Abstract
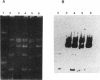
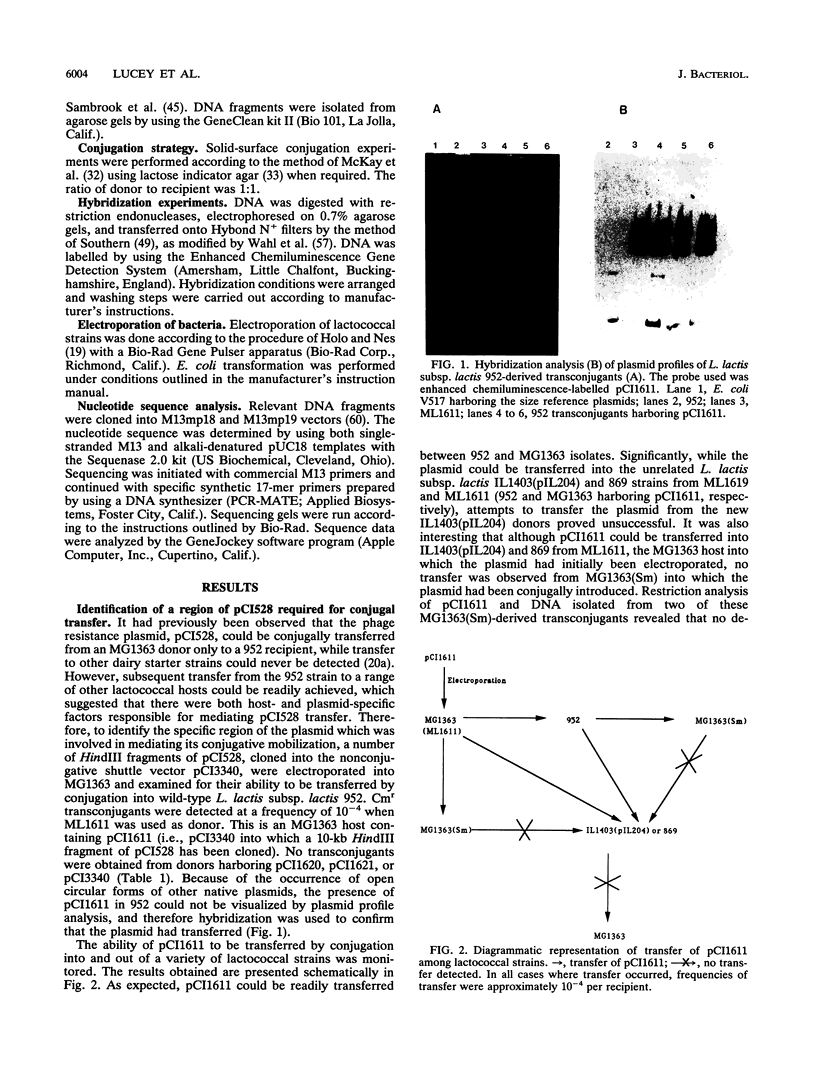
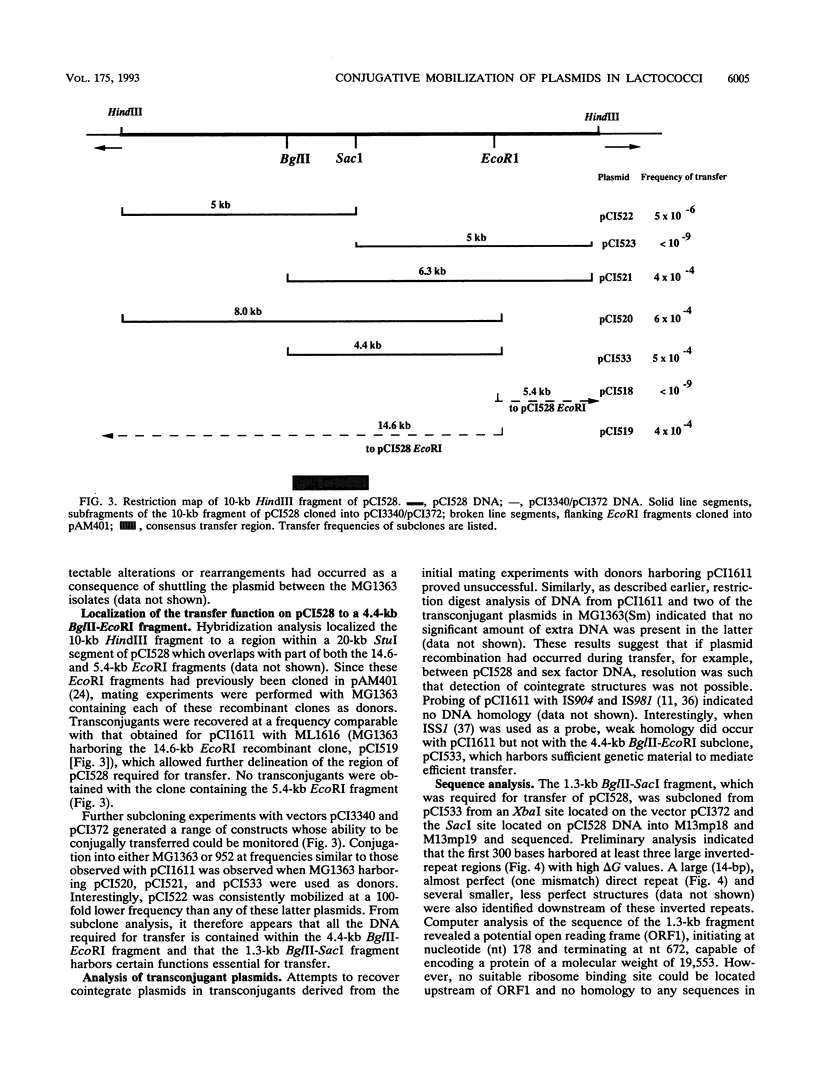
A 10-kb HindIII fragment of pCI528 cloned into the nonconjugative shuttle vector pCI3340 could be transferred by conjugative mobilization from Lactococcus lactis subsp. lactis MG1363, whereas other HindIII fragments of pCI528 or the vector alone were nonmobilizable. Subcloning of this 10-kb region identified a 4.4-kb BglII-EcoRI fragment which contained all the DNA essential for transfer. Sequence analysis of a 2-kb region within this 4.4 kb-segment revealed a region rich in inverted repeats and two potential overlapping open reading frames, one of which demonstrated homology to mobilization proteins of two nonconjugative staphylococcal plasmids.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Genetic and physical characterization of recombinant plasmids associated with cell aggregation and high-frequency conjugal transfer in Streptococcus lactis ML3. J Bacteriol. 1984 Jun;158(3):954–962. doi: 10.1128/jb.158.3.954-962.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastia D. Determination of restriction sites and the nucleotide sequence surrounding the relaxation site of ColE1. J Mol Biol. 1978 Oct 5;124(4):601–639. doi: 10.1016/0022-2836(78)90174-2. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire K. M., Hatfull G., Willetts N. Mobilization of the non-conjugative plasmid RSF1010: a genetic and DNA sequence analysis of the mobilization region. Mol Gen Genet. 1987 Jan;206(1):161–168. doi: 10.1007/BF00326552. [DOI] [PubMed] [Google Scholar]
- Derbyshire K. M., Willetts N. S. Mobilization of the non-conjugative plasmid RSF1010: a genetic analysis of its origin of transfer. Mol Gen Genet. 1987 Jan;206(1):154–160. doi: 10.1007/BF00326551. [DOI] [PubMed] [Google Scholar]
- Dodd H. M., Horn N., Gasson M. J. Analysis of the genetic determinant for production of the peptide antibiotic nisin. J Gen Microbiol. 1990 Mar;136(3):555–566. doi: 10.1099/00221287-136-3-555. [DOI] [PubMed] [Google Scholar]
- Fitzgerald G. F., Gasson M. J. In vivo gene transfer systems and transposons. Biochimie. 1988 Apr;70(4):489–502. doi: 10.1016/0300-9084(88)90085-5. [DOI] [PubMed] [Google Scholar]
- Gasson M. J. In vivo genetic systems in lactic acid bacteria. FEMS Microbiol Rev. 1990 Sep;7(1-2):43–60. doi: 10.1111/j.1574-6968.1990.tb04878.x. [DOI] [PubMed] [Google Scholar]
- Gasson M. J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983 Apr;154(1):1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennaro M. L., Kornblum J., Novick R. P. A site-specific recombination function in Staphylococcus aureus plasmids. J Bacteriol. 1987 Jun;169(6):2601–2610. doi: 10.1128/jb.169.6.2601-2610.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney D. G., Yakobson E. Location and nucleotide sequence of the transfer origin of the broad host range plasmid RK2. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3595–3598. doi: 10.1073/pnas.80.12.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F., Caplice E., McSweeney A., Fitzgerald G. F., Daly C. pAMbeta1-Associated Mobilization of Proteinase Plasmids from Lactococcus lactis subsp. lactis UC317 and L. lactis subsp. cremoris UC205. Appl Environ Microbiol. 1990 Jan;56(1):195–201. doi: 10.1128/aem.56.1.195-201.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F., Daly C., Fitzgerald G. F. Identification of the Minimal Replicon of Lactococcus lactis subsp. lactis UC317 Plasmid pCI305. Appl Environ Microbiol. 1990 Jan;56(1):202–209. doi: 10.1128/aem.56.1.202-209.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holo H., Nes I. F. High-Frequency Transformation, by Electroporation, of Lactococcus lactis subsp. cremoris Grown with Glycine in Osmotically Stabilized Media. Appl Environ Microbiol. 1989 Dec;55(12):3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippen-Ihler K. A., Minkley E. G., Jr The conjugation system of F, the fertility factor of Escherichia coli. Annu Rev Genet. 1986;20:593–624. doi: 10.1146/annurev.ge.20.120186.003113. [DOI] [PubMed] [Google Scholar]
- Kempler G. M., McKay L. L. Genetic Evidence for Plasmid-Linked Lactose Metabolism in Streptococcus lactis subsp. diacetylactis. Appl Environ Microbiol. 1979 May;37(5):1041–1043. doi: 10.1128/aem.37.5.1041-1043.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krah E. R., 3rd, Macrina F. L. Genetic analysis of the conjugal transfer determinants encoded by the streptococcal broad-host-range plasmid pIP501. J Bacteriol. 1989 Nov;171(11):6005–6012. doi: 10.1128/jb.171.11.6005-6012.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucey M., Daly C., Fitzgerald G. Identification and sequence analysis of the replication region of the phage resistance plasmid pCI528 from Lactococcus lactis subsp. cremoris UC503. FEMS Microbiol Lett. 1993 Jul 1;110(3):249–256. doi: 10.1111/j.1574-6968.1993.tb06330.x. [DOI] [PubMed] [Google Scholar]
- Ludwig W., Seewaldt E., Kilpper-Bälz R., Schleifer K. H., Magrum L., Woese C. R., Fox G. E., Stackebrandt E. The phylogenetic position of Streptococcus and Enterococcus. J Gen Microbiol. 1985 Mar;131(3):543–551. doi: 10.1099/00221287-131-3-543. [DOI] [PubMed] [Google Scholar]
- Macrina F. L., Kopecko D. J., Jones K. R., Ayers D. J., McCowen S. M. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid. 1978 Jun;1(3):417–420. doi: 10.1016/0147-619x(78)90056-2. [DOI] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Walsh P. M. Conjugal transfer of genetic information in group N streptococci. Appl Environ Microbiol. 1980 Jul;40(1):84–89. doi: 10.1128/aem.40.1.84-91.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Zottola E. A. Loss of lactose metabolism in lactic streptococci. Appl Microbiol. 1972 Jun;23(6):1090–1096. doi: 10.1128/am.23.6.1090-1096.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L. Functional properties of plasmids in lactic streptococci. Antonie Van Leeuwenhoek. 1983 Sep;49(3):259–274. doi: 10.1007/BF00399502. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Projan S. J., Rosenblum W., Edelman I. Staphylococcal plasmid cointegrates are formed by host- and phage-mediated general rec systems that act on short regions of homology. Mol Gen Genet. 1984;195(1-2):374–377. doi: 10.1007/BF00332777. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Staphylococcal plasmids and their replication. Annu Rev Microbiol. 1989;43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- Polzin K. M., McKay L. L. Identification, DNA sequence, and distribution of IS981, a new, high-copy-number insertion sequence in lactococci. Appl Environ Microbiol. 1991 Mar;57(3):734–743. doi: 10.1128/aem.57.3.734-743.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polzin K. M., Shimizu-Kadota M. Identification of a new insertion element, similar to gram-negative IS26, on the lactose plasmid of Streptococcus lactis ML3. J Bacteriol. 1987 Dec;169(12):5481–5488. doi: 10.1128/jb.169.12.5481-5488.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priebe S. D., Lacks S. A. Region of the streptococcal plasmid pMV158 required for conjugative mobilization. J Bacteriol. 1989 Sep;171(9):4778–4784. doi: 10.1128/jb.171.9.4778-4784.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Projan S. J., Archer G. L. Mobilization of the relaxable Staphylococcus aureus plasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J Bacteriol. 1989 Apr;171(4):1841–1845. doi: 10.1128/jb.171.4.1841-1845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Projan S. J., Kornblum J., Moghazeh S. L., Edelman I., Gennaro M. L., Novick R. P. Comparative sequence and functional analysis of pT181 and pC221, cognate plasmid replicons from Staphylococcus aureus. Mol Gen Genet. 1985;199(3):452–464. doi: 10.1007/BF00330758. [DOI] [PubMed] [Google Scholar]
- Projan S. J., Moghazeh S., Novick R. P. Nucleotide sequence of pS194, a streptomycin-resistance plasmid from Staphylococcus aureus. Nucleic Acids Res. 1988 Mar 25;16(5):2179–2187. doi: 10.1093/nar/16.5.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D. A., Klaenhammer T. R. Characterization of insertion sequence IS946, an Iso-ISS1 element, isolated from the conjugative lactococcal plasmid pTR2030. J Bacteriol. 1990 Aug;172(8):4151–4160. doi: 10.1128/jb.172.8.4151-4160.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanders M. E., Leonhard P. J., Sing W. D., Klaenhammer T. R. Conjugal strategy for construction of fast Acid-producing, bacteriophage-resistant lactic streptococci for use in dairy fermentations. Appl Environ Microbiol. 1986 Nov;52(5):1001–1007. doi: 10.1128/aem.52.5.1001-1007.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D., Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988 Apr;70(4):559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas W. D., Jr, Archer G. L. Identification and cloning of the conjugative transfer region of Staphylococcus aureus plasmid pGO1. J Bacteriol. 1989 Feb;171(2):684–691. doi: 10.1128/jb.171.2.684-691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts N., Wilkins B. Processing of plasmid DNA during bacterial conjugation. Microbiol Rev. 1984 Mar;48(1):24–41. doi: 10.1128/mr.48.1.24-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth R., An F. Y., Clewell D. B. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J Bacteriol. 1986 Mar;165(3):831–836. doi: 10.1128/jb.165.3.831-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- van der Lelie D., Wösten H. A., Bron S., Oskam L., Venema G. Conjugal mobilization of streptococcal plasmid pMV158 between strains of Lactococcus lactis subsp. lactis. J Bacteriol. 1990 Jan;172(1):47–52. doi: 10.1128/jb.172.1.47-52.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vossen J. M., van der Lelie D., Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl Environ Microbiol. 1987 Oct;53(10):2452–2457. doi: 10.1128/aem.53.10.2452-2457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]