Abstract
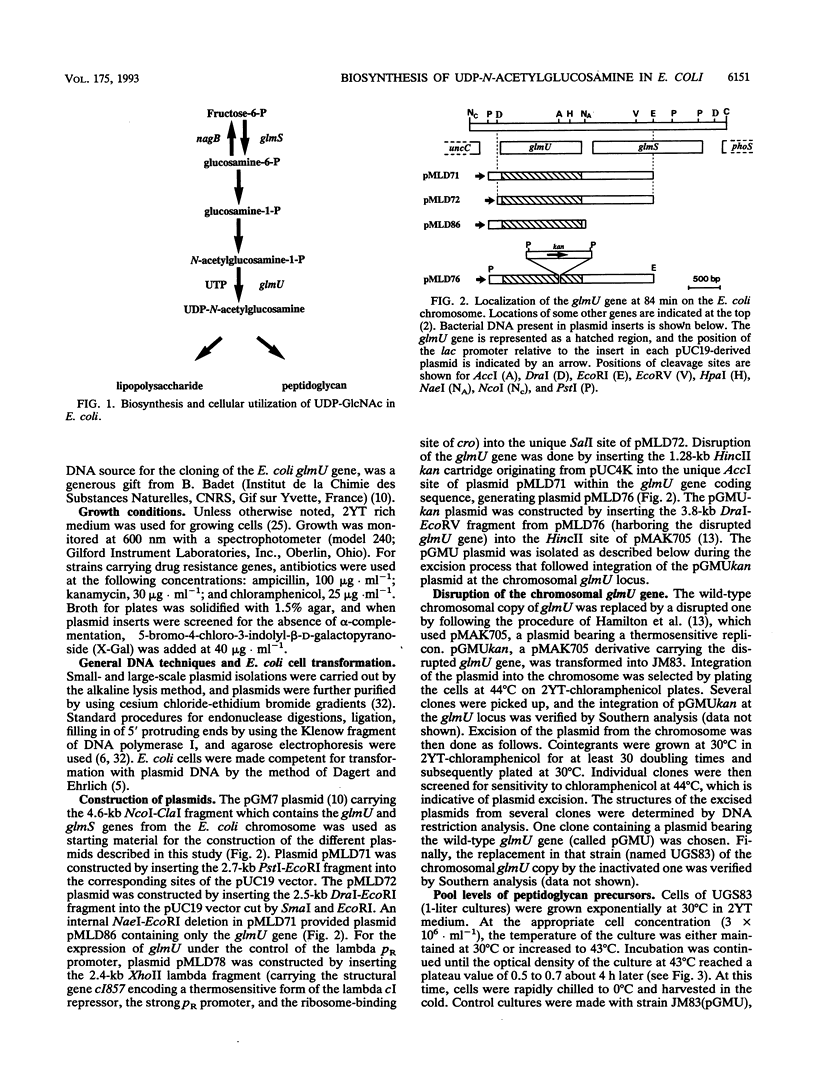
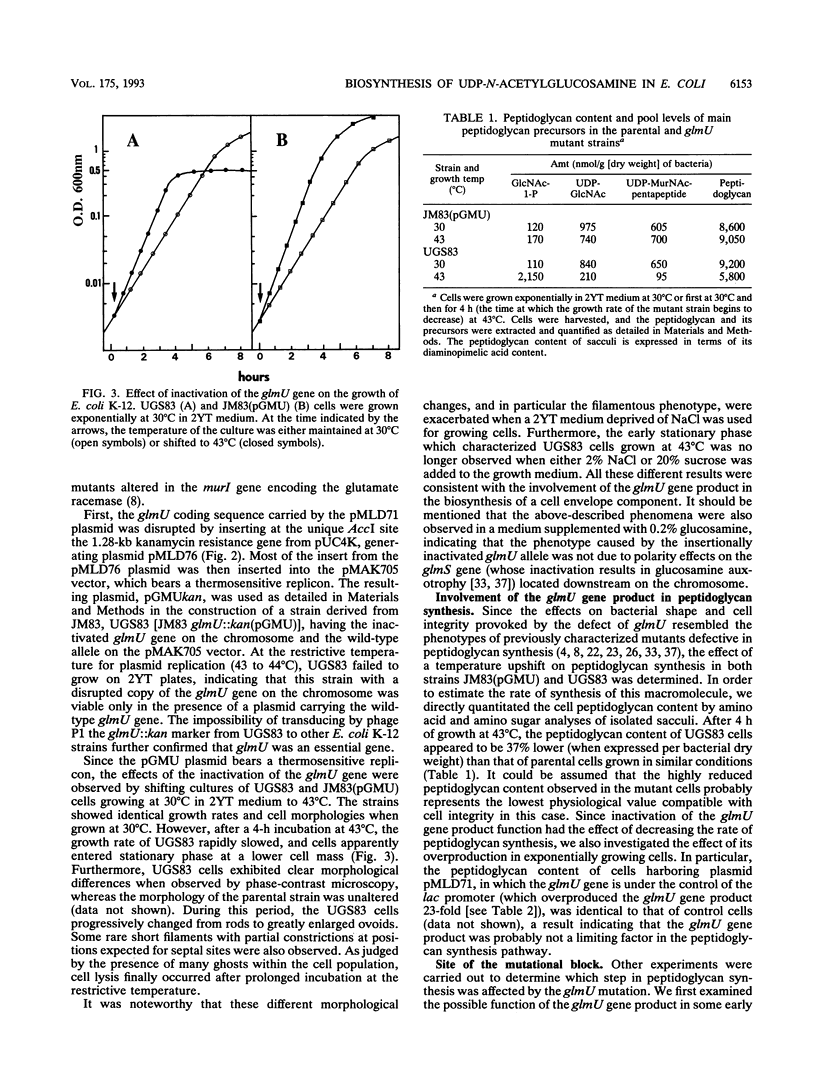
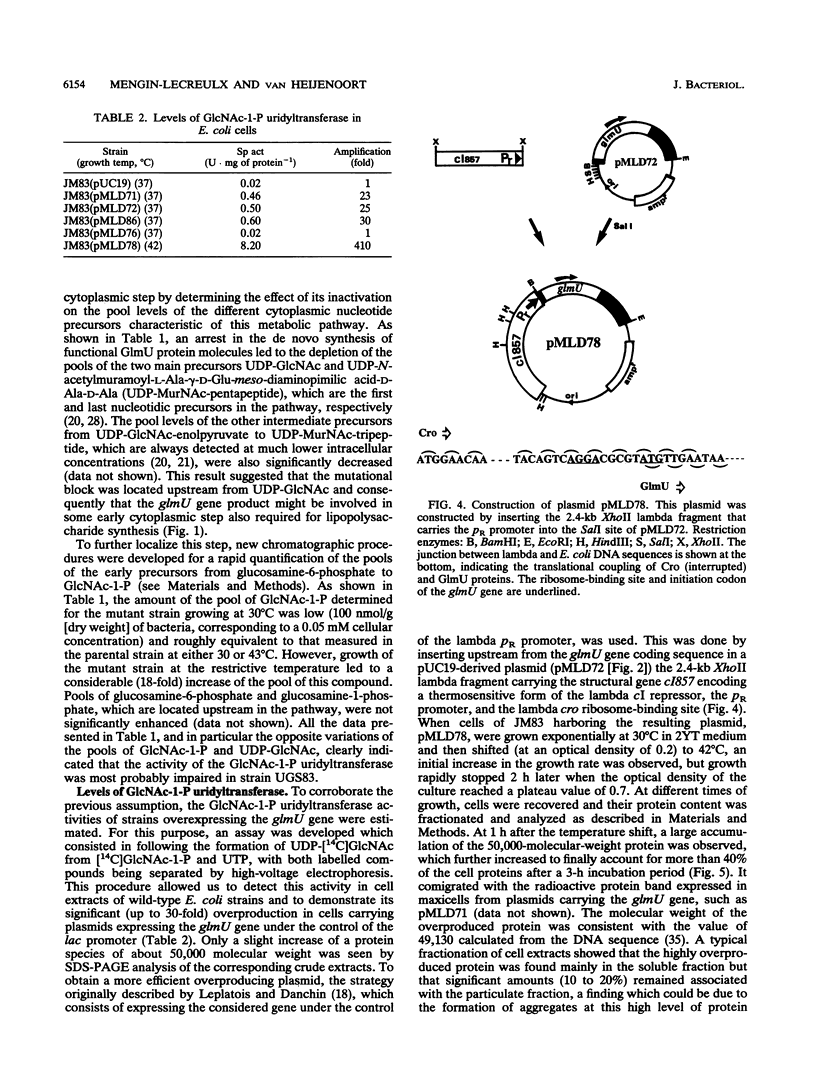
The physiological properties of the EcoURF-1 open reading frame, which precedes the glmS gene at 84 min on the Escherichia coli chromosome (J. E. Walker, N. J. Gay, M. Saraste, and A. N. Eberle, Biochem. J. 224:799-815, 1984), were investigated. A thermosensitive conditional mutant in which the synthesis of the gene product was impaired at 43 degrees C was constructed. The inactivation of the gene in exponentially growing cells rapidly inhibited peptidoglycan synthesis. As a result, various alterations of cell shape were observed, and cell lysis finally occurred when the peptidoglycan content was 37% lower than that of normally growing cells. Analysis of the pools of peptidoglycan precursors revealed a large accumulation of N-acetylglucosamine-1-phosphate and the concomitant depletion of the pools of the seven peptidoglycan nucleotide precursors located downstream in the pathway, a result indicating that the mutational block was in the step leading from N-acetylglucosamine-1-phosphate and UTP to the formation of UDP-N-acetylglucosamine. In vitro assays showed that the overexpression of this gene in E. coli cells, directed by appropriate plasmids, led to a high overproduction (from 25- to 410-fold) of N-acetylglucosamine-1-phosphate uridyltransferase activity. This allowed us to purify this enzyme to homogeneity in only two chromatographic steps. The gene for this enzyme, which is essential for peptidoglycan and lipopolysaccharide biosyntheses, was designated glmU.
Full text
PDF

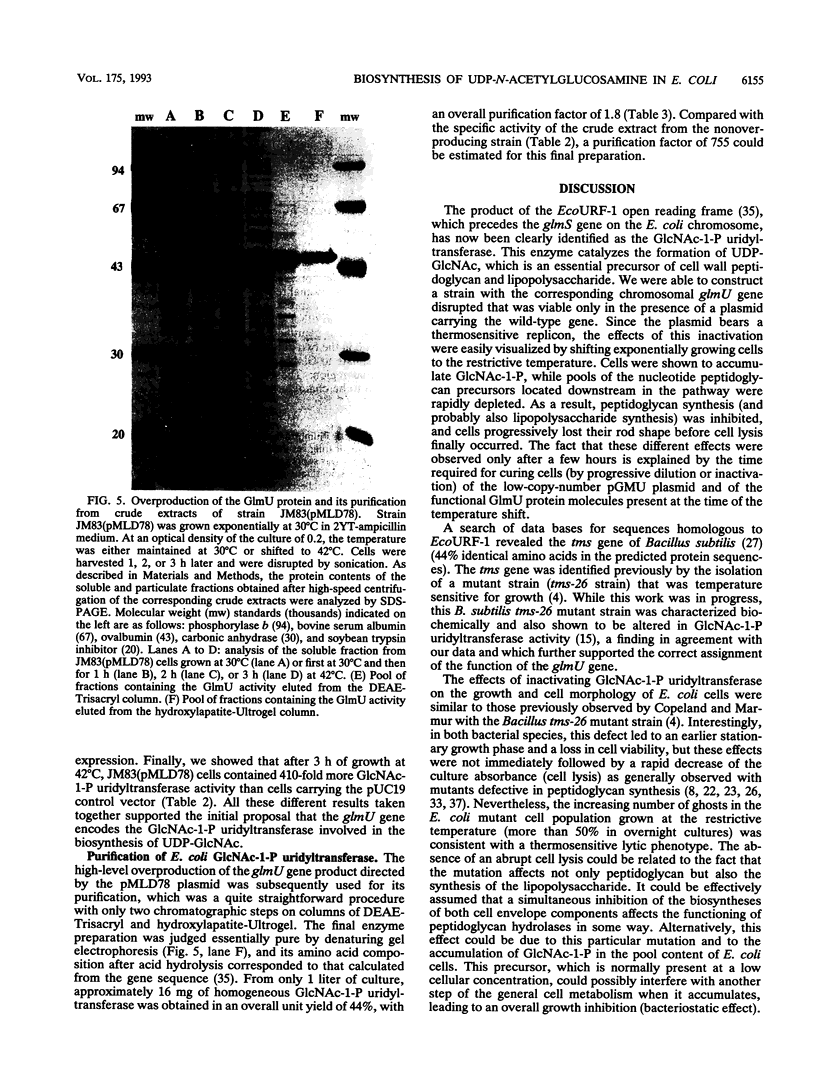


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson R. G., Douglas L. J., Hussey H., Baddiley J. The control of synthesis of bacterial cell walls. Interaction in the synthesis of nucleotide precursors. Biochem J. 1973 Dec;136(4):871–876. doi: 10.1042/bj1360871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. R., Ames B. N. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem. 1982 Aug 25;257(16):9759–9769. [PubMed] [Google Scholar]
- Copeland J. C., Marmur J. Identification of conserved genetic functions in Bacillus by use of temperature-sensitive mutants. Bacteriol Rev. 1968 Dec;32(4 Pt 1):302–312. [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Dobrogosz W. J. Effect of amino sugars on catabolite repression in Escherichia coli. J Bacteriol. 1968 Feb;95(2):578–584. doi: 10.1128/jb.95.2.578-584.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublet P., van Heijenoort J., Bohin J. P., Mengin-Lecreulx D. The murI gene of Escherichia coli is an essential gene that encodes a glutamate racemase activity. J Bacteriol. 1993 May;175(10):2970–2979. doi: 10.1128/jb.175.10.2970-2979.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublet P., van Heijenoort J., Mengin-Lecreulx D. Identification of the Escherichia coli murI gene, which is required for the biosynthesis of D-glutamic acid, a specific component of bacterial peptidoglycan. J Bacteriol. 1992 Sep;174(18):5772–5779. doi: 10.1128/jb.174.18.5772-5779.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutka-Malen S., Mazodier P., Badet B. Molecular cloning and overexpression of the glucosamine synthetase gene from Escherichia coli. Biochimie. 1988 Feb;70(2):287–290. doi: 10.1016/0300-9084(88)90073-9. [DOI] [PubMed] [Google Scholar]
- Freese E. B., Cole R. M., Klofat W., Freese E. Growth, sporulation, and enzyme defects of glucosamine mutants of Bacillus subtilis. J Bacteriol. 1970 Mar;101(3):1046–1062. doi: 10.1128/jb.101.3.1046-1062.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell E. W. Recycling of murein by Escherichia coli. J Bacteriol. 1985 Jul;163(1):305–310. doi: 10.1128/jb.163.1.305-310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton C. M., Aldea M., Washburn B. K., Babitzke P., Kushner S. R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989 Sep;171(9):4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hove-Jensen B. Identification of tms-26 as an allele of the gcaD gene, which encodes N-acetylglucosamine 1-phosphate uridyltransferase in Bacillus subtilis. J Bacteriol. 1992 Nov;174(21):6852–6856. doi: 10.1128/jb.174.21.6852-6856.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn H. M., Meier-Dieter U., Mayer H. ECA, the enterobacterial common antigen. FEMS Microbiol Rev. 1988 Sep;4(3):195–222. doi: 10.1111/j.1574-6968.1988.tb02743.x. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K., Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol. 1973 Nov 15;80(4):575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- Leplatois P., Danchin A. Vectors for high conditional expression of cloned genes. Biochimie. 1983 Jun;65(6):317–324. doi: 10.1016/s0300-9084(83)80153-9. [DOI] [PubMed] [Google Scholar]
- Mengin-Lecreulx D., Flouret B., van Heijenoort J. Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli. J Bacteriol. 1982 Sep;151(3):1109–1117. doi: 10.1128/jb.151.3.1109-1117.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengin-Lecreulx D., Flouret B., van Heijenoort J. Pool levels of UDP N-acetylglucosamine and UDP N-acetylglucosamine-enolpyruvate in Escherichia coli and correlation with peptidoglycan synthesis. J Bacteriol. 1983 Jun;154(3):1284–1290. doi: 10.1128/jb.154.3.1284-1290.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengin-Lecreulx D., Parquet C., Desviat L. R., Plá J., Flouret B., Ayala J. A., van Heijenoort J. Organization of the murE-murG region of Escherichia coli: identification of the murD gene encoding the D-glutamic-acid-adding enzyme. J Bacteriol. 1989 Nov;171(11):6126–6134. doi: 10.1128/jb.171.11.6126-6134.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengin-Lecreulx D., Texier L., Rousseau M., van Heijenoort J. The murG gene of Escherichia coli codes for the UDP-N-acetylglucosamine: N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase involved in the membrane steps of peptidoglycan synthesis. J Bacteriol. 1991 Aug;173(15):4625–4636. doi: 10.1128/jb.173.15.4625-4636.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengin-Lecreulx D., van Heijenoort J. Effect of growth conditions on peptidoglycan content and cytoplasmic steps of its biosynthesis in Escherichia coli. J Bacteriol. 1985 Jul;163(1):208–212. doi: 10.1128/jb.163.1.208-212.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa T., Matsuzawa H., Matsuhashi M., Sugino Y. Cell wall peptidoglycan mutants of Escherichia coli K-12: existence of two clusters of genes, mra and mrb, for cell wall peptidoglycan biosynthesis. J Bacteriol. 1972 Nov;112(2):950–958. doi: 10.1128/jb.112.2.950-958.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson D., Hove-Jensen B., Arnvig K. Primary structure of the tms and prs genes of Bacillus subtilis. Mol Gen Genet. 1989 Sep;218(3):565–571. doi: 10.1007/BF00332425. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. A., Cochet O., Souza J. M., Altamirano M. M., Calcagno M. L., Badet B. Coordinated regulation of amino sugar-synthesizing and -degrading enzymes in Escherichia coli K-12. J Bacteriol. 1993 Aug;175(16):4951–4956. doi: 10.1128/jb.175.16.4951-4956.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L., Pearlman-Kothencz M. Synthesis and assembly of bacterial membrane components. A lipopolysaccharide-phospholipid-protein complex excreted by living bacteria. J Mol Biol. 1969 Sep 28;44(3):477–492. doi: 10.1016/0022-2836(69)90374-x. [DOI] [PubMed] [Google Scholar]
- STROMINGER J. L., SMITH M. S. Uridine diphosphoacetylglucosamine pyrophosphorylase. J Biol Chem. 1959 Jul;234(7):1822–1827. [PubMed] [Google Scholar]
- Sarvas M. Mutant of Escherichia coli K-12 defective in D-glucosamine biosynthesis. J Bacteriol. 1971 Feb;105(2):467–471. doi: 10.1128/jb.105.2.467-471.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E., Gay N. J., Saraste M., Eberle A. N. DNA sequence around the Escherichia coli unc operon. Completion of the sequence of a 17 kilobase segment containing asnA, oriC, unc, glmS and phoS. Biochem J. 1984 Dec 15;224(3):799–815. doi: 10.1042/bj2240799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. J. Control of amino sugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem J. 1968 Feb;106(4):847–858. doi: 10.1042/bj1060847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. C., Wu T. C. Isolation and characterization of a glucosamine-requiring mutant of Escherichia coli K-12 defective in glucosamine-6-phosphate synthetase. J Bacteriol. 1971 Feb;105(2):455–466. doi: 10.1128/jb.105.2.455-466.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]