Abstract
Vigilin/Scp160p/DDP1 is a ubiquitous and highly conserved protein containing 15 related, but non-identical, K-homology (KH) nucleic acid binding domains. While its precise function remains unknown, proposed roles for vigilin include chromosome partitioning at mitosis, facilitating translation and tRNA transport, and control of mRNA metabolism, including estrogen-mediated stabilization of vitellogenin mRNA. To probe sites of vigilin action in vertebrate cells, we performed nucleic acid binding and RNA interference studies. Consistent with a potential role in chromosome partitioning, human vigilin exhibits a higher affinity for Drosophila dodecasatellite single-stranded DNA than for vitellogenin mRNA 3′-UTR. Direct observation and flow cytometry in non-mitotic, serum-starved, HeLa cells showed that RNAi-mediated vigilin knockdown is rapidly lethal, indicating an essential function for vigilin distinct from its proposed role in chromosome partitioning. Pulse labeling experiments revealed that rates of protein synthesis and degradation are unaffected by the several fold reduction in vigilin levels early in siRNA knockdown indicating that vigilin is not a global regulator of translation. These data show that vigilin is an essential protein in human cells, support the view that vigilin’s most essential functions are neither chromosome partitioning nor control of translation, and are consistent with vigilin playing a critical role in cytoplasmic mRNA metabolism.
INTRODUCTION
The K-homology, or KH, domain is a common nucleic acid binding motif with a conserved core sequence of VIGxxGxxI. Vigilin, also known as Scp160p in Saccharomyces cerevisiae and DDP1 in Drosophila, has been used as a model for this class of nucleic acid binding protein (1). Vigilin is unique in that it consists primarily of 15 related, but non-identical, KH domains. Vigilin is ubiquitous in eukaryotes and highly conserved from yeast to humans. The diversity of vigilin’s biological roles is suggested by its independent identification in several laboratories investigating different biological processes in many organisms (2–9).
Many KH domain-containing proteins bind RNA and function in RNA metabolism (10–16). Consistent with a possible role for vigilin in mRNA metabolism, vigilin is estrogen inducible in Xenopus, and it binds to a segment of the vitellogenin mRNA 3′-UTR implicated in estrogen mediated stabilization of vitellogenin mRNA against cytoplasmic degradation (2,17). In vitro binding of vigilin to this region of the vitellogenin mRNA 3′-UTR masks cleavage sites recognized by the endonuclease, polysomal messenger RNase-1 [PMR-1; (18)], and thereby protects vitellogenin mRNA from degradation by PMR-1 (19). In yeast, Scp160p/vigilin is associated with cytoplasmic mRNP particles and polysomes (20–22). Important recent microarray studies in yeast indicate that Scp160p binds to specific mRNA targets (23). Vigilin has also been reported to bind tRNA, facilitate its export from the nucleus, and control translation efficiency (4). While these data were consistent with the widely held view that KH domain proteins bind RNA and function in RNA metabolism, there were intriguing data indicating that vigilin might function primarily in nuclear events by binding to single-stranded DNA. Morphological studies suggest that Drosophila vigilin (DDP1) binds to numerous sequences in centromeric heterochromatin, and biochemical studies demonstrate that DDP1 can bind to a pyrimidine-rich, single-stranded satellite region of Drosophila heterochromatin (7,24). Binding of vigilin to the C-rich strand of Drosophila dodecasatellite DNA allows the G-rich strand to form a stable foldback structure. Consistent with a possible role for vigilin in the nucleus, the phenotype of a knockout of yeast vigilin is missegregation of chromosomes at mitosis. Although this phenotype is severe, it is not lethal (3).
To begin to explore the functions of vigilin in vertebrate cells, we used two approaches. We carried out studies comparing the ability of defined quantities of purified recombinant human vigilin to bind to the single-stranded Drosophila dodecasatellite C-strand DNA and to a segment of the vitellogenin mRNA 3′-UTR. To complement these studies, and to begin to assess the effects of loss of vigilin, it was important to obtain a vertebrate system lacking vigilin. Since vigilin is present in all vertebrate cell lines examined (25), we used RNAi to create a vigilin knockdown in human cells. In contrast to the yeast knockout, knockdown of human vigilin using vigilin-specific siRNA was lethal to both HeLa and 293 cells. Although we did not examine the pathway by which vigilin induces cell death in detail, vigilin knockdown triggers cleavage of poly(ADP-ribose) polymerase (PARP), a widely used marker for caspase-dependent apoptosis.
To assess whether vigilin is essential for cell viability because its absence results in mispartitioning of chromosomes at mitosis, we developed conditions for vigilin knockdown in serum-starved HeLa cells, that display little or no cell division. We show that vigilin knockdown is lethal in these non-dividing cells, indicating that human vigilin has an essential function independent of its potential role in chromosome partitioning at mitosis. In cells treated with vigilin-specific siRNA, vigilin disappears rapidly, well before the onset of cell death. This enabled us to examine the global effect of vigilin knockdown on translation and protein degradation. The several-fold reduction in vigilin levels early in RNAi- mediated vigilin knockdown had no effect on the overall rate of protein synthesis or degradation, suggesting that vigilin does not exert a general regulatory role in translation.
MATERIALS AND METHODS
Protein expression and purification
Full-length recombinant human FLAG epitope-tagged vigilin was expressed in baculovirus infected SF9 cells and purified to apparent homogeneity by immunoaffinity chromatography as we described (19).
Preparation of labeled probes
pBK6E215 (26) was digested with Spe I and the 145 bp Drosophila dodecasatellite DNA fragment was separated by gel electrophoresis and gel isolated. The overhangs were filled in using Klenow (Invitrogen Life Technologies, Carlsbad, CA, USA) and labeled with 80 µCi [32P]dATP per 30 µl reaction volume. The strands were denatured by incubating at 80°C for 3 min, followed by quick cooling on ice. The strands were then separated on a 3.5% native gel and the slower migrating C-strand was gel isolated, and stored in 0.1 N NaCl until use. Vitellogenin mRNA probe was prepared as described (2) with minor changes. The 120 nucleotide RNA was synthesized and radiolabeled using the SP6 expression system (Gibco BRL, Rockville, MD, USA) using 3000 mCi/mmol [32P]UTP. The RNA was then gel purified on a 5% denaturing polyacrylamide gel.
Electrophoretic mobility shift assays
Gel mobility shift assays were performed as described (2,17) with minor modifications. Equimolar concentrations (2.2 fmol per 10 µl reaction) of labeled RNA or ssDNA and FLAG epitope-tagged vigilin were incubated in buffer [6 mM Tris pH 7.6, 60 mM KCl, 1 mM EDTA, 10% glycerol, RNasin (8 U/µl), 0.6 mM DTT]. The non-specific competitors, tRNA and heparin, were each added to a final concentration of 1 ng/µl. Gel bands were visualized using a PhosphorImager and analyzed using ImageQuant software (Molecular Dynamics, Sunnyvale, CA, USA).
Cell culture
HeLa cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum. In experiments in which HeLa cells were not permitted to divide, they were cultured in DMEM and 0.5% FBS; 293 cells were maintained in Eagle’s Minimal Essential Medium (MEM), 1 mM sodium pyruvate, 0.1 mM non-essential amino acids, and supplemented with 10% heat-inactivated fetal bovine serum.
Transfection of siRNAs and western blotting
siRNA sequences were BLAST searched against the human genome to ensure that they were specific for vigilin. The two vigilin siRNA sequences showed no exact or near exact matches to any other sequences in the human genome, and are therefore vigilin-specific. siRNAs were synthesized by Dharmacon Research, Inc. (Lafayette, CO, USA). pGL3 siRNA was deprotected and annealed according to the manufacturer’s instructions. The 21-nt sequence of pGL3 siRNA was previously described [GL3; (27)]. The vigilin-specific siRNA, Vig 1, was deprotected and annealed according to the manufacturer’s instructions. Vig 1 siRNA (5′-UGGAGAGAAACUGCAAGACTT-3′) targets nucleotides 506–524 of human vigilin relative to the first nucleotide of the start codon (Acc. No. M64098). Vig 2 siRNA (5′-UCCCAACACAAGUAUGUCATT-3′) was pre-duplexed and targets nucleotides 912–930 of human vigilin.
60 000 cells/well were seeded in 12-well plates. Twenty-four hours later, the cells were transfected with 1.68 µg of siRNA per well (unless otherwise noted). Transfections were as described (28) with the following modifications. Additional Opti-Mem (Invitrogen, Carlsbad, CA, USA) was not added and medium was removed before transfection and replaced with 400 µl of Opti-Mem. Full-serum medium (unless otherwise noted) was added 3–4 h post-transfection. At the indicated times post-transfection, the cells were washed twice with PBS and then were detached from the plate with PBS-EDTA. Whole cell extract was obtained by lysing the cells with RIPA buffer containing protease inhibitors and DTT. Protein concentrations were measured using the Bradford assay. Extracts were run on 8% (12% for actin) polyacrylamide gels and transferred onto nitrocellulose membranes. The membranes were blocked overnight at 4°C in 1% non-fat dry milk (1 h at room temperature in 5% non-fat dry milk for actin and PARP western blots), then probed with either rabbit polyclonal anti-vigilin, or anti-actin (Santa Cruz Biotechnology, Santa Cruz, CA, USA), or anti-PARP (Cell Signaling, Beverly, MA, USA) antibody for 1 h at room temperature (overnight at 4°C for actin and PARP), washed and probed with a horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. Signals were detected using the ECL-Plus reagent (Amersham Biosciences, Piscataway, NJ, USA) or SuperSignal West Pico Chemiluminescent Substrate kit (Pierce, Rockford, IL, USA).
Flow cytometry and light phase microscopy
HeLa cells were transfected as described above and maintained in 0.5% heat-inactivated fetal bovine serum and 1× DMEM for 48 h at 37°C. Cells were visualized by light phase microscopy using a TE 300 Micron Eclipse inverted fluorescence microscope. For flow cytometry, HeLa cells were transfected as above and incubated for 48 h at 37°C. Cells were washed twice with PBS and harvested with PBS-EDTA, pelleted by centrifugation at 500 r.p.m. for 5 min and then resuspended in 0.5 ml PBS. DiOC6 was added to a final concentration of 20 nM. Propidium iodide was added to a final concentration of 1 µg/µl 5 min prior to FACS detection. A Coulter XL benchtop flow cytometer was used to measure relative fluorescence intensities with excitation at 488 nm and emission at 520 nm.
Metabolic labeling
HeLa cells were transfected in 12-well plates as described above and maintained in 0.5% serum medium post-transfection. At 12 and 24 h post-transfection, medium was removed and replaced with medium containing 0.1× (of the concentration in normal growth medium) methionine and 0.1× cystine for 20 min. Medium was then removed and replaced with the above medium containing 50 µCi/well of [35S]methionine and incubated at 37°C for 1 h. After incubation, the cells were washed three times with PBS containing 31.5 mg and 15 mg per 500 ml of cystine and methionine, respectively. Cells were scraped from the plates and lysed with RIPA buffer. Labeled protein was precipitated with TCA and counted in a scintillation counter. For control experiments with cycloheximide, cells were plated in normal growth medium which was then replaced with 0.5% serum medium 6 h after plating. The following day, the medium was replaced with medium containing 0.1× methionine and 0.1× cystine, and cells were incubated at 37°C for 20 min. The medium was removed and replaced with the medium described above containing 100 µCi/well of [35S]methionine; 10 µg/ml of cycloheximide was also added to half of the samples. All cells were then incubated at 37°C for 30 min. Samples were prepared as described above. For protein degradation experiments, cells were labeled as above at 12 h post-transfection. They were then incubated at 37°C for an additional 12 h followed by harvesting and lysis as described above.
RESULTS
Vigilin has a 5–10-fold higher affinity for Drosophila dodecasatellite C-strand ssDNA than for the vitellogenin mRNA 3′-UTR
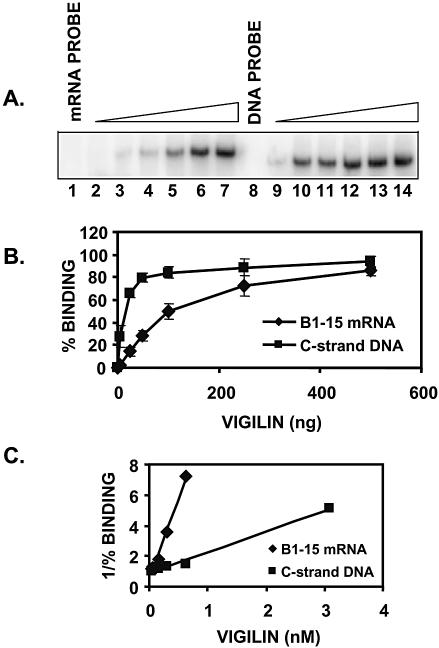
Previously, we used in vitro genetic selection to demonstrate that vigilin binds mainly to single-stranded RNAs lacking obvious secondary structure (17). Both the vitellogenin mRNA 3′-UTR and the dodecasatellite C-strand ssDNA are largely unstructured. To compare the binding of vigilin to the vitellogenin mRNA 3′-UTR and the dodecasatellite C-strand ssDNA, we carried out protein titrations using recombinant human FLAG epitope-tagged vigilin expressed in baculovirus-infected SF9 cells and purified to near homogeneity by immunoaffinity chromatography. Increasing amounts of vigilin were incubated with equal concentrations of labeled vitellogenin mRNA 3′-UTR or dodecasatellite C-strand DNA probes, and binding was determined in electrophoretic mobility shift assays. Using identical binding conditions, vigilin bound with higher affinity to the repeated dodecasatellite C-strand sequence (Fig. 1A). Quantitation of the data from three separate experiments by PhosphorImager analysis indicated that vigilin exhibits a 5–10-fold higher affinity for single-stranded C-strand DNA than for the vitellogenin mRNA 3′-UTR segment (Fig. 1B and C). The apparent dissociation constant (Kd) was 10 nM for the vigilin–vitellogenin mRNA complex and 1.4 nM for the vigilin–C-strand DNA complex (Fig. 1C).
Figure 1.
Vigilin binds with higher affinity to dodecasatellite C-strand single-stranded DNA than to a segment of the vitellogenin mRNA 3′-UTR. (A) Representative electrophoretic mobility shift assay in which increasing amounts of purified FLAG epitope-tagged human vigilin (5, 25, 50, 100, 250, 500 ng) were incubated with the same concentration of a labeled segment of the vitellogenin mRNA 3′-UTR (mRNA probe, lanes 2–7), or with labeled dodecasatellite C-strand single-stranded DNA (DNA probe, lanes 9–14). (B) The binding curves represent the average ± S.E.M. for the data from the EMSA in panel A and two additional mobility shift assays. Data were obtained by PhosphorImager analysis using ImageQuant software. (C) Double reciprocal plot of the binding data. The Kd values are corrected for the percentage of purified recombinant vigilin competent to bind nucleic acids. The 5 ng data point for vigilin binding to the vitellogenin mRNA 3′-UTR is off the trend line and is not shown.
A slower dissociation rate is primarily responsible for the higher affinity binding of vigilin to the C-strand ssDNA
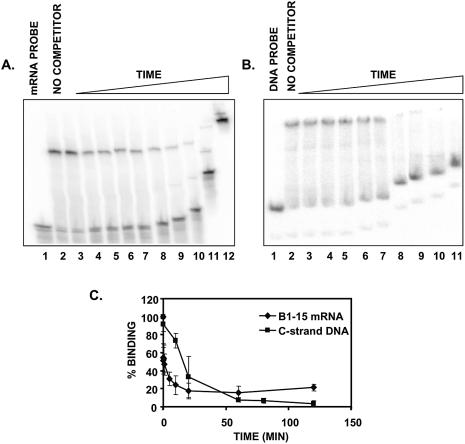
It was not clear whether vigilin exhibited higher affinity binding to the Drosophila dodecasatellite C-strand ssDNA due to a more rapid association with the ssDNA (an increased on-rate), or because of slower dissociation from the ssDNA (a reduced off-rate). Since protein–nucleic acid association rates are often extremely rapid, we determined dissociation rates for the two vigilin–nucleic acid complexes. Vigilin was incubated with equal concentrations of either the labeled vitellogenin mRNA 3′-UTR probe or with the labeled C-strand ssDNA probe. The reactions were then quenched by adding a large excess of each respective unlabeled nucleic acid. Aliquots of the samples were removed at the indicated times and loaded directly onto a 4% native polyacrylamide gel running at 4°C. Data from typical experiments are shown in Figure 2A and B. Data from three experiments were averaged and are shown in Figure 2C. There was a rapid and progressive decrease in vigilin binding over the first 20 min of the experiment. Vigilin dissociated from the vitellogenin mRNA 3′-UTR ∼5 times more rapidly than it dissociated from the dodecasatellite C-strand DNA. This approximately 5-fold difference in the dissociation rate of vigilin from the two nucleic acids indicates that a slower dissociation rate is primarily, and perhaps exclusively, responsible for the 5–10-fold higher affinity vigilin exhibits for the dodecasatellite C-strand ssDNA compared to the vitellogenin mRNA 3′-UTR. Although the binding studies suggested possible sites of vigilin action, they did not directly address potential roles for vigilin. To more directly evaluate potential roles for human vigilin, we developed a system for examining the effects of loss of intracellular vigilin.
Figure 2.
Vigilin–C-strand ssDNA complex dissociates more slowly than vigilin–vitellogenin mRNA 3′-UTR complex. (A) Labeled vitellogenin mRNA 3′-UTR was incubated with 120 ng FLAG-vigilin (to achieve ∼70% binding, lanes 2 and 3) and at time 0, a 2500-fold excess of unlabeled B1-15 mRNA was added (in preliminary competition experiments, this amount of unlabeled nucleic acid was shown to effectively abolish binding). Samples were removed at the indicated times (Lanes 4–12: 0, 15, 30 s, 1, 5, 10, 20, 60 and 120 min, respectively), and loaded onto a running 4% native polyacrylamide gel. (B) A representative mobility shift assay of labeled C-strand ssDNA incubated with 20 ng FLAG-vigilin (to achieve ∼70% binding, lane 2). At time 0, a 2500-fold excess of unlabeled C-strand ssDNA was added, samples were removed at the indicated times (Lanes 3–11: 0, 15 s, 1, 10, 20, 60, 80, 100 and 120 min, respectively), and the reaction was loaded onto a running gel. (C) The percentage of each probe bound as a function of time is plotted. For the vitellogenin mRNA 3′-UTR probe, the dissociation curve represents the average ± S.E.M. for the data shown in panel A, and two additional mobility shift assays. For the C-strand ssDNA probe, six triplicate points from panel B and additional mobility shift assays were graphed in C. Dissociation of 50% of the initial vigilin–vitellogenin B1-15 mRNA 3′-UTR complex binding was approximately five times more rapid than dissociation of 50% of the vigilin–dodecasatellite C-strand ssDNA complex.
Vigilin knockdown is rapid
To determine the importance of vigilin in vertebrate cells and to explore proposed sites of vigilin action, it was important to obtain cells containing little or no vigilin. Since every eukaryotic cell line expresses vigilin, RNA interference was used to eliminate vigilin from human cells. In preliminary experiments, HeLa cells were transfected with either of two vigilin-specific siRNAs or a non-specific siRNA (pGL3). Forty-eight hours after transfection, cell extracts were prepared and analyzed for vigilin by western blotting. Both vigilin-specific siRNAs (Vig 1 and Vig 2) elicited a vigilin knockdown (Fig. 3A). Additional experiments (data not shown) demonstrated that Vig 2 siRNA was slightly more effective than Vig 1 siRNA, and Vig 2 was therefore used in subsequent experiments. The control luciferase siRNA did not elicit vigilin knockdown across a range of siRNA concentrations (Fig. 3B, pGL3).
Figure 3.
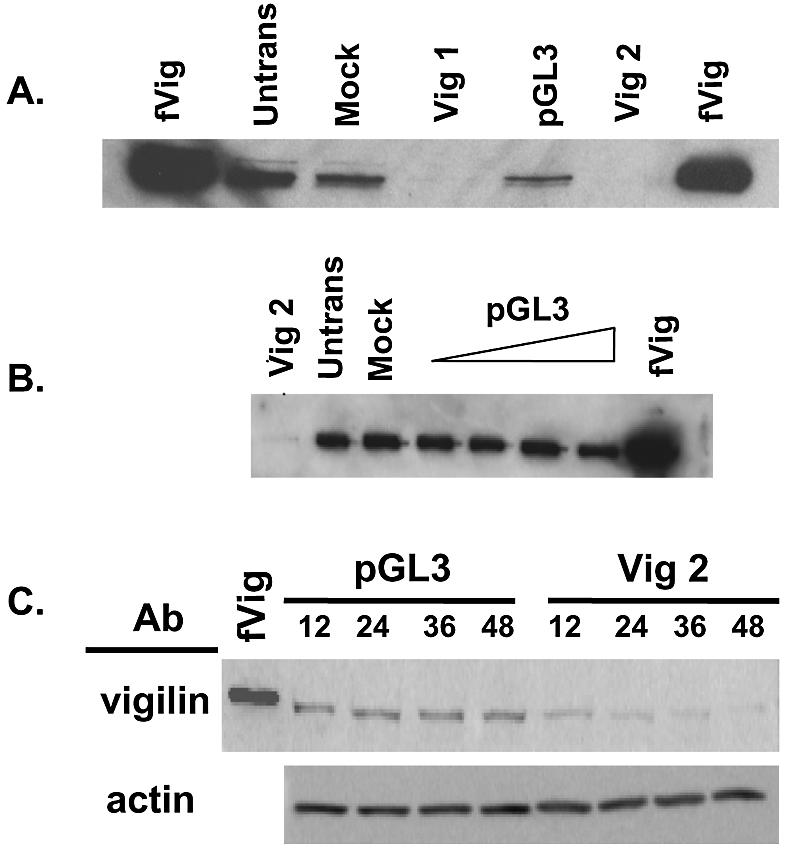
Rapid disappearance of vigilin in cells transfected with vigilin-specific siRNA. (A) HeLa cells were either not transfected (Untrans), mock transfected (Mock), transfected with either of two vigilin-specific siRNAs (Vig 1 and Vig 2), or with a non-specific pGL3 luciferase-specific siRNA (pGL3). Forty-eight hours after transfection, the cells were harvested, extracts were prepared, and 10 µg of each sample was fractionated on an 8% polyacrylamide gel and analyzed for vigilin content by western blotting with polyclonal rabbit anti-vigilin antibody. Purified recombinant FLAG epitope-tagged vigilin (fVig) was run as a control. (B) HeLa cells were left untransfected (Untrans), mock transfected (Mock), or were transfected with Vig 2 siRNA (0.84 µg) or with increasing concentrations of control pGL3 siRNA (0.14–0.84 µg). Forty-eight hours post-transfection, cell extracts were prepared, and 10 µg of each sample was fractionated by SDS–PAGE and analyzed for vigilin by western blotting. (C) HeLa cells were transfected with pGL3 or Vig 2 siRNAs, cells were harvested at 12, 24, 36 and 48 h post-transfection, extracts were prepared, and 30 µg of each sample was fractionated on an 8% polyacrylamide gel and analyzed for vigilin by western blotting. As a control for the specificity of the siRNAs, 20 µg of each extract was run on a 12% SDS–PAGE gel and analyzed for actin content by western blotting.
While these studies demonstrated effective knockdown of vigilin 48 h post-transfection, the time course of vigilin knockdown was unknown. The rate of turnover of vigilin had not been studied, and given its abundance, it was possible that vigilin was relatively stable. The rate of disappearance of vigilin after transfection with vigilin-specific siRNA might provide initial information about vigilin’s stability. Direct visual observation indicated that some of the cells transfected with the vigilin-specific siRNA died within the 48 h time period used in our initial experiments (data not shown). Since vigilin knockdown should precede cell death, we determined vigilin levels 12, 24, 36 and 48 h after transfection with vigilin specific and control siRNAs. The specificity of the RNAi knockdown was shown by the absence of knockdown of actin with the Vig 2 siRNA and by the inability of the control siRNA to knockdown vigilin levels over the course of the experiment (Fig. 3C, actin and pGL3, respectively). Despite substantial cell death by 48 h, the apparent level of actin did not decline. Although total protein per plate declined as the cells died, each sample loaded onto the gel contained a constant amount of total cell protein (30 µg). The ratio of actin to total cell protein was apparently unchanged over the course of the experiment, resulting in constant levels of actin. In cells transfected with Vig 2 siRNA, vigilin levels were significantly reduced 12 h after transfection, and vigilin had nearly disappeared from the cells 48 h post-transfection (Fig. 3C, Vig 2). These data suggest that vigilin has a relatively short half-life and demonstrate that vigilin knockdown is specific, rapid and progressive.
Vigilin is essential for viability of dividing and non-dividing human cells
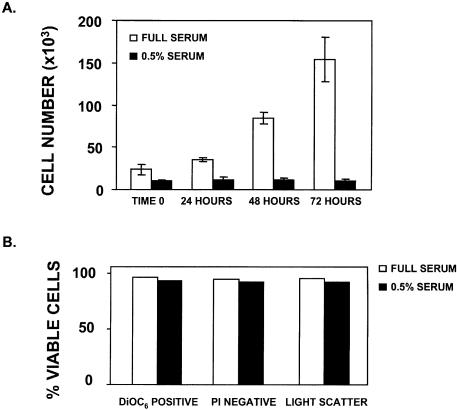
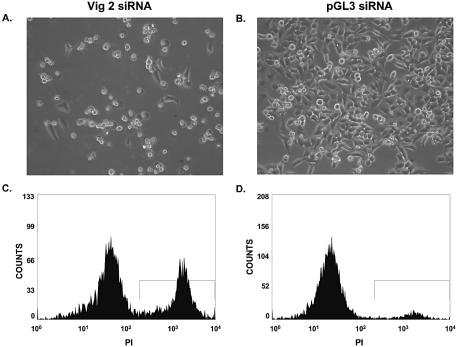
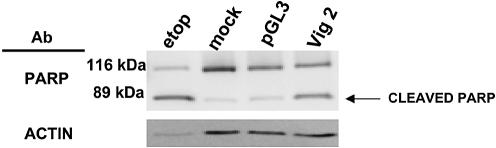
Direct observation of the HeLa cells exhibiting vigilin knockdown indicated that most of the cells died. Similarly, vigilin knockdown caused cell death in 293 cells (human embryonic kidney cells; data not shown). Since the cells were maintained in standard culture medium in which control HeLa cells divide every 18–24 h, the data were compatible with an essential role for vigilin in each of the three proposed functions of vigilin including segregation of chromosomes at mitosis, control of translation and regulation of the stability of specific mRNAs. To begin to distinguish between these possibilities, we identified low serum conditions in which HeLa cells remained viable for up to several weeks, but exhibited little or no cell division. Serum starvation has been widely used to arrest cells at G0 (29–31). When we maintained HeLa cells in medium containing 0.5% FBS, the number of cells was essentially constant, and the cells exhibited negligible growth over 72 h (Fig. 4A). HeLa cells maintained in 10% FBS exhibited robust growth (doubling time ∼20 h) and were approaching confluency by 72 h. While the number of HeLa cells did not increase over 72 h in our reduced serum medium, these data did not exclude the possibility that there was substantial cell growth in the reduced serum medium, with the number of dividing cells equaling the number of cells that were dying. To test this possibility we compared the percentage of cells undergoing cell death in the standard medium containing 10% FBS and in the reduced serum medium containing 0.5% FBS. We compared the cells in normal and reduced serum medium using three different assays for cell death: mitochondrial dysfunction evaluated by DiOC6, propidium iodide staining of DNA, and light scattering. DiOC6 provides a measure of mitochondrial membrane potential. Damaged mitochondria of cells undergoing apoptosis or necrosis exhibit decreased retention of the strong cationic dye DiOC6 and are visualized as a distinct sub-population on flow cytometry. Propidium iodide diffuses through the damaged plasma membranes of non-viable cells and intercalates into their DNA, where it fluoresces and is detected by flow cytometry. Finally, light scattering can detect the altered morphology of sick and dying cells. Analysis of the two cell populations by fluorescence activated cell sorting (FACS) using the three methods mentioned above showed that there was no significant difference in cell viability between the serum starved cells and those grown in normal growth medium containing 10% FBS (Fig. 4B). We therefore conclude that the reason the HeLa cells did not increase in number in the 0.5% serum medium is because they were not undergoing mitosis and dividing. If the cells maintained in 0.5% serum still died following vigilin knockdown, cell death would not result from chromosome missegregation or mispartitioning. HeLa cells were transfected with Vig 2 siRNA or pGL3 siRNA and were maintained in medium containing 0.5% serum to prevent cell division following transfection. To help visualize the difference between the number of surviving cells in the control cells transfected with pGL3 siRNA and the cells transfected with the Vig 2 siRNA, the cells were transfected at a high density. After 48 h, there were only a few cells left on the plate transfected with the vigilin-specific, Vig 2 siRNA, while the cells transfected with the control pGL3 siRNA were nearly confluent and exhibited typical cell morphology (Fig. 5A and B). Most of the small number of cells that remained on the dish transfected with Vig 2 siRNA were rounded up, extensively vacuolized, and appeared to be in the early stages of cell death. A few cells, which were probably untransfected HeLa cells, appeared normal. To more directly assess cell death, we treated the transfected cells with propidium iodide 48 h after transfection and analyzed the cell populations by flow cytometry. A strong peak indicating the presence of non-viable cells (approximately 38% non-viable, Fig. 5C) was observed in the cells transfected with Vig 2 siRNA, but not in the cells transfected with the control pGL3 siRNA (Fig. 5D, approximately 9% non-viable). Propidium iodide fluorescence only identifies the fraction of cells in a population that have traversed enough of the death pathway for their membranes to become permeable to propidium iodide, but not those cells that are so far down the death pathway that the cells have largely been resorbed and their DNA extensively fragmented. Another widely used marker for cell death is the caspase-dependent cleavage of poly(ADP-ribose) polymerase (PARP) into 89 and 24 kDa fragments (32). HeLa cells were treated with etoposide, a widely used inducer of caspase-dependent apoptosis, mock transfected, or transfected with either pGL3 or Vig 2 siRNAs. Etoposide treatment and vigilin knockdown in the cells transfected with Vig 2 siRNA resulted in PARP cleavage, while extracts from the mock transfected cells and cells transfected with pGL3 siRNA did not exhibit PARP cleavage (Fig. 6). An actin western blot was performed as a loading control. Taken together, the data from light microscopy, propidium iodide staining with flow cytometry, and PARP cleavage clearly indicate that knockdown of vigilin triggers death of non-mitotic cells and demonstrate that vigilin is essential for the viability of human cells.
Figure 4.
Serum starvation results in a decrease in cell division but not cell death. (A) HeLa cells were plated and maintained in either 10% or 0.5% FBS-supplemented DMEM and counted at time 0 (4 h after plating), 24, 48 and 72 h after time 0. (B) HeLa cells were maintained in either full growth medium or 0.5% FBS supplemented medium for 48 h. Cells were stained with DiOC6 and PI and analyzed by FACS for cell viability as described in Materials and Methods.
Figure 5.
Vigilin is essential for viability of non-dividing human cells. HeLa cells were transfected with either Vig 2 siRNA (A) or pGL3 siRNA (B) and visualized by phase contrast microscopy 48 h post-transfection. The same samples were then stained with propidium iodide and fluorescence intensity was detected by flow cytometry. The histogram derived from the Vig 2 siRNA-transfected cells is shown in (C) and the histogram from the pGL3 siRNA-transfected cells is shown in (D).
Figure 6.
Vigilin knockdown in HeLa cells results in programmed cell death. HeLa cells were treated with 6.25 nM etoposide (a standard inducer of programmed cell death), mock transfected, or transfected with pGL3 or Vig 2 siRNAs. A western blot was performed 48 h post-transfection using a polyclonal antibody to poly(ADP-ribose) polymerase (PARP). A western blot using a polyclonal antibody to actin was also performed as a loading standard. Because the etoposide-treated cells were in the late stages of apoptosis, they exhibited reduced levels of actin. Levels of actin in the pGL3 and Vig 2-treated cells were similar and the ratio of the cleaved PARP to uncleaved PARP is dramatically increased in the Vig 2-treated cells relative to the mock or pGL3-treated cells.
Protein synthesis and degradation are unaffected by knockdown of vigilin
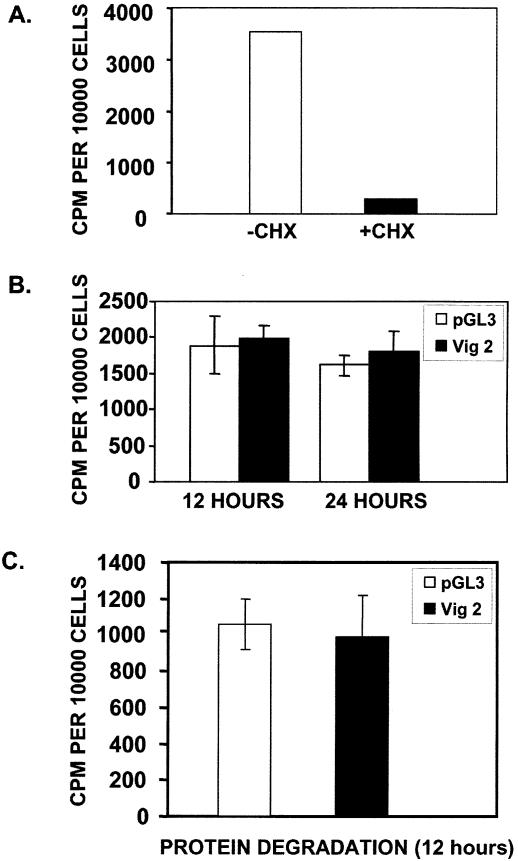
In addition to a potential role in chromosome partitioning, a role for vigilin in the control of translation has been proposed (6). Vigilin is reported to bind tRNA and to be part of a tRNA-elongation factor 1α (EF-1α) complex in human cells (5,6). Also, vigilin’s levels are elevated during periods of active protein synthesis (6). However, our data indicate that vigilin preferentially binds relatively unstructured RNAs (17), and tRNA is highly structured. Creating vigilin-free cells allows us to perform the first direct in vivo test of the hypothesis that vigilin is important in translation. We measured the incorporation of [35S]methionine into newly synthesized protein in cells that contain either wild-type levels of vigilin or several-fold less vigilin. Control cells transfected with pGL3 siRNA and cells transfected with Vig 2 siRNA to elicit vigilin knockdown were pulse labeled with [35S]methionine at 12 and 24 h post-transfection. These times were chosen because vigilin knockdown is readily apparent at 12 h and is progressive through 24 h post-transfection (Fig. 3C). At these early times, there is not much evidence of cell death, and potential effects should not result from secondary changes in the cells due to the initiation of cell death. The cells were pulse-labeled, washed, harvested, lysed and total protein was isolated by TCA precipitation and counted. We used cycloheximide to demonstrate that the assay was actually measuring radioactive amino acid incorporated into protein, and not free amino acid. Addition of the protein synthesis inhibitor, cycloheximide, to the culture medium completely blocked the production of labeled protein (Fig. 7A). At both 12 and 24 h post-transfection, there was essentially no change in the incorporation of [35S]methionine into protein in the pGL3 siRNA transfected cells that contain vigilin compared to the Vig 2 siRNA transfected cells, in which vigilin levels have been knocked down (Fig. 7B). Since vigilin knockdown does not result in a reduced overall rate of protein synthesis, the data clearly demonstrate that vigilin is not a global activator of translation. To test the possibility that vigilin exerts effects on overall rates of protein degradation, HeLa cells were transfected with Vig 2 or pGL3 siRNA, pulse labeled at 12 h post-transfection and harvested 12 h after labeling. Again, the amount of labeled protein in the cells containing wild-type levels of vigilin or greatly reduced levels of vigilin was similar (Fig. 7C).
Figure 7.
Vigilin has no effect on translation or protein degradation. (A) HeLa cells were placed in medium containing 0.5% serum and then pulse labeled with [35S]methionine ± 10 µg/ml cycloheximide. (B) HeLa cells were transfected with either pGL3 or Vig 2 siRNAs. Medium containing 0.5% serum was added 3 h post-transfection. At 12 and 24 h post-transfection, the cells were pulse labeled with [35S]methionine for 1 h at 37°C. The cells were then washed with PBS containing methionine and cystine and immediately harvested. Proteins were precipitated with TCA and counted. For each siRNA, three wells of cells for each time point were counted on a hemocytometer and averaged. The counts were normalized to the number of cells in each sample. The data represent the average of three samples ± S.E.M. (C) HeLa cells were transfected as described in panel A. Twelve hours post-transfection, the cells were pulse labeled, and the label was chased as described above. The cells were subsequently maintained in culture medium at 37°C for an additional 12 h. The cells were then harvested, lysed, precipitated and counted as described for panel B.
DISCUSSION
We used nucleic acid binding studies and elimination of intracellular vigilin as complementary approaches to begin to probe the functions of vertebrate vigilin. Since one proposed function of vigilin was binding to dodecasatellite DNA and controlling chromosome partitioning at mitosis, we compared the binding of human vigilin to single-stranded DNA and mRNA probes. Vigilin and other KH domain-containing proteins were generally considered to be primarily RNA binding proteins (12). However, recent studies describe the structures of KH3 and KH4 of FBP (FUSE binding protein) and of KH3 of hnRNP K bound to single-stranded DNA (33,34). Vertebrate vigilin was initially shown to bind to segments of mRNAs (2). However, immunohistochemistry revealed that Drosophila vigilin (DDP1) bound at several sites in Drosophila heterochromatin (7), and binding studies suggested that vigilin binds well to single-stranded multimers of a repeated sequence in Drosophila dodecasatellite DNA. Since those studies were done using ∼155 kDa Drosophila DDP1 expressed in E.coli, with an unknown amount of DDP1 in the binding reactions, and without non-specific competitor DNA present (24), we carried out binding studies under conditions similar to those we previously used to analyze vigilin binding to RNAs (17,35). Since the primary focus of our work was the function of human vigilin, we used human vigilin to examine binding to Drosophila dodecasatellite C-strand DNA and Xenopus vitellogenin mRNA. Binding studies using human vigilin should yield representative data since vigilins exhibit high homology between species. In previous work, we found that Xenopus and human vigilin exhibited similar binding to the vitellogenin mRNA 3′-UTR segment we employed (2,17,19,35). Further, Cortes et al. have shown that Drosophila vigilin (DDP1) effectively complements a yeast SCP160 knockout (7). Purified vigilin exhibited higher affinity binding to the Drosophila dodecasatellite C-strand ssDNA than to a segment (of similar size) of the vitellogenin mRNA 3′-UTR. While it might seem that higher affinity binding to the ssDNA sequence suggests that this is more likely to be a physiologically relevant binding site for vigilin, the moderately high affinity binding of vigilin to the vitellogenin mRNA 3′-UTR segment is also consistent with its proposed role in binding to this region of the mRNA and mediating estrogen control of mRNA stability. In our model, in the absence of estrogen, vigilin levels in Xenopus hepatocytes are low, and there is insufficient vigilin to occupy the vitellogenin mRNA 3′-UTR. This region of the mRNA contains two sites at which polysomal mRNase I (PMR-1) preferentially cleaves mRNA (18). When vigilin levels are low, these sites will be exposed, enabling PMR-1 to cleave vitellogenin mRNA and remove its poly(A) tail. This results in rapid degradation of the mRNA (18,19). When estrogen is present, vigilin is induced (35), binds to this sequence, and protects vitellogenin mRNA against cleavage by PMR-1 (12,19). Extremely high affinity binding of vigilin to the vitellogenin mRNA 3′-UTR is therefore incompatible with the type of regulated binding required to control mRNA degradation.
Slower dissociation is primarily responsible for the higher affinity binding of vigilin to the C-strand ssDNA than to the vitellogenin mRNA 3′-UTR. The stability of the vigilin–C-strand ssDNA complex may be due in part to the highly repetitive nature of this DNA. Drosophila dodecasatellite C-strand DNA is composed of 12 repeats of a 12-nucleotide sequence [5′-TCGGTCCCGTAC-3′ (26)]. The vigilin–C-strand DNA complex is most likely formed by a tight interaction between many of vigilin’s 15 KH domains, and most or all of the 12 repeats in the dodecasatellite ssDNA. The large number of potential contacts between the ssDNA and vigilin makes dissociation of vigilin less likely than for a sequence containing a single high-affinity binding site. When random thermal motion results in loss of interaction between some KH domains and dodecasatellite repeats, the newly dissociated KH domain(s) and the repeats they dissociated from remain in very close proximity, resulting in an extremely high local concentration of vigilin and ssDNA. Rebinding of dissociated vigilin KH domains to the repeats will therefore be very rapid and much more likely to occur than following complete dissociation of the remaining repeat–KH domain interactions. In contrast to the 12 repeats in the dodecasatellite ssDNA, only one region near the center of the vitellogenin mRNA 3′-UTR segment provides a high affinity binding site for vigilin (2,17).
The binding data were consistent with a nuclear role for vigilin in control of chromosome segregation at mitosis and possibly a functional role in cytoplasmic mRNA metabolism. Since these studies did not address a proposed role for vigilin in translation, or the importance of vigilin to vertebrate cells, we carried out RNAi knockdown experiments.
The rapid disappearance of vigilin protein following siRNA-mediated degradation of vigilin mRNA requires that vigilin be a short-lived protein. Many tightly regulated proteins exhibit rapid turnover enabling them to respond to changes in the cell environment by rapid changes in protein level. Our knockdown data suggest that vigilin has the potential to change its level rapidly in response to environmental changes. Although little is known regarding factors regulating human vigilin levels, Xenopus vigilin levels are regulated by estrogen and testosterone in different tissues (36). These data are consistent with a new picture of vigilin’s action in which it mediates critical functions in vertebrate cells through tight regulation of its level.
Whether or not vigilin has a primarily nuclear function centered around chromosome segregation is controversial. While vigilin’s subcellular localization might help to determine whether vigilin functions primarily in the nucleus or cytosol, one study found vigilin in both locations (5), while other work localized yeast Scp160p/vigilin primarily to the cytosol (20,21). Although a yeast Scp160p/vigilin knockout was not lethal, the cells exhibited a reasonably severe phenotype with evidence of missegregation of chromosomes at mitosis. However, this phenotype is proposed to be an indirect effect resulting from altered cytoplasmic mRNA metabolism (21). The fact that SCP160 null yeast remain viable allowed further studies of mRNA and protein localization (3,20). The binding data were consistent with potential roles for vigilin in chromosome segregation and in mRNA stability. To begin to probe these potential sites of human vigilin action, we used RNA interference.
An important issue in studies using RNA interference is whether the siRNA is specific for the target mRNA. To ensure that our knockdowns were vigilin-specific, we used two siRNAs. Searching the human genome database demonstrated that the sequences of both siRNAs were unique to human vigilin and that there were no human sequences that were nearly identical to the vigilin siRNAs. Since Elbashir and co-workers demonstrated that a single central mismatch in an siRNA can abolish RNA interference (37), the phenotype we observe arises from vigilin knockdown and not from siRNA-mediated knockdown of a family of mRNAs encoding diverse KH domain-containing proteins.
Our initial data demonstrating that vigilin knockdown resulted in the death of both HeLa and 293 cells were surprising since the researchers who carried out the initial Scp160p/vigilin disruption in yeast reported, ‘Disruption of the SCP160 gene is not lethal…’ (3). The difference between the phenotypes of the yeast and human cells lacking Scp160p/vigilin may reflect either binding of Scp160p/vigilin to different mRNA targets in the two cell types or different functional consequences resulting from the absence of Scp160p/vigilin.
Our data demonstrating that knockdown of vigilin was lethal in human cells did not discriminate between these potential sites of vigilin action. To resolve this issue, we developed conditions for the long-term culture of HeLa cells with little or no cell division. In reduced serum medium, HeLa cells exhibited long-term viability without significant cell division (Fig. 4A), or cell death (Fig. 4B). Virtually all of these non-mitotic cells died within 48 h after transfection with vigilin-specific siRNA (Fig. 5). Since these cells are not dividing and still die rapidly following vigilin knockdown, missegregation of chromosomes at mitosis cannot be responsible for their death.
Kruse and co-workers reported that vertebrate vigilin can bind tRNA, exists in a complex with EF-1α and proposed that vigilin regulates translation (5,6). This hypothesis predicts that the loss of vigilin should result in a reduced rate of protein synthesis. However, our metabolic labeling experiments indicate that cells in which vigilin has been knocked down several fold, and the control siRNA transfected cells, show similar levels of incorporation of [35S]methionine into newly synthesized protein. Since vigilin is knocked down within 12 h after transfection (Fig. 3C), and there was no change in the overall rate of protein synthesis, even 24 h after transfection when vigilin levels have been knocked-down by several fold, it is clear that at normal levels vigilin does not have an essential function in translation. Rapid cell death following vigilin knockdown required that these studies be performed in the time period after significant vigilin knockdown and before the onset of cell death. Thus, the experiments had to be performed when vigilin levels were knocked down several fold, but while some vigilin was still detectable. We therefore cannot formally exclude the possibility that human cells contain a large excess of vigilin and that vigilin is still in excess after a several-fold decline in its level. However, this seems unlikely because stimulation of cell division results in a prompt increase in vigilin levels (6), and vigilin levels are tightly regulated by hormones in several cell types (2,17,19,35,36). Further, we would expect at least a modest effect of a several-fold vigilin knockdown on total protein synthesis, and we observe no detectable change in protein synthesis as vigilin levels decline (Fig. 7B).
Our research and that of Li and co-workers suggest a role for vigilin in the metabolism of a subset of cytoplasmic mRNAs. We found that vigilin bound with high affinity to the vitellogenin and dystrophin mRNA 3′-UTRs, but did not bind well to segments of albumin mRNA and transferrin receptor mRNA (17,19). Recently, Li et al. reported that the yeast orthologue of vertebrate vigilin, Scp160p, binds to approximately 1% of yeast mRNAs (23). A selective effect of Scp160p/vigilin on the metabolism of such a small subset of mRNAs is consistent with the absence of a global effect of vigilin knockdown on the overall rate of protein synthesis.
Most early work on vigilin was consistent with the view that vigilin was a generic, low-specificity nucleic acid binding protein. Until recently, it was thought that most KH domain proteins bound to RNA with little sequence specificity. However, studies from our own and several other laboratories demonstrate that several KH domain proteins, including vigilin and Nova-1, bind to specific RNAs (14,17,23). Consistent with the view that vigilin was unlikely to play an essential role in nucleic acid metabolism or function, the phenotype of a yeast vigilin knockout was not lethal.
Our data indicate that vigilin is essential for the viability of dividing and non-dividing human cells, and that it does not play a critical role in translation. There is currently no direct evidence supporting a role for vigilin in chromosome partitioning in vertebrate cells. In addition, our data show that vigilin has an essential function in non-dividing cells that cannot be due to chromosome partitioning. The data presented here are consistent with the third proposed role for vigilin functioning as a trans-acting factor important in the metabolism of specific mRNAs. This view of vigilin/Scp160p action is supported by our earlier data describing a role for vigilin in the estrogen-mediated control of vitellogenin mRNA stability and more recently by the important demonstration that Scp160p binds to a small subset of yeast polysomal mRNAs (23). Vigilin/Scp160p’s sequence-specific interaction with a subset of mRNAs and the death of non-dividing human cells following vigilin knockdown are consistent with a critical role for vigilin in mRNA metabolism.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr R. Dodson of this laboratory for advice on gel shift assays and to Dr A. Villasante (Madrid, Spain) for the gift of the dodecasatellite plasmid. This work was supported by NIH grant DK 50080.
REFERENCES
- 1.Musco G., Stier,G., Joseph,C., Castiglione Morelli,M.A., Nilges,M., Gibson,T.J. and Pastore,A. (1996) Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome. Cell, 85, 237–245. [DOI] [PubMed] [Google Scholar]
- 2.Dodson R.E. and Shapiro,D.J. (1994) An estrogen-inducible protein binds specifically to a sequence in the 3′ untranslated region of estrogen-stabilized vitellogenin mRNA. Mol. Cell. Biol., 14, 3130–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wintersberger U., Kuhne,C. and Karwan,A. (1995) Scp160p, a new yeast protein associated with the nuclear membrane and the endoplasmic reticulum, is necessary for maintenance of exact ploidy. Yeast, 11, 929–944. [DOI] [PubMed] [Google Scholar]
- 4.Kruse C., Willkomm,D.K., Grunweller,A., Vollbrandt,T., Sommer,S., Busch,S., Pfeiffer,T., Brinkmann,J., Hartmann,R.K. and Muller,P.K. (2000) Export and transport of tRNA are coupled to a multi-protein complex. Biochem. J., 346, 107–115. [PMC free article] [PubMed] [Google Scholar]
- 5.Kruse C., Grunweller,A., Willkomm,D.K., Pfeiffer,T., Hartmann,R.K. and Muller,P.K. (1998) tRNA is entrapped in similar, but distinct, nuclear and cytoplasmic ribonucleoprotein complexes, both of which contain vigilin and elongation factor 1 alpha. Biochem. J., 329, 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kruse C., Grunweller,A., Notbohm,H., Kugler,S., Purschke,W.G. and Muller,P.K. (1996) Evidence for a novel cytoplasmic tRNA–protein complex containing the KH-multidomain protein vigilin. Biochem. J., 320, 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortes A., Huertas,D., Fanti,L., Pimpinelli,S., Marsellach,F.X., Pina,B. and Azorin,F. (1999) DDP1, a single-stranded nucleic acid-binding protein of Drosophila, associates with pericentric heterochromatin and is functionally homologous to the yeast Scp160p, which is involved in the control of cell ploidy. EMBO J., 18, 3820–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKnight G.L., Reasoner,J., Gilbert,T., Sundquist,K.O., Hokland,B., McKernan,P.A., Champagne,J., Johnson,C.J., Bailey,M.C., Holly,R. et al. (1992) Cloning and expression of a cellular high density lipoprotein-binding protein that is up-regulated by cholesterol loading of cells. J. Biol. Chem., 267, 12131–12141. [PubMed] [Google Scholar]
- 9.Plenz G., Gan,Y., Raabe,H.M. and Muller,P.K. (1993) Expression of vigilin in chicken cartilage and bone. Cell Tissue Res., 273, 381–389. [DOI] [PubMed] [Google Scholar]
- 10.Grishin N.V. (2001) KH domain: one motif, 2-folds. Nucleic Acids Res., 29, 638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adinolfi S., Bagni,C., Musco,G., Gibson,T., Mazzarella,L. and Pastore,A. (1999) Dissecting FMR1, the protein responsible for fragile X syndrome, in its structural and functional domains. RNA, 5, 1248–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dodson R.E. and Shapiro,D.J. (2002) Regulation of pathways of mRNA destabilization and stabilization. Prog. Nucleic Acid Res. Mol. Biol., 72, 129–164. [DOI] [PubMed] [Google Scholar]
- 13.Siomi H., Choi,M., Siomi,M.C., Nussbaum,R.L. and Dreyfuss,G. (1994) Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell, 77, 33–39. [DOI] [PubMed] [Google Scholar]
- 14.Buckanovich R.J. and Darnell,R.B. (1997) The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol. Cell. Biol., 17, 3194–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishizuka A., Siomi,M.C. and Siomi,H. (2002) A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev., 16, 2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caudy A.A., Myers,M., Hannon,G.J. and Hammond,S.M. (2002) Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev., 16, 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanamori H., Dodson,R.E. and Shapiro,D.J. (1998) In vitro genetic analysis of the RNA binding site of vigilin, a multi-KH-domain protein. Mol. Cell. Biol., 18, 3991–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dompenciel R.E., Garnepudi,V.R. and Schoenberg,D.R. (1995) Purification and characterization of an estrogen-regulated Xenopus liver polysomal nuclease involved in the selective destabilization of albumin mRNA. J. Biol. Chem., 270, 6108–6118. [DOI] [PubMed] [Google Scholar]
- 19.Cunningham K.S., Dodson,R.E., Nagel,M.A., Shapiro,D.J. and Schoenberg,D.R. (2000) Vigilin binding selectively inhibits cleavage of the vitellogenin mRNA 3′-untranslated region by the mRNA endonuclease polysomal ribonuclease 1. Proc. Natl Acad. Sci. USA, 97, 12498–12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang B.D. and Fridovich-Keil,J.L. (2000) Scp160p, a multiple KH-domain protein, is a component of mRNP complexes in yeast. Nucleic Acids Res., 28, 1576–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang B.D., Li,A., Black-Brewster,H.D. and Fridovich-Keil,J.L. (2001) The brefeldin A resistance protein Bfr1p is a component of polyribosome-associated mRNP complexes in yeast. Nucleic Acids Res., 29, 2567–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frey S., Pool,M. and Seedorf,M. (2001) Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J. Biol. Chem., 276, 15905–15912. [DOI] [PubMed] [Google Scholar]
- 23.Li A.M., Watson,A. and Fridovich-Keil,J.L. (2003) Scp160p associates with specific mRNAs in yeast. Nucleic Acids Res., 31, 1830–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortes A. and Azorin,F. (2000) DDP1, a heterochromatin-associated multi-KH-domain protein of Drosophila melanogaster, interacts specifically with centromeric satellite DNA sequences. Mol. Cell. Biol., 20, 3860–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neu-Yilik G., Zorbas,H., Gloe,T.R., Raabe,H.M., Hopp-Christensen,T.A. and Muller,P.K. (1993) Vigilin is a cytoplasmic protein. A study on its expression in primary cells and in established cell lines of different species. Eur. J. Biochem., 213, 727–736. [DOI] [PubMed] [Google Scholar]
- 26.Abad J.P., Carmena,M., Baars,S., Saunders,R.D., Glover,D.M., Ludena,P., Sentis,C., Tyler-Smith,C. and Villasante,A. (1992) Dodeca satellite: a conserved G+C-rich satellite from the centromeric heterochromatin of Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 89, 4663–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 28.Harborth J., Elbashir,S.M., Bechert,K., Tuschl,T. and Weber,K. (2001) Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci., 114, 4557–4565. [DOI] [PubMed] [Google Scholar]
- 29.Griffin M.J. (1976) Synchronization of some human cell strains by serum and calcium starvation. In Vitro, 12, 393–398. [DOI] [PubMed] [Google Scholar]
- 30.Brooks R.F. (1976) Regulation of fibroblast cell cycle by serum. Nature, 260, 248–250. [DOI] [PubMed] [Google Scholar]
- 31.Knehr M., Poppe,M., Enulescu,M., Eickelbaum,W., Stoehr,M., Schroeter,D. and Paweletz,N. (1995) A critical appraisal of synchronization methods applied to achieve maximal enrichment of HeLa cells in specific cell cycle phases. Exp. Cell Res., 217, 546–553. [DOI] [PubMed] [Google Scholar]
- 32.Oliver F.J., de la Rubia,G., Rolli,V., Ruiz-Ruiz,M.C., de Murcia,G. and Murcia,J.M. (1998) Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis. Lesson from an uncleavable mutant. J. Biol. Chem., 273, 33533–33539. [DOI] [PubMed] [Google Scholar]
- 33.Braddock D.T., Baber,J.L., Levens,D. and Clore,G.M. (2002) Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: solution structure of a complex between hnRNP K KH3 and single-stranded DNA. EMBO J., 21, 3476–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braddock D.T., Louis,J.M., Baber,J.L., Levens,D. and Clore,G.M. (2002) Structure and dynamics of KH domains from FBP bound to single-stranded DNA. Nature, 415, 1051–1056. [DOI] [PubMed] [Google Scholar]
- 35.Dodson R.E. and Shapiro,D.J. (1997) Vigilin, a ubiquitous protein with 14 K homology domains, is the estrogen-inducible vitellogenin mRNA 3′-untranslated region-binding protein. J. Biol. Chem., 272, 12249–12252. [DOI] [PubMed] [Google Scholar]
- 36.Dodson R.E., Acena,M.R. and Shapiro,D.J. (1995) Tissue distribution, hormone regulation and evidence for a human homologue of the estrogen-inducible Xenopus laevis vitellogenin mRNA binding protein. J. Steroid Biochem. Mol. Biol., 52, 505–515. [DOI] [PubMed] [Google Scholar]
- 37.Elbashir S.M., Martinez,J., Patkaniowska,A., Lendeckel,W. and Tuschl,T. (2001) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J., 20, 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]