Abstract
Cofilin stimulates actin filament disassembly and accelerates actin filament turnover. Cofilin is also involved in stimulus-induced actin filament assembly during lamellipodium formation. However, it is not clear whether this occurs by replenishing the actin monomer pool, through filament disassembly, or by creating free barbed ends, through its severing activity. Using photoactivatable Dronpa-actin, we show that cofilin is involved in producing more than half of all cytoplasmic actin monomers and that the rate of actin monomer incorporation into the tip of the lamellipodium is dependent on the size of this actin monomer pool. Finally, in cofilin-depleted cells, stimulus-induced actin monomer incorporation at the cell periphery is attenuated, but the incorporation of microinjected actin monomers is not. We propose that cofilin contributes to stimulus-induced actin filament assembly and lamellipodium extension by supplying an abundant pool of cytoplasmic actin monomers.
Introduction
Actin filament dynamics are essential for various cell activities, including cell migration, morphological change, and polarity formation. These events are regulated by a variety of actin-binding proteins, which cooperatively act in the assembly/disassembly and reorganization of actin filaments in cells (Pollard and Borisy, 2003; Revenu et al., 2004). Cofilin/actin-depolymerizing factor (ADF) family proteins, ubiquitously expressed in eukaryotes, are key regulators of actin filament dynamics (Moon and Drubin, 1995; Welch et al., 1997; Bamburg et al., 1999; Pantaloni et al., 2001). In vitro studies have demonstrated that cofilin stimulates actin filament disassembly by accelerating the off rate of actin monomers from the pointed ends of actin filaments (depolymerization) and by severing actin filaments (Carlier et al., 1997; Rosenblatt et al., 1997; Lappalainen and Drubin, 1997; Maciver, 1998). Depletion or inactivation of cofilin in Drosophila melanogaster or mammalian cells results in aberrant F-actin accumulation, implicating cofilin in actin filament disassembly in the cell (Gunsalus et al., 1995; Arber et al., 1998; Yang et al., 1998; Chen et al., 2001; Hotulainen et al., 2005; Nishita et al., 2005).
Conversely, cofilin is required for actin filament assembly in the cell, as seen in the case of stimulus-induced lamellipodium formation (Chan et al., 2000; Zebda et al., 2000; Ghosh et al., 2004). The observations that cofilin preferentially binds to the ADP-bound actin in filaments and enhances actin filament disassembly from the pointed ends in the rear of the lamellipodium have led to the treadmilling model, where cofilin contributes to actin filament assembly by replenishing actin monomers for polymerization (Bamburg et al., 1999; Pantaloni et al., 2001; Pollard and Borisy, 2003). An alternative model has recently proposed that cofilin is involved in stimulus-induced actin filament assembly by severing actin filaments to create free barbed ends that are used as nucleation sites for actin polymerization (Condeelis, 2001; Ghosh et al., 2004; DesMarais et al., 2005). This model is based on the observation that in MTLn3 mammary adenocarcinoma cells cofilin inactivation inhibited EGF-induced barbed end formation and lamellipodium extension in the cell periphery, without changing the G/F-actin ratio in the cell (Chan et al., 2000; Zebda et al., 2000). However, in other types of cells, conflicting results have suggested that the G/F-actin ratio decreases after cofilin inactivation (Chen et al., 2001; Hotulainen et al., 2005). Thus, it remains unclear whether cofilin contributes to stimulus-induced actin filament assembly in the cells by supplying actin monomers through its depolymerizing/severing activity, creating free barbed ends through its severing activity, or both of these two processes. To define the extent to which cofilin plays a role in these two possible processes in stimulus-induced actin filament assembly, it is essential to determine the G/F-actin ratio quantitatively in both cofilin-active and -inactive cells.
In this study, we have assessed the actin monomer pool in the cytoplasm of living cells by measuring the fluorescence decay of Dronpa (Dp)-labeled actin photoactivated in a small region of the cytoplasm. Dp is a GFP-like protein whose fluorescence can be reversibly switched off and on by photobleaching and photoactivation, respectively (Ando et al., 2004). Together with F-actin sedimentation assays, we provide evidence that cofilin is involved in the generation of more than half of the actin monomers in the cytoplasm. Using cofilin mutants, we also show that the severing activity, rather than the depolymerization activity, of cofilin is predominantly involved in maintaining the actin monomer pool in the cell. We also demonstrate that actin monomers in the cytoplasm are incorporated into the tip of the lamellipodium at the rate dependent on the actin monomer pool size in the cytoplasm. Furthermore, in cofilin-inactivated or -depleted cells, in which >80% of total cofilin is converted to the inactive phosphocofilin by LIM-kinase overexpression or total cofilin expression is decreased ∼80% by RNA interference, stimulus-induced actin monomer incorporation at the cell periphery is attenuated, but the incorporation of microinjected actin monomers is not. Our results suggest that cofilin contributes to stimulus-induced actin filament assembly in the cell periphery by supplying an abundant pool of actin monomers to the cytoplasm.
Results
Assessment of actin monomer population in the cytoplasm of living cells by measuring fluorescence decay of Dp-actin
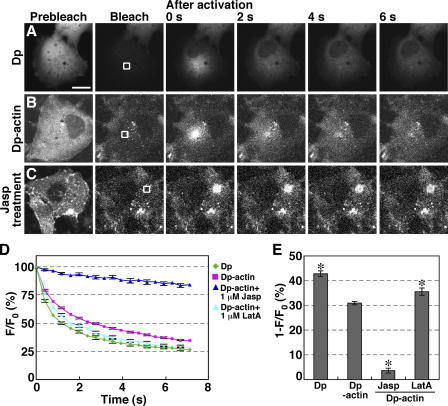
To assess actin monomer pool size in the cytoplasm of living cells, we expressed Dp-labeled actin in COS7 cells and measured the fluorescence decay of Dp-actin in the cytoplasm. The level of Dp-actin expressed in COS7 cells was <1%, compared with endogenous actin, and the expression had no apparent effect on F-actin assembly (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200610005/DC1). After the fluorescence of the whole cell was photobleached to the background level, Dp or Dp-actin was locally photoactivated in a small square region of the cytoplasm, and fluorescence images were acquired every 0.4 s. Both Dp (Fig. 1 A and Video 1) and Dp-actin (Fig. 1 B and Video 2 A) rapidly diffused from the photoactivated region throughout the cytoplasm. Quantitative analyses of the time-dependent changes in fluorescence intensity in the photoactivated region (Fig. 1 D) and the fluorescence decay at 0.8 s after photoactivation (Fig. 1 E) indicate that the rate of fluorescence decay of Dp-actin is slightly slower than that of Dp.
Figure 1.
The rate of fluorescence decay of photoactivated Dp-actin reflects the actin monomer population in the cytoplasm. (A–C) Fluorescence decay of Dp or Dp-actin. COS7 cells were transfected with Dp (A) or Dp-actin (B and C). Whole cells were photobleached, and a 5.8-μm2 region (white box) was photoactivated. Fluorescence images were acquired every 0.4 s for 7.2 s at 37°C using a laser-scanning confocal imaging system (Videos 1 and 2 A, available at http://www.jcb.org/cgi/content/full/jcb.200610005/DC1). (C) COS7 cells expressing Dp-actin were pretreated with 1 μM Jasp for 20 min before photoactivation (Video 2 B). Bar, 20 μm. (D) Time course of the fluorescence decay of Dp and Dp-actin in the photoactivated region with correction for photobleaching. The mean fluorescence intensity immediately after photoactivation (t = 0 s) was set to 100%. Data are means ± SEM of 24 (Dp), 68 (Dp-actin), 23 (Dp-actin; Jasp), and 22 cells (Dp-actin; LatA) from at least three independent experiments. (E) Fluorescence decay at 0.8 s after photoactivation from data in D. *, P < 0.05, compared with the decay of Dp-actin in untreated cells.
To determine whether the rate of fluorescence decay of Dp-actin is related to the actin monomer pool in the cytoplasm, we examined the effects of actin-modulating drugs on the diffusion of Dp-actin. When COS7 cells were pretreated with jasplakinolide (Jasp), a drug that induces actin polymerization, Dp-actin in the photoactivated region was almost immobile (Fig. 1 C and Video 2 B), and the rate of fluorescence decay was correspondingly reduced (Fig. 1, D and E). In contrast, treatment of the cell with latrunculin A (LatA), a drug that induces actin filament disassembly, slightly accelerated the fluorescence decay rate of Dp-actin, compared with untreated cells (Fig. 1, D and E). These results suggest that the rate of Dp-actin fluorescence decay reflects the relative levels of G/F-actin in the cytoplasm, with fast decay rates corresponding to a high G-actin population. Although the rate of Dp-actin fluorescence decay theoretically also depends on the turnover rate of actin filaments, the half-life of actin filaments in the cell ranges from tens of seconds (in the leading edge of migrating keratocytes) to several minutes (in stress fibers; Amato and Taylor, 1986; Theriot and Mitchison, 1991, 1992; McGrath et al., 1998). Therefore, the rates of fluorescence decay of Dp-actin measured in this study, at least in the initial phase after photoactivation (less than a few seconds), appear to reflect primarily the G/F-actin ratio in the cytoplasm. Thus, the level of G-actin relative to total actin in the cytoplasm of living cells can be estimated by measuring the rate of fluorescence decay of Dp-actin immediately after photoactivation.
Cofilin inactivation or knockdown markedly decreases the cytoplasmic actin monomer pool
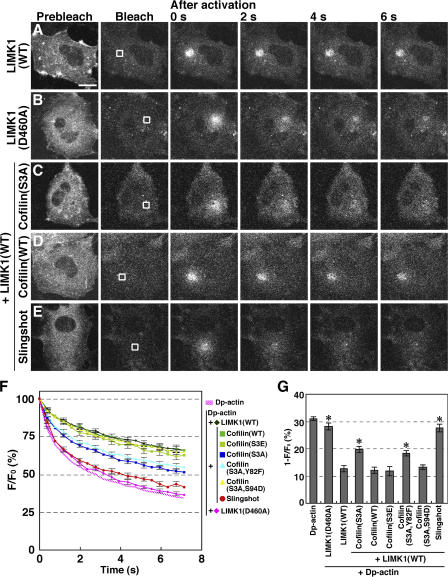
To examine the role of cofilin in regulating the actin monomer pool in the cytoplasm, we analyzed the effect of overexpression of LIM-kinase 1 (LIMK1), which inactivates cofilin by phosphorylation of Ser-3 (Yang et al., 1998), on the fluorescence decay of Dp-actin in COS7 cells. By expression of wild-type (WT) LIMK1, >80% of total cofilin was converted to phosphocofilin in COS7 cells (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200610005/DC1). Expression of LIMK1 (WT) clearly reduced the mobility of photoactivated Dp-actin in the cytoplasm and the rate of fluorescence decay in the photoactivated region (Fig. 2, A and F; and Video 2 C). In contrast, expression of kinase-dead LIMK1(D460A) had no apparent effect on fluorescence decay (Fig. 2, B and F; and Video 2 D). These results suggest that LIMK1-mediated cofilin inactivation markedly reduced the G-actin pool in the cytoplasm. The fluorescence decay of Dp-actin in LIMK1-expressing cells was reduced by 59%, compared with control cells expressing Dp-actin alone (Fig. 2 G), which indicates that cofilin contributes to the production of more than half of the G-actin pool in control cells. When a nonphosphorylatable, constitutively active cofilin(S3A) mutant was coexpressed with LIMK1, it partially rescued the mobility of Dp-actin and increased the fluorescence decay rate, compared with cells expressing LIMK1 alone (Fig. 2, C, F, and G; and Video 3 A). The partial rescue is probably due to the low actin-disassembling activity of cofilin(S3A) mutant (Fig. S3 B). In contrast, coexpression of cofilin(WT) had no apparent effect (Fig. 2, D, F, and G; and Video 3 B), most likely because of its phosphorylation and inactivation by LIMK1. Similarly, the phosphorylation-mimic cofilin(S3E) mutant had no effect (Fig. 2, F and G). Furthermore, coexpression of the cofilin phosphatase Slingshot-1, which neutralizes LIMK1 activity (Niwa et al., 2002), almost completely blocked the inhibitory effect of LIMK1 on fluorescence decay (Fig. 2, E, F, and G; and Video 3 C). These observations suggest that the LIMK1-induced decrease in Dp-actin fluorescence decay is primarily caused by cofilin phosphorylation and inactivation and that the cytoplasmic G-actin pool depends largely on cofilin activity.
Figure 2.
Effects of expression of LIMK1, cofilin, or their mutants on the fluorescence decay of Dp-actin. COS7 cells were cotransfected with Dp-actin and LIMK1 (A), LIMK1(D460A) (B), LIMK1 + cofilin(S3A) (C), LIMK1 + cofilin(WT) (D), or LIMK1 + Slingshot-1 (E). Photobleaching, photoactivation, and fluorescence microscopy were conducted as in Fig. 1. White boxes indicate the photoactivated regions. See Videos 2 and 3 (available at http://www.jcb.org/cgi/content/full/jcb.200610005/DC1). Bar, 20 μm. (F) Time course of the fluorescence decay of Dp-actin in the photoactivated region, measured as in Fig. 1 D. Data are means ± SEM of 44 (LIMK1), 33 (LIMK1[D460A]), 55 (LIMK1 + cofilin[S3A]), 30 (LIMK1 + cofilin[WT]), 35 (LIMK1 + cofilin[S3E]), 47 (LIMK1 + cofilin[S3A,Y82F]), 57 (LIMK1 + cofilin[S3A,S94D]), and 30 cells (LIMK1 + Slingshot-1) from at least three independent experiments. (G) Fluorescence decay at 0.8 s after photoactivation from data in F. *, P < 0.05, compared with cells expressing LIMK1(WT).
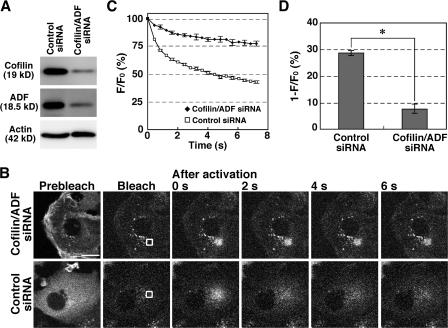
We also examined the effect of knock down of cofilin/ADF expression using siRNA in MCF-7 human breast carcinoma cells. Similar to LIMK1 overexpression in COS7 cells, double knock down of cofilin and ADF remarkably reduced the mobility of Dp-actin and the rate of fluorescence decay of Dp-actin in the cytoplasm of MCF-7 cells (Fig. 3 and Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200610005/DC1), which further supports the theory that cofilin plays a critical role in maintaining the G-actin pool in the cytoplasm.
Figure 3.
Cofilin/ADF double knockdown decreases the rate of fluorescence decay of Dp-actin. (A) Suppression of endogenous cofilin and ADF expression by siRNA. MCF-7 cells were cotransfected with cofilin and ADF siRNA plasmids or transfected with control siRNA plasmid. After 4 d of culture, cell lysates were analyzed by immunoblotting with antibodies specific for cofilin, ADF, and actin. (B) Effects of cofilin/ADF double knockdown on the fluorescence decay of Dp-actin. MCF-7 cells were cotransfected with Dp-actin and cofilin/ADF siRNAs (top) or control siRNA (bottom). Photobleaching, photoactivation, and fluorescence microscopy were conducted as in Fig. 1. White boxes indicate the photoactivated regions. See also Video 4 (available at http://www.jcb.org/cgi/content/full/jcb.200610005/DC1). Bar, 20 μm. (C) Time course of the fluorescence decay of Dp-actin in the photoactivated region, measured as in Fig. 1 D. Data are means ± SEM of 24 cells (cofilin/ADF siRNA) and 25 (control siRNA) from three independent experiments. (D) Fluorescence decay at 0.8 s after photoactivation from data in C. *, P < 0.05, compared with control cells.
The actin filament–severing activity of cofilin plays a dominant role in increasing the actin monomer pool in the cytoplasm
Cofilin stimulates depolymerization (dissociation of actin monomers from the pointed end) and severing of actin filaments. To elucidate which of these two activities plays a dominant role in increasing the G-actin pool in the cytoplasm, we expressed LIMK1 with (S3A,Y82F)- or (S3A,S94D)-cofilin mutants in COS7 cells and examined the effects of these mutants on the LIMK1-mediated inhibition of Dp-actin diffusion. Similar to the reported characteristics of Y82F- and S94D-cofilin mutants (Moriyama and Yahara, 1999, 2002), the in vitro analyses showed that cofilin(S3A,Y82F), but not cofilin(S3A,S94D), has severing activity (Fig. S3, C and D), whereas both mutants retain the ability to promote dilution-induced F-actin disassembly (which reflects the combined depolymerization and severing activities; Fig. S3 B). Cofilin(S3A,Y82F), which retains the severing activity, significantly recovered the LIMK1-mediated inhibition of the fluorescence decay of Dp-actin, to the extent similar to cofilin(S3A) (Fig. 2, F and G; and Video 3 D). In contrast, cofilin(S3A,S94D), which exhibits no severing activity, did not block LIMK1 inhibition of fluorescence decay (Fig. 2, F and G; and Video 3 E). These results suggest that the severing activity of cofilin plays a dominant role in increasing the actin monomer pool in the cytoplasm.
Quantification of the G/F-actin ratio by F-actin sedimentation assays
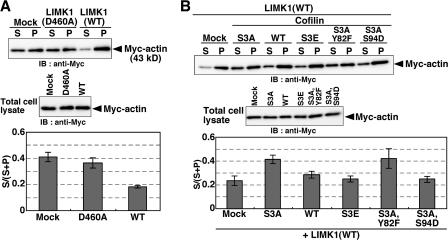
We also analyzed the effects of LIMK1(WT or D460A) expression on the G/F-actin ratio in cells using F-actin sedimentation assays. COS7 cells were cotransfected with plasmids for Myc-actin and LIMK1 at a molar ratio of 1:5, to ensure the coexpression of LIMK1 in almost all Myc-actin–expressing cells. Cell lysates were centrifuged to separate G- and F-actin and analyzed by anti-Myc immunoblotting. Expression of LIMK1(WT), but not LIMK1(D460A), markedly reduced the ratio of G- to F-actin, compared with mock-transfected cells (Fig. 4 A). G-actin accounted for 41 and 18% of total actin (G- plus F-actin) in mock-transfected and LIMK1-expressing cells, respectively, thus indicating that 56% of the G-actin in control cells was shifted to F-actin by LIMK1 expression (Fig. 4 A). These data are consistent with results from the fluorescence decay of Dp-actin (Fig. 2 G). The LIMK1-induced reduction in actin monomer content was substantially blocked by coexpression of cofilin(S3A) or cofilin(S3A,Y82F), but not by cofilin(WT), cofilin(S3E), or cofilin(S3A,S94D) (Fig. 4 B). Together with data from the Dp-actin fluorescence decay assays, these results strongly suggest that cofilin contributes to the production of more than half of the G-actin in the cell and that the severing activity of cofilin plays a dominant role in this process.
Figure 4.
Effects of expression of LIMK1, cofilin, or their mutants on the G/F-actin ratio, as measured by F-actin sedimentation assays. Myc-actin was cotransfected into COS7 cells with empty vector (Mock), LIMK1(D460A), LIMK1(WT) (A), or LIMK1 (WT) plus cofilin mutants (B), as indicated. Cell lysates were centrifuged, and the amount of Myc-actin recovered in the supernatant (S) and pellet (P) was analyzed by immunoblotting with an anti-Myc antibody (A and B, top). To analyze the total amount of Myc-actin, total cell lysates were analyzed by immunoblotting with anti-Myc antibody (A and B, middle). The amounts of G-actin (S) and F-actin (P) were determined by densitometric analysis of immunoblots, and the ratios of G-actin (S) to total actin (S + P) were calculated (A and B, bottom). Data are means ± SEM of four independent experiments.
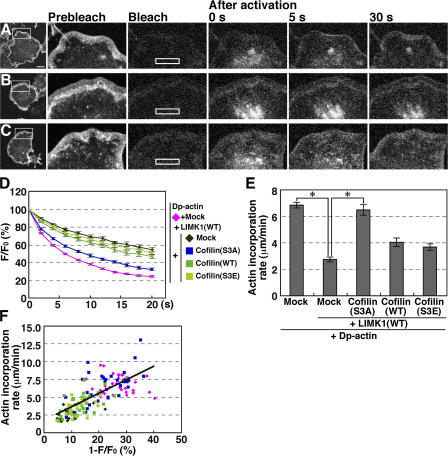
The rate of actin monomer incorporation into the tip of the lamellipodium is correlated to the actin monomer pool in the cytoplasm
We next examined whether the G-actin population in the cytoplasm is correlated to actin filament assembly in the lamellipodium. We analyzed the fluorescence decay of Dp-actin in the cytoplasm and its incorporation into the lamellipodium in the same cell. We used N1E-115 cells because they stably produced lamellipodia by expression of active Rac(V12). After cotransfection of the cells with Dp-actin and Rac(V12) and cell-wide photobleaching, Dp-actin was photoactivated in a rectangular region in the cytoplasm (Fig. 5 A). Dp-actin photoactivated in the cytoplasm was continuously incorporated into the tip of the lamellipodium and then flowed retrogradely toward the cell body (Fig. 5 A and Video 5 A, available at http://www.jcb.org/cgi/content/full/jcb.200610005/DC1). Expression of LIMK1 slowed the fluorescence decay of the photoactivated Dp-actin in the cytoplasm and repressed its incorporation into the lamellipodium (Fig. 5 B and Video 5 B). Fig. 5 D shows the time courses of Dp-actin fluorescence decay in the photoactivated region. Fig. 5 E shows the rate of Dp-actin incorporation into the lamellipodium, measured as the length of Dp-actin fluorescence from the tip of the lamellipodium to the cell center per unit time after photoactivation. These analyses revealed that LIMK1 expression suppresses both the fluorescence decay of Dp-actin in the cytoplasm and its incorporation into the lamellipodium. The inhibitory effects of LIMK1 were substantially blocked by coexpression of cofilin(S3A) (Fig. 5 C and Video 5 C), but not by cofilin(WT) or cofilin(S3E) (Fig. 5, D and E). The rate of Dp-actin incorporation into the lamellipodium is linearly correlated with the fluorescence decay in the cytoplasm in each cell (Fig. 5 F), which indicates that actin monomer assembly at the tip of the lamellipodium is highly dependent on the cytoplasmic actin monomer pool size. Because cofilin elevates the cytoplasmic G-actin levels, cofilin likely enhances actin filament assembly at the tip of the lamellipodium by increasing the G-actin level in the cytoplasm.
Figure 5.
Dp-actin photoactivated in the cytoplasm is efficiently incorporated into the lamellipodium, and the rate of incorporation is dependent on the G-actin pool in the cytoplasm. (A–C) Dp-actin photoactivated in the cytoplasm is incorporated into the lamellipodium. NIE-115 cells were cotransfected with Dp-actin and RacV12 (A); Dp-actin, RacV12, and LIMK1 (B); or Dp-actin, RacV12, LIMK1, and cofilin(S3A) (C). After cell-wide photobleaching, a 14.25- × 2.85-μm rectangular region (white box) was photoactivated, and fluorescence images were acquired every 2 s for 20 s at 37°C using a laser-scanning confocal imaging system to measure the fluorescence decay of Dp-actin (shown in D). The same cells were photobleached, and the same rectangular region was again photoactivated. Fluorescence images were acquired every 5 s for 40 s to measure the incorporation of Dp-actin into the tip of the lamellipodium (A–C; see Video 5, available at http://www.jcb.org/cgi/content/full/jcb.200610005/DC1). Bar, 20 μm. (D) Time course of the fluorescence decay of Dp-actin in the photoactivated region. Plasmids transfected into the cells are indicated on the right. (E) The rate of Dp-actin incorporation into the lamellipodium, measured as the distance of Dp-actin fluorescence advanced from the tip of the lamellipodium toward the cytoplasm for 20 or 40 s after photoactivation. Data in D and E are means ± SEM of 36 (Dp-actin), 20 (Dp-actin; LIMK1[WT]), 33 (Dp-actin; LIMK1[WT]; cofilin[S3A]), 26 (Dp-actin; LIMK1[WT]; cofilin[WT]), and 23 cells (Dp-actin; LIMK1[WT]; cofilin[S3E]). *, P < 0.05, compared with cells expressing LIMK1(WT) alone. (F) The correlation between the fluorescence decay of Dp-actin at 2 s after photoactivation and the rate of Dp-actin incorporation into the lamellipodium. Each point represents an individual cell transfected with the plasmids indicated in D.
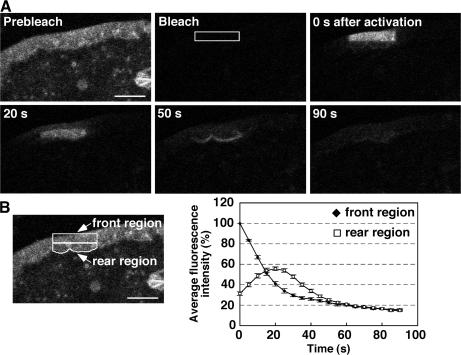
When Dp-actin in the front region of the lamellipodium was photoactivated, it flowed retrogradely and then rapidly diffused throughout the cytoplasm (Fig. 6 A and Video 6, available at http://www.jcb.org/cgi/content/full/jcb.200610005/DC1). The fluorescence intensity in the front region declined continuously until it reached a plateau at 15%, whereas the intensity in the rear of the lamellipodium initially increased, reached a maximum level at 20 s after photoactivation, and declined to a level similar to that in the front region (Fig. 6 B). These results suggest that most of the actin monomers that disassembled from the rear of the lamellipodium diffused into the cytoplasm.
Figure 6.
Dp-actin photoactivated in the front region of the lamellipodium flows retrogradely and diffuses into the cytoplasm. (A) Fluorescence images of Dp-actin photoactivated in the front region of the lamellipodium in NIE-115 cells expressing Dp-actin and RacV12. After cell-wide photobleaching, a 14.25- × 2.85-μm rectangular region (white box) was photoactivated, and fluorescence images were acquired every 5 s for 90 s at 37°C using a laser-scanning confocal imaging system. See Video 6 (available at http://www.jcb.org/cgi/content/full/jcb.200610005/DC1). Bar, 10 μm. (B) Time-dependent changes in fluorescence intensity of Dp-actin in the front and the rear regions of the lamellipodium (left) with correction for photobleaching. The mean fluorescence intensity in the front region immediately after photoactivation (t = 0 s) was set to 100%. Data are means ± SEM of 25 cells from three independent experiments.
Cofilin is required for EGF-induced actin filament assembly in the cell periphery
Previous studies have shown that EGF induced actin filament assembly in the periphery of MTLn3 carcinoma cells (Chan et al., 1998, 2000). As this EGF-induced assembly was suppressed by cofilin inactivation without changing the G/F-actin ratio in the cell, it was suggested that cofilin promotes actin filament assembly by creating new barbed ends through its severing activity (Chan et al., 2000; Zebda et al., 2000). We have shown here, however, that LIMK1-mediated inactivation or knock down of cofilin/ADF led to a large decrease in the actin monomer population in the cytoplasm. We also showed that the rate of actin monomer incorporation into the cell periphery depends on cofilin activity, as well as the cytoplasmic actin monomer population. We therefore hypothesized that cofilin is involved in stimulus-induced actin filament assembly in the cell periphery by supplying actin monomers in the cytoplasm and that cofilin inactivation inhibits actin filament assembly by decreasing the cytoplasmic actin monomer concentration to levels at which actin assembly is not feasible. To test this hypothesis, we analyzed the effects of LIMK1 overexpression on EGF-induced actin filament assembly by measuring the incorporation of Alexa Fluor 546–labeled actin (Alexa546-actin) monomers into actin filaments in the cell periphery by two distinct experimental procedures. First, Alexa546-actin was added to the cell after EGF stimulation using a cell permeabilization approach; second, Alexa546-actin was microinjected into the cell at the time of EGF stimulation.
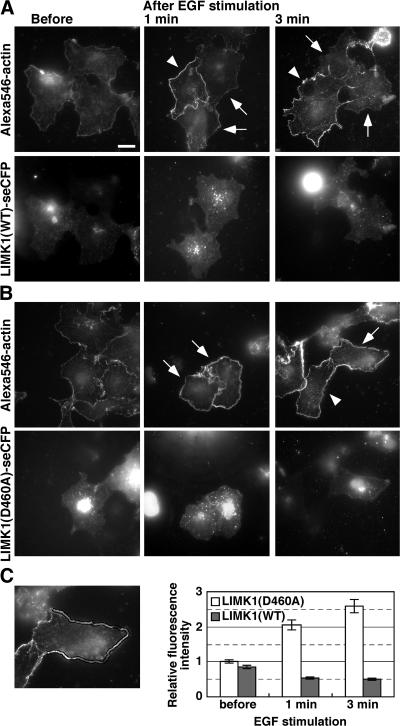
We first analyzed changes in the number of barbed ends in COS7 cells before and after EGF stimulation, by applying Alexa546-actin monomers and saponin (cell-permeabilizing agent) to the outside of the cells, according to previously reported procedures (Chan et al., 1998, 2000). COS7 cells expressing super-enhanced CFP (SECFP)-tagged LIMK1(WT) or LIMK1(D460A) were left unstimulated or stimulated with EGF for 1–3 min and then incubated with cell-permeabilization buffer containing 0.025% saponin and 0.45 μM Alexa546-actin monomers. At a concentration of 0.45 μM, Alexa546-actin monomers were incorporated only into barbed ends (Chan et al., 1998). In response to EGF stimulation, Alexa546-actin incorporation at the cell periphery markedly increased in cells not expressing LIMK1 (Fig. 7 A, arrowheads) and in cells expressing LIMK1(D460A) (Fig. 7 B, arrows), indicating that EGF stimulation increased the number of barbed ends at the cell periphery of cofilin-active cells. In contrast, in cells expressing LIMK1(WT), EGF-induced Alexa546-actin incorporation at the cell periphery was not observed (Fig. 7 A, arrows). Quantitative analysis revealed that Alexa546-actin incorporated into the cell margin increased 2.1–2.6-fold after EGF stimulation in cells expressing LIMK1(D460A), but no increase was observed in cells expressing LIMK1(WT) (Fig. 7 C). Thus, the number of actin filament barbed ends at the cell periphery was considerably increased after EGF stimulation of cofilin-active cells, but not of cofilin-inactive cells, which suggests that cofilin is required for EGF-induced barbed end formation at the cell periphery.
Figure 7.
Cofilin inactivation suppresses EGF-induced barbed end formation in the cell periphery. (A and B) COS7 cells transfected with LIMK1(WT)-SECFP (A) or LIMK1(D460A)-SECFP (B) were left unstimulated or stimulated with EGF for 1–3 min. Cells were permeabilized with nucleation buffer containing 0.025% saponin and 0.45 μM Alexa546-actin monomers, washed three times, and fixed to analyze Alexa546-actin (top) and LIMK1-SECFP fluorescence (bottom) using fluorescence microscopy. Arrows indicate LIMK1(WT)- or LIMK1(D460A)-expressing cells, and arrowheads indicate nonexpressing cells, assessed by SECFP fluorescence. Bar, 20 μm. (C) Quantitative analysis of Alexa546-actin fluorescence in the cell periphery. Incorporation of Alexa546-actin into the cell periphery was measured as the mean fluorescence intensity in a region 2 μm from the cell edge, using ImageJ. The left image shows the region measured. Data are means ± SEM of 38–42 cells from three independent experiments, with the mean fluorescence in unstimulated LIMK1(D460A)-expressing cells normalized to 1.0.
Microinjected actin monomers are effectively incorporated into the cell periphery after EGF stimulation in both cofilin-active and -inactive cells
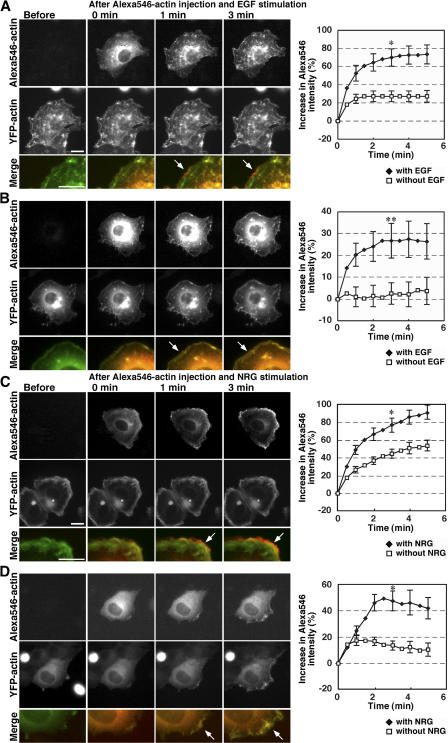
Previous studies have suggested that cell stimulation activates the Arp2/3 complex, which enhances de novo nucleation and arborization of actin filaments to generate dendritic actin filament structures (Takenawa and Miki, 2001; Pantaloni et al., 2001; Pollard and Borisy, 2003). Arp2/3 complex–mediated dendritic actin structure formation exponentially increases the number of barbed ends, and this process requires actin monomers for polymerization. To examine whether cofilin contributes to EGF-induced barbed end formation directly, by severing actin filaments, or indirectly, by supplying actin monomers for Arp2/3-mediated dendritic actin structure formation, we analyzed the effect of microinjection of Alexa546-actin monomers into cells expressing LIMK1(WT) or LIMK1(D460A). If cofilin primarily contributes to EGF-induced barbed end formation by severing actin filaments, injected Alexa546-actin will be incorporated into the periphery of control cells, but not into the periphery of cells expressing LIMK1(WT), after stimulation with EGF. Alternatively, if cofilin contributes to barbed end formation indirectly by supplying actin monomers, microinjected Alexa546-actin monomers will be incorporated into the periphery of both control and LIMK1(WT)-expressing cells in response to EGF stimulation. As shown in Fig. 8 A (top) and in Video 7 A (available at http://www.jcb.org/cgi/content/full/jcb.200610005/DC1), Alexa546-actin monomers injected into cells expressing LIMK1(WT) were initially diffuse in the cytoplasm and then incorporated into the distal cell margin 1–3 min after EGF stimulation. The YFP-actin distribution remained largely unchanged after EGF stimulation, probably because most YFP-actin was already assembled into F-actin as a result of cofilin inactivation (Fig. 8 A, middle; and Video 7 A). Alexa546- actin injected into control cells expressing LIMK1(D460A) was similarly incorporated into the cell periphery after EGF stimulation (Fig. 8 B, top; and Video 8 A). In this experiment, YFP-actin was also incorporated into the cell periphery after EGF stimulation, because YFP-actin monomers were abundant in the cell (Fig. 8 B, middle). Quantitative analysis of Alexa546-actin fluorescence intensity in the cell margin revealed that Alexa546- actin incorporation into the cell margin significantly increased with time after EGF stimulation in both LIMK1(WT)- and LIMK1(D460A)-expressing cells (Fig. 8, A and B, right). The time course of Alexa546-actin incorporation in LIMK1(WT)-expressing cells was comparable to that of LIMK1(D460A)- expressing cells.
Figure 8.
Microinjected actin monomers are incorporated at the cell periphery after cell stimulation in both cofilin-active and -inactive cells. COS7 cells were cotransfected with YFP-actin and LIMK1(WT) (A) or LIMK1(D460A) (B). MCF-7 cells were cotransfected with YFP-actin and cofilin/ADF siRNA (C) or control siRNA (D). Cells were microinjected with 24 μM Alexa546-actin monomers. Immediately after microinjection, cells were treated with EGF (A and B) or NRG (C and D) or left untreated (see Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200610005/DC1), and fluorescence images of Alexa546-actin and YFP-actin were acquired every 30 s for 9.5 min at 30°C using fluorescence microscopy. Merged images of Alexa546-actin (red) and YFP-actin (green) are shown in the bottom panels. Arrows indicate stimulus-induced Alexa546-actin incorporation at the cell periphery. See Videos 7 A, 8 A, 9 A, and 10 A. The graphs at the right quantify the Alexa546-actin incorporation into the cell periphery over time, measured as the percentage of increase in fluorescence intensity of Alexa546-actin in a region 2 μm interior to the cell edge, using ImageJ, with the mean fluorescence intensity at Alexa546-actin microinjection at time 0 normalized to 100%. Data are means ± SEM of 19 (LIMK1[WT] with EGF), 23 (LIMK1[WT] without EGF), 19 (LIMK1[D460A] with EGF), 14 (LIMK1[D460A] without EGF), 16 (cofilin/ADF siRNA with NRG), 18 (cofilin/ADF siRNA without NRG), 11 (control siRNA with NRG), and 14 cells (control siRNA without NRG). *, P < 0.05; **, P < 0.1, compared with unstimulated cells. Bars, 20 μm.
To further examine the role of cofilin in stimulus-induced actin assembly, we performed Alexa546-actin microinjection studies on neuregulin (NRG)-stimulated MCF-7 cells, in which cofilin/ADF were knocked down by siRNA. Alexa546-actin monomers injected were efficiently incorporated into the cell periphery of both cofilin/ADF and control siRNA MCF-7 cells in response to NRG stimulation (Fig. 8, C and D; and Videos 9 A and 10 A, available at http://www.jcb.org/cgi/content/full/jcb.200610005/DC1). Similar to the case of LIMK1 overexpression, YFP-actin was incorporated into the cell periphery in control siRNA cells, but it was almost unchanged in cofilin/ADF siRNA cells. In both LIMK1(WT)-expressing COS7 cells and cofilin/ADF siRNA MCF-7 cells, injected Alexa546-actin monomers were also incorporated into the cell periphery even in the absence of cell stimulation, although the level of incorporation was substantially lower than in stimulated cells (Fig. 8, A and C, graphs; Fig. S4, A and C; and Videos 7 and 9). This behavior probably reflects the fact that actin monomers are depleted in cofilin-inactive cells, and therefore, the ratio of the injected Alexa546-actin monomers is relatively high in the actin monomer pool available for polymerization. Thus, we conclude that actin monomers injected into the cytoplasm were effectively incorporated into actin filaments in the cell periphery in response to cell stimulation in both cofilin-active and -inactive cells. These results suggest that cofilin is required for stimulus-induced barbed end formation and actin filament assembly in the cell periphery primarily by supplying actin monomers.
Discussion
Cofilin is known to stimulate actin filament disassembly by depolymerizing and severing actin filaments both in vitro and in vivo (Bamburg et al., 1999; Pantaloni et al. 2001). In contrast, it has also been reported that cofilin is involved in actin filament polymerization within the cell, as in the case of stimulus-induced lamellipodium formation (Chan et al., 2000; Ghosh et al., 2004). Because the rate of actin filament assembly at the leading edge depends on the concentrations of actin monomers and free barbed ends, and cofilin can both increase actin monomers by disassembling actin filaments and create free barbed ends by severing the filaments, the precise role of cofilin in actin filament assembly and disassembly in the cell is unclear. This study was conducted to understand comprehensively the cellular role of cofilin in actin filament assembly and disassembly, especially in stimulus-induced lamellipodium formation. Our results demonstrate that (1) inactivation or knock down of cofilin remarkably decreased the actin monomer pool in the cytoplasm, indicating that cofilin is responsible for the generation of the bulk of the actin monomer pool in the cytoplasm; (2) actin monomers in the cytoplasm were efficiently incorporated into the tip of the lamellipodium, and incorporation was dependent on both cofilin activity and the cytoplasmic actin monomer population; and (3) cofilin inactivation suppressed stimulus-induced actin monomer incorporation at the cell periphery, but microinjection of actin monomers into cofilin-inactivated cells rescued actin incorporation into the cell periphery, which indicates that defective actin monomer incorporation in cofilin-inactivated cells is due to the lack of actin monomers, not to the lack of free barbed ends. Together, these findings suggest that cofilin contributes to stimulus-induced actin filament assembly in the cell periphery by supplying a large amount of actin monomers to the cytoplasm.
Role of cofilin in controlling the G-actin population in the cytoplasm
In unstimulated cells, actin filaments are continuously assembled and disassembled at rates considerably faster than those of actin filament treadmilling in vitro (Pollard, 1986; Theriot and Mitchison, 1991). Cofilin is probably involved in this rapid turnover of actin filaments by stimulating actin filament disassembly. To understand the role of cofilin in actin filament dynamics within the cell, it is essential to quantify the G/F-actin ratios in cofilin-active and -inactive cells, but there have been only a few controversial reports measuring these ratios. Previous studies have reported that no significant change in F-actin content was observed after cofilin inactivation by LIMK1 expression or anti-cofilin antibody injection, by measuring the fluorescence intensity of MTLn3 cells stained by fluorescently labeled phalloidin, although they observed thick stress fibers and F-actin aggregates in cofilin-inactivated cells (Chan et al., 2000; Zebda et al., 2000). Based on these observations that cofilin inactivation did not change the G/F-actin ratio in the cell, these researchers proposed the barbed end creation model. In contrast, Hotulainen et al. (2005) analyzed the relative ratio of G/F-actin in NIH3T3 and B16F1 cells by estimating the content of G-actin, F-actin, and total actin using the fluorescence intensities of DNase I, phalloidin, and anti–β-actin antibody staining, respectively, and showed that the G-actin content decreased substantially in cofilin knockdown cells. However, phalloidin staining of F-actin is affected by association with actin-binding proteins, and the anti–β-actin antibody used to measure total actin does not stain stress fibers (Mies et al., 1998). Thus, more precise and quantitative analyses are needed to determine the G/F-actin ratio in cells. In this study, we assessed the actin monomer pool size in living cells by measuring the fluorescence decay of Dp-actin photoactivated in the cytoplasm. Using this method and F-actin sedimentation assay, we showed that cofilin inactivation or knock down markedly reduced the G-actin content in the cell. Thus, we conclude that cofilin critically contributes to the production of actin monomers in cells; more than half of the G-actin pool is generated by the action of cofilin. Considering the very low content of actin monomers in cofilin-inactivated cells, cofilin inactivation probably inhibits stimulus-induced actin filament assembly and lamellipodium formation by the lack of actin monomers required for polymerization.
Although the dendritic nucleation/array treadmill model proposes that actin monomers disassembled from the pointed ends of actin filaments by the help of cofilin activity are recycled for polymerization at the tip of the lamellipodium (Cramer, 1999; Pollard and Borisy, 2003), it has not been clarified whether actin monomers incorporated into the tip of the lamellipodium are self-sufficiently supplied from the pointed ends of actin filaments within the lamellipodium or indirectly via the cytoplasmic actin monomer pool. Here, we showed that actin monomers in the cytoplasm are efficiently incorporated into the tip of the lamellipodium at the rate correlated with the cytoplasmic actin monomer pool size. We also observed that Dp-actin photoactivated in the lamellipodium flowed retrogradely and rapidly diffused into the cytoplasm. Thus, most of the actin monomers disassembled from the lamellipodium are probably diffused into the cytoplasm, reserved in the cytoplasmic actin monomer pool, and reused for polymerization. As the cytoplasmic actin monomer pool size is controlled by cofilin activity, cofilin plays a role in actin filament assembly at the tip of the lamellipodium by increasing the G-actin pool in the cytoplasm by disassembling the preexisting actin filaments.
We have shown that the severing activity of cofilin plays a dominant role in the generation of actin monomers. However, how the severing activity of cofilin generates actin monomers is not clear. The severing activity of cofilin may promote actin monomer production by generating short actin filaments that are quickly broken down by the action of decorated cofilin and/or by exposing free actin pointed ends for depolymerization. Using similar cofilin mutants, several studies showed that the severing activity of cofilin is essential for various cell events (Moriyama and Yahara, 2002; Endo et al., 2003).
Role of cofilin in stimulus-induced actin filament assembly
During cell migration, actin filaments are assembled to form the lamellipodial protrusion at the leading edge of the cell. Addition of actin monomers to the barbed ends of actin filaments at the tip of the lamellipodium gives the force to propel the membrane forward. A previous study showed that EGF induces a large generation of actin barbed ends at the leading edge and cofilin inactivation inhibited EGF-induced barbed end formation, lamellipodium extension, and cell migration (Chan et al., 2000). By measuring the incorporation of extracellularly added Alexa546-actin into the periphery of permeabilized COS7 cells, we also observed that cofilin inactivation suppressed EGF- induced barbed end formation. Thus, cofilin plays a critical role in stimulus-induced barbed end formation at the cell periphery. In general, free barbed ends can be generated by severing or decapping of preexisting actin filaments or de novo synthesis (and branching) of actin filaments. It is theoretically possible that cofilin is involved in barbed end formation either directly, by severing actin filaments, or indirectly, by supporting formation of branched actin filaments by increasing the actin monomer pool in the cell. Studies on EGF-stimulated MTLn3 carcinoma cells proposed the former model where cofilin creates new barbed ends by directly severing actin filaments (Condeelis, 2001; Ghosh et al., 2004; DesMarais et al., 2005). If cofilin dominantly acts by directly creating new barbed ends, microinjected actin monomers would not be effectively incorporated into the cell periphery of cofilin-inactivated cells after EGF stimulation, because the barbed end creation is blocked by cofilin inactivation. If cofilin dominantly acts by supplying actin monomers, injection of actin monomers would recover the inhibitory effect of cofilin inactivation on stimulus-induced actin filament assembly at the cell periphery. We demonstrated that Alexa546-actin monomers injected were effectively incorporated into the cell periphery after cell stimulation, in both control and cofilin-depleted cells, which suggests that cofilin contributes to stimulus-induced actin filament assembly by supplying actin monomers.
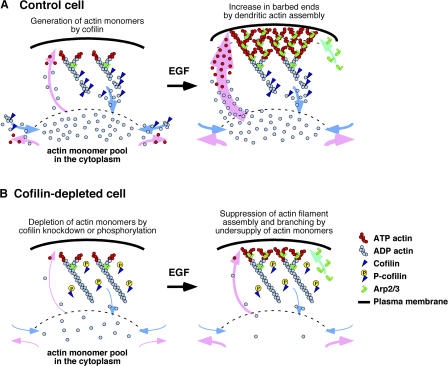
Based on these observations, we propose a model for the role of cofilin in stimulus-induced lamellipodium formation (Fig. 9). In resting cells, cofilin plays a critical role in maintaining the actin monomer pool in the cytoplasm by stimulating actin filament disassembly, thereby creating an abundant supply of actin monomers. Stimulation of the cell induces activation of the Arp2/3 complex, via Rac and WAVE activation (Takenawa and Miki, 2001), which stimulates de novo synthesis and arborization of actin filaments to form the dendritic actin structures. Cofilin contributes to this process by supplying actin monomers for polymerization. Arp2/3-assisted dendritic growth of actin filaments will exponentially increase the number of free barbed ends. In contrast, in cofilin-depleted cells, the actin monomer pool is substantially lower in resting cells; therefore, stimulus-induced dendritic actin polymerization and barbed end formation in the cell periphery are inhibited even when the Arp2/3 complex is activated. In conclusion, we have provided evidence that cofilin contributes to stimulus-induced actin filament assembly by supplying an abundant pool of actin monomers to the cytoplasm and that its severing activity is predominantly involved in this process.
Figure 9.
A model for the role of cofilin in stimulus-induced lamellipodium formation. (A) In control cofilin-active cells, cofilin contributes to production of the bulk of actin monomers in the cytoplasm and enhances actin filament turnover by stimulating actin filament disassembly. Stimulation of the cell with factors (such as EGF) activates the Arp2/3 complex and thereby stimulates the formation of dendritic actin filament structures, resulting in an increase in the number of barbed ends. Actin assembly onto the barbed ends near the cell edge pushes the membrane forward, leading to lamellipodial extension. Actin monomers required for dendritic actin filament assembly are supplied from the cytoplasmic actin monomer pool, which is maintained by cofilin activity. (B) In cofilin-inactivated or -depleted cells, actin monomers in the cytoplasm are decreased. Stimulation of the cell activates the Arp2/3 complex, but dendritic actin filament assembly is suppressed because sufficient actin monomers are not available. When actin monomers are exogenously injected into the cytoplasm, they are efficiently incorporated into the cell periphery, and actin filament assembly occurs even under conditions of cofilin depletion. P-cofilin, Ser-3–phosphorylated cofilin.
Materials and methods
Materials
Jasp, LatA, and Alexa Fluor 546 were purchased from Invitrogen. EGF and NRG were purchased from Sigma-Aldrich and R&D Systems, respectively. Actin was purified from rabbit skeletal muscle. G-actin was labeled with Alexa Fluor 546 C5 maleimide, according to the manufacturer's protocols (Invitrogen).
Plasmid construction
Plasmids coding for Dp and SECFP were provided by A. Miyawaki (Riken, Wako, Japan). Expression plasmids for GFP (pEGFP-C1) and DsRed-monomer (mDsRed-C1) were purchased from CLONTECH Laboratories, Inc. Plasmids for YFP-actin and CFP-SSH-1L were constructed as described previously (Endo et al., 2003; Kaji et al., 2003). Expression plasmids for Dp-actin, LIMK1-SECFP, LIMK1-mDsRed, and cofilin-mDsRed were constructed by subcloning PCR-amplified Dp, SECFP, mDsRed, β-actin, LIMK1, or cofilin cDNA into the pEGFP-C1 vector. The plasmid for Myc-actin was constructed by inserting β-actin cDNA containing a Myc epitope tag into the pEGFP-C1 vector. Human cofilin siRNA plasmid (target sequence GGAGGATCTGGTGTTTATC), human ADF siRNA plasmid (target sequence GCAAATGGACCAGAAGATC), and control siRNA plasmid (target sequence TCTTCCCCCAAGAAAGATA, corresponding to the mutated human SSH-1L oligo) were constructed as described previously (Nishita et al., 2005).
Cell culture and transfection
Cells were cultured in DME supplemented with 10% (COS7 and MCF-7 cells) or 15% fetal calf serum (N1E-115 cells). Cells were transfected with expression plasmids using Lipofectamine 2000 (Invitrogen). Cells were used for various assays after being cultured for 18–24 h (COS7 and N1E-115 cells) or 4 d (MCF-7 cells) after transfection.
Photoactivation and time-lapse fluorescence microscopy
Photoactivation of Dp or Dp-actin and fluorescence imaging were performed using a laser-scanning confocal imaging system (LSM 510; Carl Zeiss MicroImaging, Inc.) equipped with a PL Apo 63× oil-immersion objective lens (NA 1.4) at 37°C in DME (without phenol red) supplemented with 10% fetal calf serum. The pinhole was set to 5.2 Airy units to obtain a greater depth of field. After photobleaching the whole cell by intense irradiation with a 488-nm laser, Dp or Dp-actin was photoactivated in a 5.8-μm2 region (400 pixels, with no obvious F-actin accumulation) in the cytoplasm by intense irradiation with a 458-nm laser for 1 s. Immediately after photoactivation, fluorescence images were automatically acquired every 0.4 s for 7.2 s by weak irradiation with the 488-nm laser. In pharmacological experiments, cells were pretreated with 1 μM Jasp for 20 min or 1 μM LatA for 2 min on the microscopic stage before photobleaching. For N1E-115 cells, Dp-actin was photoactivated for 1 s in a 14.25- × 2.85-μm rectangular region (4,500 pixels) in the cytoplasm, and fluorescence images were acquired every 2 s for 20 s to measure the fluorescence decay of Dp-actin in the photoactivated region. The whole cell was again photobleached, and Dp-actin was again photoactivated in the same region for 1.8 s, and fluorescence images were acquired every 5 s for 40 s at the maximum gain level of the photomultiplier to measure Dp-actin incorporation into the lamellipodium. The fluorescence images were analyzed by ImageJ (http://rsb.info.nih.gov/ij/). The rate of Dp-actin incorporation was measured as the distance Dp-actin moved from the tip of the lamellipodium toward the cytoplasm during 20 or 40 s after photoactivation. The time course of fluorescence decay in the photoactivated region was calculated as the mean fluorescence intensity in the region versus time. The mean fluorescence intensity in the same region just before photoactivation was subtracted as the background. The fluorescence decay includes a contribution of photobleaching over the time of imaging after photoactivation. The contribution of photobleaching was reduced by minimizing the excitation laser power. A photobleaching correction was performed by capturing images without photobleaching and photoactivation under the same laser-scanning conditions. The time course of photobleaching was well approximated by a single exponential decay function using Excel (Microsoft), and the fluorescence decay data were corrected for photobleaching by the exponential decay function.
F-actin sedimentation assay
COS7 cells were transfected with plasmids for Myc-actin and LIMK1-SECFP at a molar ratio of 1:5 or with plasmids for Myc-actin, LIMK1-SECFP, and cofilin-mDsRed in a molar ratio of 1:5:10. After 24 h, cells were lysed in lysis buffer (50 mM Hepes, pH 7.4, 100 mM NaCl, 1 mM MgCl2, 0.2 mM CaCl2, 1 mM dithiothreitol, 0.2 mM ATP, 1% NP-40, and 2 μM phalloidin). Cell lysates were centrifuged at 100,000 g for 30 min at 4°C. Equal amounts of pellet and supernatant were subjected to SDS-PAGE and analyzed by immunoblotting with an anti-Myc antibody (9E10; Roche).
Visualization of free barbed ends using a cell-permeabilization assay
COS7 cells expressing LIMK1(WT or D460A)-SECFP were stimulated with 100 ng/ml EGF, permeabilized for 1 min with 0.025% saponin and 0.45 μM Alexa546-actin monomers, and fixed with formaldehyde, according to the method described previously (Chan et al., 1998, 2000). The images were obtained using a fluorescence microscope (DMIRBE; Leica) equipped with a PL APO 63× oil-immersion objective lens (NA 1.3; Leica) and a cooled charge-coupled device camera (CoolSNAP HQ; Roper Scientific) driven by Q550FW Imaging Software (Leica). Incorporation of Alexa546-actin into the cell periphery was quantified by measuring the mean fluorescence intensity in a region 2 μm from the cell edge, using a customized macro in ImageJ (Fig. 7 C).
Visualization of Alexa546-actin monomer incorporation after microinjection
COS7 cells cotransfected with plasmids for YFP-actin and LIMK-SECFP at a molar ratio of 1:1 were cultured for 24 h and placed in DME (without phenol red) with 10% fetal bovine serum for 3 h before microinjection. MCF-7 cells cotransfected with plasmids for YFP-actin, cofilin siRNA, and ADF siRNA at a molar ratio of 1:2:2, or with plasmids for YFP-actin and control siRNA at a molar ratio of 1:4, were cultured for 4 d and placed in DME (without phenol red) without serum for 3 h before microinjection. 24 μM of Alexa546-actin monomers were injected into cells using a microinjection system (FemtoJet; Eppendorf) equipped with Femtotips 2. Immediately after the injection, cells were treated with 100 ng/ml EGF or 50 ng/ml NRG, and fluorescence images were acquired every 30 s for 9.5 min at 30°C. The images were obtained using the aforementioned fluorescence microscope equipped with a PL FLUOTAR 40× dry objective lens (NA 0.75; Leica). Incorporation of Alexa546-actin into the cell periphery was quantified by measuring the mean fluorescence intensity in a region 2 μm from the cell edge using ImageJ.
Online supplemental material
Fig. S1 shows the expression level and localization of Dp-actin in COS7 cells. Fig. S2 shows the level of P-cofilin in LIMK1-expressing COS7 cells. Fig. S3 shows the biochemical characterization of cofilin mutants. Fig. S4 shows the fluorescence images of Alexa546-actin microinjected into COS7 cells expressing LIMK1(WT or D460A) or MCF-7 cells transfected with cofilin/ADF siRNA or control siRNA, without cell stimulation. Videos 1 and 2 show the time-lapse fluorescence of Dp or Dp-actin in COS7 cells. Video 3 shows the time-lapse fluorescence of Dp-actin in COS7 cells coexpressing LIMK1(WT) with cofilin mutants or SSH-1. Video 4 shows the time-lapse fluorescence of Dp-actin in the MCF-7 cells transfected with cofilin/ADF or control siRNA. Videos 5 and 6 show the time-lapse fluorescence of Dp-actin photoactivated in the cytoplasm or in the lamellipodium in RacV12-expressing N1E-115 cells. Videos 7 and 8 show the time-lapse fluorescence of Alexa546-actin and YFP-actin in COS7 cells expressing LIMK1(WT) (Video 7) or LIMK1(D460A) (Video 8) with or without EGF stimulation. Videos 9 and 10 show the time-lapse fluorescence of Alexa546-actin and YFP-actin in MCF-7 cells transfected with cofilin/ADF siRNA (Video 9) or control siRNA (Video 10) with or without NRG stimulation. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200610005/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Atsushi Miyawaki for plasmids for Dp and SECFP and Sachiko Fujiwara for technical assistance.
This work was supported by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (18013007 and 17049002).
Abbreviations used in this paper: ADF, actin-depolymerizing factor; Dp, Dronpa; Jasp, jasplakinolide; LatA, latrunculin A; LIMK1, LIM-kinase 1; NRG, neuregulin; SECFP, super-enhanced CFP; WT, wild-type.
References
- Amato, P.A., and D.L. Taylor. 1986. Probing the mechanism of incorporation of fluorescently labeled actin into stress fibers. J. Cell Biol. 102:1074–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando, R., H. Mizuno, and A. Miyawaki. 2004. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 306:1370–1373. [DOI] [PubMed] [Google Scholar]
- Arber, S., F.A. Barbayannis, H. Hanser, C. Schneider, C.A. Stanyon, O. Bernard, and P. Caroni. 1998. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 393:805–809. [DOI] [PubMed] [Google Scholar]
- Bamburg, J.R., A. McGough, and S. Ono. 1999. Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol. 9:364–370. [DOI] [PubMed] [Google Scholar]
- Carlier, M.-F., V. Laurent, J. Santolini, R. Melki, D. Didry, G.X. Xia, Y. Hong, N.H. Chua, and D. Pantaloni. 1997. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136:1307–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, A.Y., S. Raft, M. Bailly, J.B. Wyckoff, J.E. Segall, and J.S. Condeelis. 1998. EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells. J. Cell Sci. 111:199–211. [DOI] [PubMed] [Google Scholar]
- Chan, A.Y., M. Bailly, N. Zebda, J.E. Segall, and J.S. Condeelis. 2000. Role of cofilin in epidermal growth factor–stimulated actin polymerization and lamellipod protrusion. J. Cell Biol. 148:531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., D. Godt, K. Gunsalus, I. Kiss, M. Goldberg, and F.A. Laski. 2001. Cofilin/ADF is required for cell motility during Drosophila ovary development and oogenesis. Nat. Cell Biol. 3:204–209. [DOI] [PubMed] [Google Scholar]
- Condeelis, J. 2001. How is actin polymerization nucleated in vivo? Trends Cell Biol. 11:288–293. [DOI] [PubMed] [Google Scholar]
- Cramer, L.P. 1999. Roles of actin-filament disassembly in lamellipodium protrusion in motile cells revealed using the drug jasplakinolide. Curr. Biol. 9:1095–1105. [DOI] [PubMed] [Google Scholar]
- DesMarais, V., M. Ghosh, R. Eddy, and J.S. Condeelis. 2005. Cofilin takes the lead. J. Cell Sci. 118:19–26. [DOI] [PubMed] [Google Scholar]
- Endo, M., K. Ohashi, Y. Sasaki, Y. Goshima, R. Niwa, T. Uemura, and K. Mizuno. 2003. Control of growth cone motility and morphology by LIM kinase and Slingshot via phosphorylation and dephosphorylation of cofilin. J. Neurosci. 23:2527–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, M., X. Song, G. Mouneimne, M. Sidani, D.S. Lawrence, and J.S. Condeelis. 2004. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 304:743–746. [DOI] [PubMed] [Google Scholar]
- Gunsalus, K.C., S. Bonaccorsi, E. Williams, F. Verni, M. Gatti, and M.L. Goldberg. 1995. Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J. Cell Biol. 131:1243–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen, P., E. Paunolla, M.K. Vartiainen, and P. Lappalainen. 2005. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol. Biol. Cell. 16:649–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji, N., K. Ohashi, M. Shuin, R. Niwa, T. Uemura, and K. Mizuno. 2003. Cell cycle-associated changes in Slingshot phosphatase activity and roles in cytokinesis in animal cells. J. Biol. Chem. 278:33450–33455. [DOI] [PubMed] [Google Scholar]
- Lappalainen, P., and D.G. Drubin. 1997. Cofilin promotes rapid actin filament turnover in vivo. Nature. 388:78–82. [DOI] [PubMed] [Google Scholar]
- Maciver, S.K. 1998. How ADF/cofilin depolymerizes actin filaments. Curr. Opin. Cell Biol. 10:140–144. [DOI] [PubMed] [Google Scholar]
- McGrath, J.L., Y. Tardy, C.F. Dewey Jr., J.J. Meister, and J.H. Hartwig. 1998. Simultaneous measurements of actin filament turnover, filament fraction, and monomer diffusion in endothelial cells. Biophys. J. 75:2070–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mies, B., K. Rottner, and J.V. Small. 1998. Multiple immunofluorescence microscopy of the cytoskeleton. In Cell Biology: A Laboratory Handbook, 2nd ed. J.E. Celis, editor. Academic Press, New York. 469–476.
- Moon, A., and D.G. Drubin. 1995. The ADF/cofilin proteins: stimulus-responsive modulators of actin dynamics. Mol. Biol. Cell. 6:1423–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, K., and I. Yahara. 1999. Two activities of cofilin, severing and accelerating directional depolymerization of actin filaments, are affected differentially by mutations around the actin-binding helix. EMBO J. 18:6752–6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, K., and I. Yahara. 2002. The actin-severing activity of cofilin is exerted by the interplay of three distinct sites on cofilin and essential for cell viability. Biochem. J. 365:147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishita, M., C. Tomizawa, M. Yamamoto, Y. Horita, K. Ohashi, and K. Mizuno. 2005. Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration. J. Cell Biol. 171:349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa, R., K. Nagata-Ohashi, M. Takeichi, K. Mizuno, and T. Uemura. 2002. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 108:233–246. [DOI] [PubMed] [Google Scholar]
- Pantaloni, D., C. Le Clainche, and M.-F. Carlier. 2001. Mechanism of actin based motility. Science. 292:1502–1506. [DOI] [PubMed] [Google Scholar]
- Pollard, T.D. 1986. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 103:2747–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T.D., and G.G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. [DOI] [PubMed] [Google Scholar]
- Revenu, C., R. Athman, S. Robine, and D. Louvard. 2004. The co-workers of actin filaments: from cell structures to signals. Nat. Rev. Mol. Cell Biol. 5:635–646. [DOI] [PubMed] [Google Scholar]
- Rosenblatt, J., B.J. Agnew, H. Abe, J.R. Bamburg, and T.J. Mitchison. 1997. Xenopus actin depolymerizing factor/cofilin (XAC) is responsible for the turnover of actin filaments in Listeria monocytogenes tails. J. Cell Biol. 136:1323–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa, T., and H. Miki. 2001. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci. 114:1801–1809. [DOI] [PubMed] [Google Scholar]
- Theriot, J.A., and T.J. Mitchison. 1991. Actin microfilament dynamics in locomoting cells. Nature. 352:126–131. [DOI] [PubMed] [Google Scholar]
- Theriot, J.A., and T.J. Mitchison. 1992. Comparison of actin and cell surface dynamics in motile fibroblasts. J. Cell Biol. 118:367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, M.D., A. Mallavarapu, J. Rosenblatt, and T.J. Mitchison. 1997. Actin dynamics in vivo. Curr. Opin. Cell Biol. 9:54–61. [DOI] [PubMed] [Google Scholar]
- Yang, N., O. Higuchi, K. Ohashi, K. Nagata, A. Wada, K. Kangawa, E. Nishida, and K. Mizuno. 1998. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 393:809–812. [DOI] [PubMed] [Google Scholar]
- Zebda, N., O. Bernard, M. Bailly, S. Welti, D.S. Lawrence, and J.S. Condeelis. 2000. Phosphorylation of ADF/cofilin abolishes EGF-induced actin nucleation at the leading edge and subsequent lamellipod extension. J. Cell Biol. 151:1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.