1996). It has been suggested that the cells have a sensor that responds directly to altered fluidity or flexibility by activating the expression of genes needed to modify membrane composition (Vigh et al., 1993).
Cells lacking Apq12 are cold sensitive (Fig. 1 C) and display more severe defects in NE morphology and nuclear transport when incubated at lower temperatures. The finding that BA, a known membrane fluidizer, can prevent the mislocalization of Nups and suppress the mRNA export defect (Fig. 7) suggests that apq12Δ cells may be defective in the ability to adjust membrane lipid composition so as to retain the appropriate fluidity and flexibility. This hypothesis is supported by the observed genetic interactions between apq12Δ and mtr7-1 (Fig. 7). It also noteworthy that apq12Δ has a strong genetic interaction with ndc1-39 and pom152Δ (Table I), two transmembrane Nups. Although apq12Δ has genetic interactions with several nonmembrane Nups, there are many Nups with which it shows little or no genetic interaction.
It is interesting that BA was able to partially suppress the Nup mislocalization defect of prp20-G282S. This defect is unique to this particular allele of PRP20, which was isolated in a screen for mutants defective in NPC assembly that identified mutants based on Nup mislocalization. Prp20 is not thought to be associated directly with the NE. BA did not lead to any change in the NPC clustering seem in nup120Δ cells (unpublished data). This indicates that the absence of Nup120 does not affect the NE in the same way that the absence of Apq12 does even though nup120Δ cells have abnormalities of the NE, including protrusions and invaginations. There was also no effect of BA on Nup mislocalization in brr6-1 cells. Brr6 is an NE protein, and the phenotypes of brr6-1 cells are similar to those of apq12Δ cells.
Defects in nuclear transport, nuclear morphology, and in the NE were also observed in npl4 mutant strains (DeHoratius, and Silver, 1996). Npl4 along with Cdc48 and Ufd1 form a complex that is involved in the ER-associated proteolysis of some ubiquitinated proteins (Hitchcock et al., 2003). Among the substrates of this complex are the subunits of a heterodimeric transcription factor, Mga2 and Spt23. These proteins are released from a membrane-bound state and are thereby activated by the Npl4–Cdc48–Ufd1 complex (Hitchcock et al., 2001). One of the targets of Mga2/Spt23 is Ole1, which encodes the sole fatty acid desaturase in yeast, and this protein cannot be produced in npl4 mutant strains. This suggests that the nuclear membrane defects of npl4 cells arise as a consequence of their inability to activate OLE1 and that the nuclear transport defects of these cells are an indirect consequence of defects in the nuclear membrane.
Why would a protein that affects membrane dynamics have such a dramatic defect in NPC biogenesis? As shown recently by electron tomography, there are intimate contacts between the NE and NPCs. NPCs have a flexible structure that undergoes conformational and positional changes within the NE during transport (Stoffler et al., 2003; Beck et al., 2004; Melcak et al., 2007). Although the details of NPC biogenesis remain unknown, a critical event is the fusion of the INM and ONM, resulting in the formation of a protein-lined membrane tunnel connecting the nucleus and the cytoplasm. Formation of a membranous tunnel represents a dramatic reorganization of the INM and ONM, and one might expect the formation of NPCs to be quite sensitive to altered biophysical properties of the nuclear membranes.
Nuclear membrane dynamics and cell division
Another study reported that cells lacking Apq12 had defects in the completion of cytokinesis and showed that these defects were independent of apq12Δ's mRNA export defect (Montpetit et al., 2005). They identified apq12Δ in a screen for mutants that are synthetically lethal with ts alleles affecting SPB proteins. In contrast to what was observed for NPCs, we did not detect the mislocalization of any of the several SPB components we examined (unpublished data).
Moreover, through the study of an apq12Δ/nup170Δ double mutant, we were able to separate apq12Δ's defects in cell division and mRNA export (Figs. 8 and S1). Nup170 has been linked previously to chromosome segregation (Kerscher et al., 2001). Combining the two deletion mutations exacerbated the defects in cell division and NE morphology, whereas the mRNA export and NPC biogenesis defects shown by apq12Δ cells were not increased. One possibility is that when Nup170 is absent, less physical stress is placed upon the nuclear membranes as they undergo fusion. Little is known about the steps of NPC biogenesis, but several Nups may associate with the INM and ONM before fusion. In the absence of Nup170, perhaps less bending of the membranes is required in forming the NPC tunnel, and this would account for reduced Nup mislocalization in apq12Δ/nup170Δ cells. If this were the case, one would expect that mRNA export would be less defective in the double mutant than in apq12Δ cells, as we observed. Entirely different demands are likely placed upon the NE during cell division than during NPC biogenesis. The presence of a less flexible NE could readily enhance the cell division defect in apq12Δ/nup170Δ cells as compared with the defect of nup170Δ cells.
We speculate that the cell cycle defects of apq12Δ are also a consequence of altered nuclear membrane properties. Yeast cells undergo a closed mitosis, and NE breakdown does not occur. Instead, after chromosome duplication, cells undergo karyokinesis before cytokinesis. This involves movement of the nucleus to the bud neck and elongation of the nucleus into the bud followed by severe constriction of the NE as karyokinesis proceeds and the mother and daughter cells begin to become separated by cytokinesis. This dramatic remodeling of the NE is likely to be impacted by mutations that affect NE flexibility.
Interestingly, mutations in ACC1/MTR7 also cause defects in mitosis, but acc1/mtr7mutants are sensitive to elevated as well as reduced temperatures. At nonpermissive temperature, acc1/mtr7 cells arrest at the G2/M boundary with a single nucleus and a short spindle, and this could reflect an inability of the NE to undergo the changes in nuclear shape required for karyokinesis (Al-Feel et al., 2003).
Collectively, these data support the hypothesis that the biophysical properties of the nuclear membrane are affected by the loss of Apq12, and this leads to defects in NPC biogenesis. The reported defects in nuclear transport and RNA metabolism are consequences of the defects in NPC biogenesis. Although a detailed mechanistic role for Apq12 has not been defined, there are several possibilities for how it may be acting. Because Apq12 is present in the NE, it could contribute directly to the biophysical properties of the nuclear membranes as a structural component. Alternatively, Apq12 could be required to sense the physical properties of the NE so that the cell is signaled to modify its membranes after temperature changes. Also, Apq12 might function downstream of a sensor to recruit enzymes involved in lipid biosynthesis or modification. These include biosynthetic enzymes such as Acc1/Mtr7 and the desaturase Ole1. Defining the precise role played by Apq12 will be the focus of future studies.
Materials and methods
Yeast strains and plasmids
Yeast strains and plasmids used for these studies are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200702120/DC1). All strains were grown and media was prepared using standard methods (Burke et al., 2000). For growth assays, strains were grown overnight and diluted back to OD600 = 0.3. Strains were then serially diluted 1:10, and 3 μl were plated of each dilution. Plasmid pCSNup49-GFP-1 was created by the digestion of GFP-NUP49 plasmid (gift of V. Doye, Institut Curie, Paris, France) with SacI and BamHI and ligation into YIplac211. The plasmid was then linearized for genomic integration with SwaI.
Microscopy
All live cell fluorescent microscopy was performed using cells grown and mounted in synthetic complete plus dextrose media (Burke et al., 2000). Images were acquired using a microscope (TE2000-E; Nikon) fitted with a 100× NA 1.4 plan Apochromat oil objective (Nikon), CCD camera (Orca-ER; Hamamatsu), and Phylum Live software version 3.5.1 (Improvision). GFP and RFP were visualized by using an X-cite 120-UV lamp and Chroma filter sets. Images were processed using Photoshop 7.0 (Adobe). DiOC6 staining was performed as previously described (Koning et al., 1993). In brief, cells in midlog phase were stained with 1 μg/ml DiOC6 (Invitrogen) using a 0.1-mg/ml ethanol stock. Indirect IF using α-Nup159/Rat7, α-Gle1/Rss1, or α-RL1 and FISH were performed as described previously (Gorsch et al., 1995; Cole et al., 2002). α-Nup159/Rat7 antibody was used at a 1:3,000 dilution, and α-RL1 (Affinity BioReagents, Inc.) was used at a 1:500 dilution. Indirect IF using α-Pom152 antibodies (Strambio-de-Castillia et al., 1995) was performed as described previously (Wente et al., 1992) using a 1:2 antibody dilution.
Membrane association assay
100 ml Apq12-GFP cells were grown to an OD600 = 0.6 and were used for semiintact cell preparation. In brief, cells were washed once in 100 mM Tris-HCl, pH 9.4, and 5 mM DTT and resuspended in 10 mL lyticase buffer (0.7 M sorbitol, 0.5% dextrose, 10 mM Tris-HCl, pH 7.5, and 1 mM DTT). 250 μl lyticase was added, and cells were gently rotated for 15 min, at which time the reaction was stopped by adding 90 mL lyticase buffer and spinning down cells. The pellet was resuspended in 1.8 ml lysis buffer (0.4 M sorbitol, 20 mM Hepes, 150 mM KOAc, and 2 mM MgOAc). 35 μl of the semiintact cells were treated with 115 μl of buffer A (20 mM Hepes, pH 7.0, 150 mM KOAc, and 2 mM EDTA), buffer A plus 1% Triton X-100, or 0.1 M sodium carbonate, pH 11.0, for 5 min and spun at 60 K for 12 min. The supernatant was then removed, the pellet was resuspended, and the fractions were analyzed via Western blotting.
Electron microscopy
Electron microscopy was performed as previously described (Goldstein et al., 1996) with some modifications. In brief, the cells were grown to an OD600 of 0.5–1.0 in YPD media, pelleted, and resuspended in 0.1 M cacodylate buffer, pH 6.8. Primary fixation was performed with 3% glutaraldehyde and 0.1% tannic acid in 0.1 M cacodylate buffer, pH 6.8, at room temperature for 1 h and then overnight at 4°C. Cells were washed twice with 0.1 M cacodylate buffer, pH 6.8, and twice with 0.1 M phosphate buffer, pH 7.5, and treated with zymolyase (10 mg/ml 100T) to produce spheroplasts. After washing with phosphate buffer and cacodylate buffer, pH 6.8, the cells were retreated with 3% glutaraldehyde and 0.1% tannic acid in 0.1 M cacodylate buffer, pH 6.8, for 1 h at room temperature, washed three times with 0.1 M cacodylate buffer, pH 6.8, and embedded in low melting temperature agarose (SeaPrep; FMC Corp.). Postfixation was performed with 2% osmium tetroxide in 0.1 M cacodylate buffer, pH 6.8, for 1 h on ice. Subsequently, the cells were washed in cacodylate buffer, pH 6.8, and deionized water, en bloc stained with 0.5% uranyl acetate overnight, dehydrated with ethanol, and embedded in Spurr's resin (medium grade). Thin sections were cut on an ultramicrotome (MT5000; Sorvall) with a section thickness of 100 nm. Sections were poststained with uranyl acetate and Venable and Coggleshell's lead citrate and examined on a transmission electron microscope (JEM 1010; JEOL) at 100 kV.
Online supplemental material
Fig. S1 shows growth assays for some of the double mutants whose growth behavior is summarized in Table I and a comparison of the protein levels of various Nup-GFP fusion proteins in WT and apq12Δ cells. Fig. S2 documents that apq12Δ cells do not have an ER-trafficking defect, as analyzed by comparing both the maturation of carboxypeptidase Y and the induction of the UPR in WT and apq12Δ cells. Fig. S3 contains fluorescence micrographs that document the degree of colocalization of multiple Nups that mislocalize to cytoplasmic foci in the absence of Apq12. Fig. S4 shows how BA affects the distribution of Sec63-RFP and Nup82-GFP in apq12Δ cells, Nup188-GFP in apq12Δ cells, Nup170-GFP and Nic96-GFP in prp20G280S cells, and Nup159/Rat7 in brr6-1 cells. Table S1 lists the strains and plasmids used in these studies. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200702120/DC1.
Supplementary Material
Acknowledgments
The authors wish to thank the following colleagues for sharing yeast strains, plasmids, and/or antibodies: Susan Wente, Mark Winey, Francoise Stutz, Alan Tartakoff, William Prinz, James Falvo, Valerie Doye, Shingo Izawa, and Rick Wozniak. We also thank Josh Wilson and Cathie Heath for technical assistance and Susan Wente for comments on the manuscript.
This work was supported by a grant from the National Institutes of Health (R01 GM-33998) to C.N. Cole.
Abbreviations used in this paper: BA, benzyl alcohol; DiOC6, 3,3′- dihexyloxacarbocyanine iodide; IF, immunofluorescence; INM, inner nuclear membrane; NE, nuclear envelope; NPC, nuclear pore complex; Nup, nucleoporin; ONM, outer nuclear membrane; SPB, spindle pole body; UPR, unfolded protein response; UPRE, UPR element; WT, wild type.
References
- Aitchison, J., M. Rout, M. Marelli, G. Blobel, and R. Wozniak. 1995. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 131:1133–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Feel, W., J.C. DeMar, and S.J. Wakil. 2003. A Saccharomyces cerevisiae mutant strain defective in acetyl-CoA carboxylase arrests at the G2/M phase of the cell cycle. Proc. Natl. Acad. Sci. USA. 100:3095–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba, T., A. Jacq, E. Brickman, J. Beckwith, T. Taura, C. Ueguchi, Y. Akiyama, and K. Ito. 1990. Characterization of cold-sensitive secY mutants of Escherichia coli. J. Bacteriol. 172:7005–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, K.E., J. Coller, and R. Parker. 2004. The yeast Apq12 protein affects nucleocytoplasmic mRNA transport. RNA. 10:1352–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, M., F. Forster, M. Ecke, J. Plitzko, F. Melchior, G. Gerisch, W. Baumeister, and O. Medalia. 2004. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 306:1387–1390. [DOI] [PubMed] [Google Scholar]
- Burke, D., D. Dawson, and T. Stearns. 2000. Methods in Yeast Genetics: a Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 205 pp.
- Cole, C.N., C.V. Heath, C.A. Hodge, C.M. Hammell, and D.C. Amberg. 2002. Analysis of RNA export. Methods Enzymol. 351:568–587. [DOI] [PubMed] [Google Scholar]
- Colley, C., and J. Metcalfe. 1972. The localisation of small molecules in lipid bilayers. FEBS Lett. 24:241–246. [DOI] [PubMed] [Google Scholar]
- Copeland, C.S., and M. Synder. 1993. Nuclear pore complex antigens delineate nuclear envelope dynamics in vegetative and conjugating Saccharomyces cerevisiae. Yeast. 9:235–249. [DOI] [PubMed] [Google Scholar]
- Cox, J., C. Shamu, and P. Walter. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 73:1197–1206. [DOI] [PubMed] [Google Scholar]
- de Bruyn Kops, A., and C. Guthrie. 2001. An essential nuclear envelope integral membrane protein, Brr6p, required for nuclear transport. EMBO J. 20:4183–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeHoratius, C., and P. Silver. 1996. Nuclear transport defects and nuclear envelope alterations are associated with mutation of the Saccharomyces cerevisiae NPL4 gene. Mol. Biol. Cell. 7:1835–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning, D.P., S.S. Patel, V. Uversky, A.L. Fink, and M. Rexach. 2003. Disorder in the nuclear pore complex: The FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. USA. 100:2450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, T., K. Strasser, A. Racz, S. Rodriguez-Navarro, M. Oppizzi, P. Ihrig, J. Lechner, and E. Hurt. 2002. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 21:5843–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A.L., C.A. Snay, C.V. Heath, and C.N. Cole. 1996. Pleiotropic nuclear defects associated with a conditional allele of the novel nucleoporin Rat9p/Nup85p. Mol. Biol. Cell. 7:917–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, L., R. Sauerheber, J. Esgate, I. Dipple, R. Marchmont, and M. Houslay. 1980. The increase in bilayer fluidity of rat liver plasma membranes achieved by the local anesthetic benzyl alcohol affects the activity of intrinsic membrane enzymes. J. Biol. Chem. 255:4519–4527. [PubMed] [Google Scholar]
- Gorsch, L.C., T.C. Dockendorff, and C.N. Cole. 1995. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J. Cell Biol. 129:939–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath, C., C. Copeland, D. Amberg, V. Del Priore, M. Snyder, and C. Cole. 1995. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J. Cell Biol. 131:1677–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus, H., M.C. Yu, and P.A. Silver. 2004. Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev. 18:2652–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock, A.L., H. Krebber, S. Frietze, A. Lin, M. Latterich, and P. Silver. 2001. The conserved Npl4 protein complex mediates proteasome- dependent membrane-bound transcription factor activation. Mol. Biol. Cell. 12:3226–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock, A.L., K. Auld, S. Gygi, and P. Silver. 2003. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc. Natl. Acad. Sci. USA. 100:12735–12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge, C.A., H.V. Colot, P. Stafford, and C.N. Cole. 1999. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. EMBO J. 18:5778–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W.K., J.V. Falvo, L.C. Gerke, A.S. Carroll, R.W. Howson, J.S. Weissman, and E.K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691. [DOI] [PubMed] [Google Scholar]
- Izawa, S., R. Takemura, and Y. Inoue. 2004. Gle2p is essential to induce adaptation of the export of bulk poly(A)+ mRNA to heat shock in Saccharomyces cerevisiae. J. Biol. Chem. 279:35469–35478. [DOI] [PubMed] [Google Scholar]
- Kadowaki, T., S. Chen, M. Hitomi, E. Jacobs, C. Kumagai, S. Liang, R. Schneiter, R. Singleton, J. Wisniewska, and A.M. Tartakoff. 1994. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J. Cell Biol. 126:649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher, O., P. Hieter, M. Winey, and M.A. Basrai. 2001. Novel role for a Saccharomyces cerevisiae nucleoporin, Nup170p, in chromosome segregation. Genetics. 157:1543–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning, A.J., P.Y. Lum, J.M. Williams, and R. Wright. 1993. DiOC6 staining reveals organelle structure and dynamics in living yeast cells. Cell Motil. Cytoskeleton. 25:111–128. [DOI] [PubMed] [Google Scholar]
- Krogh, A., B. Larsson, G. von Heijne, and E.L.L. Sonnhammer. 2001. Predicting transmembrane topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580. [DOI] [PubMed] [Google Scholar]
- Lau, C.K., T.H. Giddings Jr., and M. Winey. 2004. A novel allele of Saccharomyces cerevisiae NDC1 reveals a potential role for the spindle pole body component Ndc1p in nuclear pore assembly. Eukaryot. Cell. 3:447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid, A.S., J. Mancuso, W.Z. Cande, and K. Weis. 2006. The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J. Cell Biol. 173:361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcak, I., A. Hoelz, and G. Blobel. 2007. Structure of Nup58/45 suggests flexible nuclear pore diameter by intermolecular sliding. Science. 315:1729–1732. [DOI] [PubMed] [Google Scholar]
- Miao, M., K. Ryan, and S. Wente. 2006. The integral membrane protein Pom34 functionally links nucleoporin subcomplexes. Genetics. 172:1441–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montpetit, B., K. Thorne, I. Barrett, K. Andrews, R. Jadusingh, P. Hieter, and V. Measday. 2005. Genome-wide synthetic lethal screens identify an interaction between the nuclear envelope protein, Apq12p, and the kinetochore in Saccharomyces cerevisiae. Genetics. 171:489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, K., A. Sant, K. Kohno, K. Normington, M. Gething, and J. Sambrook. 1992. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 11:2583–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida, I., and N. Murata. 1996. Chilling sensitivity in plants and cyanobacteria: the crucial contribution of membrane lipids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47:541–568. [DOI] [PubMed] [Google Scholar]
- Pollard, M., K. Travers, and J. Weissman. 1998. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol. Cell. 1:171–182. [DOI] [PubMed] [Google Scholar]
- Prinz, W.A., L. Grzyb, M. Veenhuis, J.A. Kahana, P.A. Silver, and T.A. Rapoport. 2000. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150:461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, M.P., J.D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B.T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, K.J., J.M. McCaffery, and S.R. Wente. 2003. The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J. Cell Biol. 160:1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, K.J., Y. Zhou, and S.R. Wente. 2006. The karyopherin Kap95 regulates nuclear pore complex assembly into intact nuclear envelopes in vivo. Mol. Biol. Cell. 18:886–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter, R., M. Hitomi, A. Ivessa, E. Fasch, S. Kohlwein, and A. Tartakoff. 1996. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol. Cell. Biol. 16:7161–7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref, A., K. Sharma, V. Doye, A. Hellwig, J. Huber, R. Luhrmann, and E. Hurt. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16:3256–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski, C., R. Chapman, and P. Walter. 1998. The unfolded protein response: an intracellular signaling pathway with many surprising features. Trends Cell Biol. 8:245–249. [DOI] [PubMed] [Google Scholar]
- Stoffler, D., B. Feja, B. Fahrenkrog, J. Walz, D. Typke, and U. Aebi. 2003. Cryo-electron tomography provides novel insights into nuclear pore architecture: implications for nucleocytoplasmic transport. J. Mol. Biol. 328:119–130. [DOI] [PubMed] [Google Scholar]
- Strambio-de-Castillia, C., G. Blobel, and M.P. Rout. 1995. Isolation and characterization of nuclear envelopes from the yeast Saccharomyces. J. Cell Biol. 131:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia, C., G. Blobel, and M.P. Rout. 1999. Proteins connecting the nuclear pore complex with the nuclear interior. J. Cell Biol. 144:839–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, E.J., and S.R. Wente. 2006. Dynamic nuclear pore complexes: life on the edge. Cell. 125:1041–1053. [DOI] [PubMed] [Google Scholar]
- Travers, K., C. Patil, L. Wodicka, D. Lockhart, J. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 101:249–258. [DOI] [PubMed] [Google Scholar]
- Vigh, L., D. Los, I. Horvath, and N. Murata. 1993. The primary signal in the biological perception of temperature: Pd-catalyzed hydrogenation of membrane lipids stimulated the expression of the desA gene in synechocystis PCC6803. Proc. Natl. Acad. Sci. USA. 90:9090–9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich, C.S., J.P. Erzberger, J.M. Berger, and K. Weis. 2004. The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol. Cell. 16:749–760. [DOI] [PubMed] [Google Scholar]
- Wente, S.R., and G. Blobel. 1993. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J. Cell Biol. 123:275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente, S.R., M.P. Rout, and G. Blobel. 1992. A new family of yeast nuclear pore complex proteins. J. Cell Biol. 119:705–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J.D., Y. Liu, C. Bentivoglio, and C. Barlowe. 2006. Sel1p/Ubx2p participates in a distinct Cdc48-dependent endoplasmic reticulum-associated degradation pathway. Traffic. 7:1213–1223. [DOI] [PubMed] [Google Scholar]
- Zabel, U., V. Doye, H. Tekotte, R. Wepf, P. Grandi, and E. Hurt. 1996. Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p. J. Cell Biol. 133:1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.