Abstract
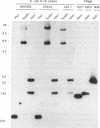
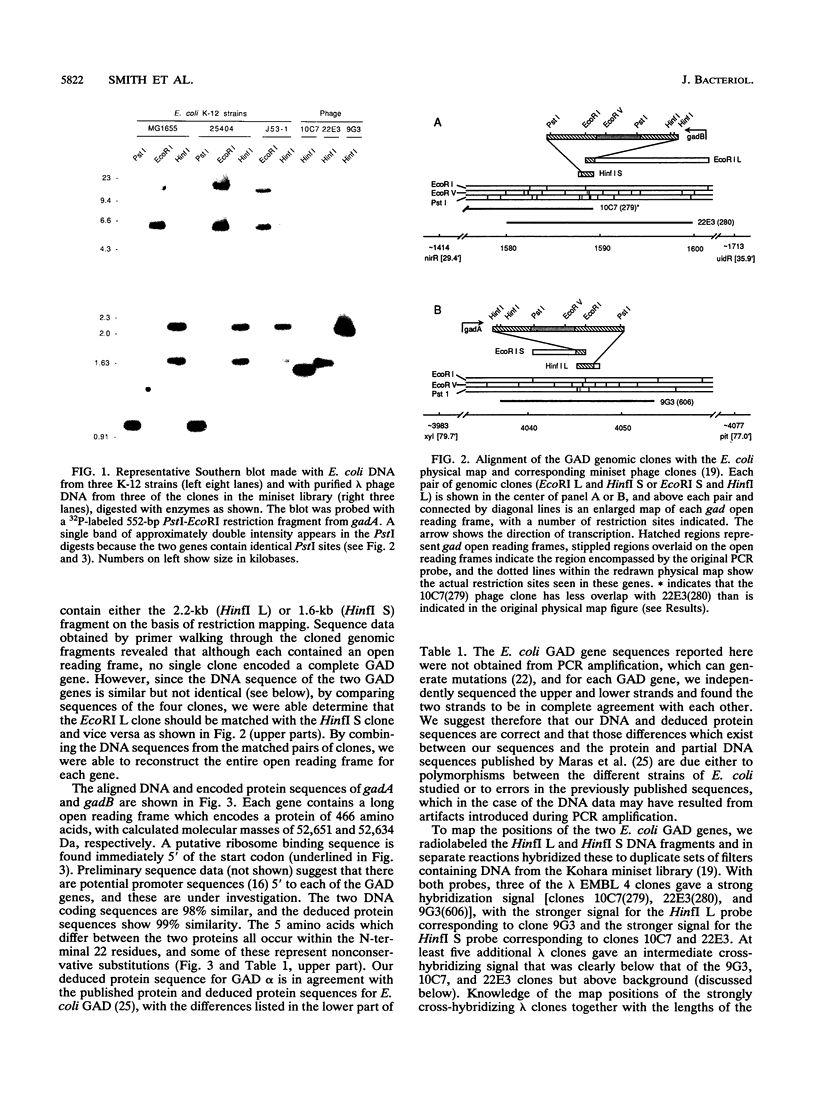
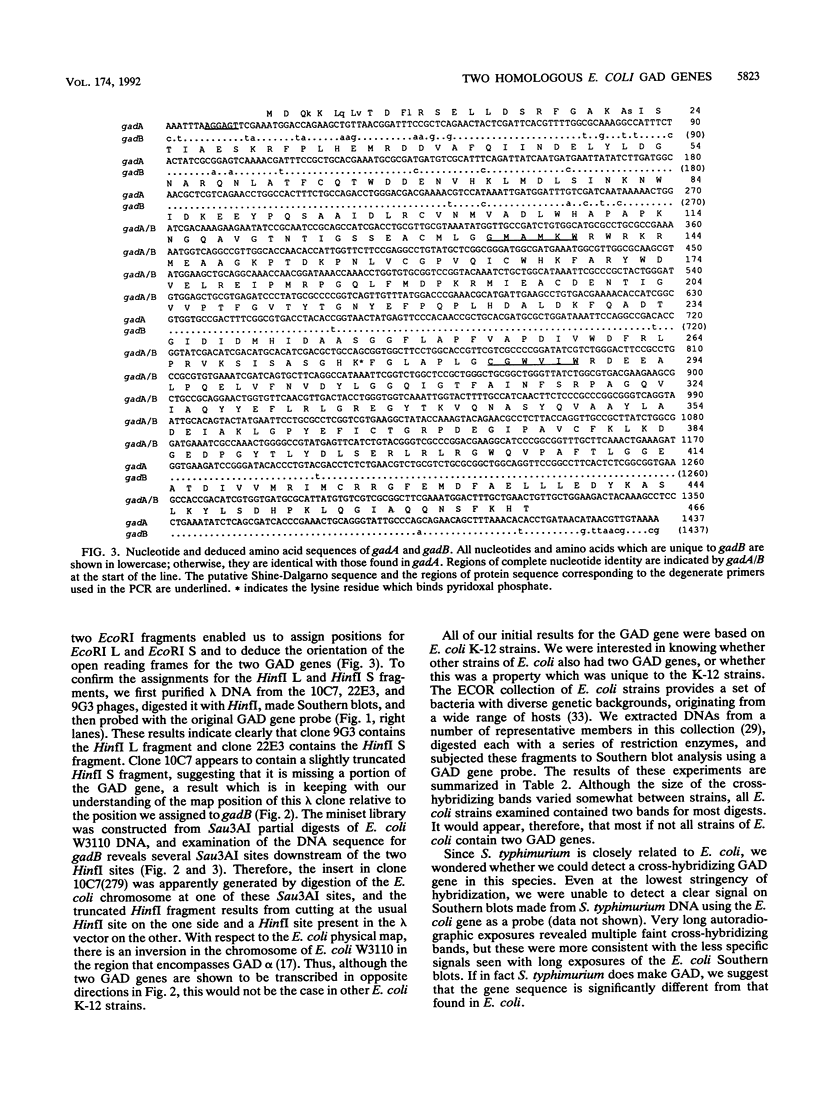
Degenerate oligonucleotides based on the published Escherichia coli glutamate decarboxylase (GAD) protein sequence were used in a polymerase chain reaction to generate a DNA probe for the E. coli GAD structural gene. Southern blots showed that there were two cross-hybridizing GAD genes, and both of these were cloned and sequenced. The two GAD structural genes, designated gadA and gadB, were found to be 98% similar at the nucleotide level. Each gene encoded a 466-residue polypeptide, named, respectively, GAD alpha and GAD beta, and these differed by only five amino acids. Both GAD alpha and GAD beta contain amino acid residues which are highly conserved among pyridoxal-dependent decarboxylases, but otherwise the protein sequences were not homologous to any other known proteins. By restriction mapping and hybridization to the Kohara miniset library, the two GAD genes were located on the E. coli chromosome. gadA maps at 4046 kb and gadB at 1588 kb. Neither of these positions is in agreement with the current map position for gadS as determined by genetic means. Analysis of Southern blots indicated that two GAD genes were present in all E. coli strains examined, including representatives from the ECOR collection. However, no significant cross-hybridizing gene was found in Salmonella species. Information about the DNA sequences and map positions of gadA and gadB should facilitate a genetic approach to elucidate the role of GAD in E. coli metabolism.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almazov V. P., Morozov Iu V., Savin F. A., Sukhareva B. S. Model' fermentativnogo dekarboksilirovaniia glutaminovoi kisloty. Mol Biol (Mosk) 1985 Mar-Apr;19(2):359–370. [PubMed] [Google Scholar]
- An G., Friesen J. D. The nucleotide sequence of tufB and four nearby tRNA structural genes of Escherichia coli. Gene. 1980 Dec;12(1-2):33–39. doi: 10.1016/0378-1119(80)90013-x. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencini D. A., Houghton J. E., Hoover T. A., Foltermann K. F., Wild J. R., O'Donovan G. A. The DNA sequence of argI from Escherichia coli K12. Nucleic Acids Res. 1983 Dec 10;11(23):8509–8518. doi: 10.1093/nar/11.23.8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu D. F., Erlander M. G., Hitz B. C., Tillakaratne N. J., Kaufman D. L., Wagner-McPherson C. B., Evans G. A., Tobin A. J. Two human glutamate decarboxylases, 65-kDa GAD and 67-kDa GAD, are each encoded by a single gene. Proc Natl Acad Sci U S A. 1992 Mar 15;89(6):2115–2119. doi: 10.1073/pnas.89.6.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS B. D., MINGIOLI E. S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950 Jul;60(1):17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca V., Marineau C., Brisson N. Molecular cloning and analysis of cDNA encoding a plant tryptophan decarboxylase: comparison with animal dopa decarboxylases. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2582–2586. doi: 10.1073/pnas.86.8.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988 Jul 11;16(13):6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott J. F., Albrecht G. R., Gilladoga A., Handunnetti S. M., Neequaye J., Lallinger G., Minjas J. N., Howard R. J. Genes for Plasmodium falciparum surface antigens cloned by expression in COS cells. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6363–6367. doi: 10.1073/pnas.87.16.6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlander M. G., Tobin A. J. The structural and functional heterogeneity of glutamic acid decarboxylase: a review. Neurochem Res. 1991 Mar;16(3):215–226. doi: 10.1007/BF00966084. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Furano A. V. The elongation factor Tu coded by the tufA gene of Escherichia coli K-12 is almost identical to that coded by the tufB gene. J Biol Chem. 1977 Mar 25;252(6):2154–2157. [PubMed] [Google Scholar]
- Guyer M. S., Reed R. R., Steitz J. A., Low K. B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. W., Harnish B. W. Inversions between ribosomal RNA genes of Escherichia coli. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7069–7072. doi: 10.1073/pnas.78.11.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson F. R. Prokaryotic and eukaryotic pyridoxal-dependent decarboxylases are homologous. J Mol Evol. 1990 Oct;31(4):325–329. doi: 10.1007/BF02101126. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Legrain C., Halleux P., Stalon V., Glansdorff N. The dual genetic control of ornithine carbamolytransferase in Escherichia coli. A case of bacterial hybrid enzymes. Eur J Biochem. 1972 May;27(1):93–102. doi: 10.1111/j.1432-1033.1972.tb01814.x. [DOI] [PubMed] [Google Scholar]
- Legrain C., Stalon V., Glansdorff N. Escherichia coli ornithine carbamolytransferase isoenzymes: evolutionary significance and the isolation of lambdaargF and lambdaargI transducing bacteriophages. J Bacteriol. 1976 Oct;128(1):35–38. doi: 10.1128/jb.128.1.35-38.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg K. S., Shoemaker D. D., Adams M. W., Short J. M., Sorge J. A., Mathur E. J. High-fidelity amplification using a thermostable DNA polymerase isolated from Pyrococcus furiosus. Gene. 1991 Dec 1;108(1):1–6. doi: 10.1016/0378-1119(91)90480-y. [DOI] [PubMed] [Google Scholar]
- Lupo M., Halpern Y. S. Gene controlling L-glutamic acid decarboxylase synthesis in Escherichia coli K-12. J Bacteriol. 1970 Aug;103(2):382–386. doi: 10.1128/jb.103.2.382-386.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus M., Halpern Y. S. Genetic analysis of glutamate transport and glutamate decarboxylase in Escherichia coli. J Bacteriol. 1967 Apr;93(4):1409–1415. doi: 10.1128/jb.93.4.1409-1415.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus M., Halpern Y. S. Genetic and physiological analysis of glutamate decarboxylase in Escherichia coli K-12. J Bacteriol. 1969 Mar;97(3):1509–1510. doi: 10.1128/jb.97.3.1509-1510.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. L. A comparison of the activities of the products of the two genes for elongation factor Tu. Mol Gen Genet. 1978 Feb 7;159(1):57–62. doi: 10.1007/BF00401748. [DOI] [PubMed] [Google Scholar]
- Mishin A. A., Sukhareva B. S. Glutamatdekarboksilaza iz Escherichia coli: kataliticheskaia rol' ostatka gistidina. Dokl Akad Nauk SSSR. 1986;290(5):1268–1271. [PubMed] [Google Scholar]
- Ochman H., Selander R. K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984 Feb;157(2):690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S., Blumenthal R. M., Reeh S., Russell L. B., Lemaux P., Laursen R. A., Nagarkatti S., Friesen J. D. A mutant of Escherichia coli with an altered elongation factor Tu. Proc Natl Acad Sci U S A. 1976 May;73(5):1698–1701. doi: 10.1073/pnas.73.5.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S., Reeh S. V. Analysis of the proteins synthesized in ultraviolet light-irradiated Escherichia coli following infection with the bacteriophages lambdadrifd 18 and lambdadfus-3. Mol Gen Genet. 1976 Mar 30;144(3):339–343. doi: 10.1007/BF00341733. [DOI] [PubMed] [Google Scholar]
- SHUKUYA R., SCHWERT G. W. Glutamic acid decarboxylase. I. Isolation procedures and properties of the enzyme. J Biol Chem. 1960 Jun;235:1649–1652. [PubMed] [Google Scholar]
- SHUKUYA R., SCHWERT G. W. Glutamic acid decarboxylase. II. The spectrum of the enzyme. J Biol Chem. 1960 Jun;235:1653–1657. [PubMed] [Google Scholar]
- SHUKUYA R., SCHWERT G. W. Glutamic acid decarboxylase. III. The inactivation of the enzyme at low temperatures. J Biol Chem. 1960 Jun;235:1658–1661. [PubMed] [Google Scholar]
- Schubert R., Esanu J. G., Schäfer V. Der Glutaminsäuredecarboxylase-Plättchen-Test. Ein Ansatz zur Vereinfachung und Beschleunigung des E. coli-Nachweises. Zentralbl Bakteriol Mikrobiol Hyg B. 1988 Dec;187(2):107–111. [PubMed] [Google Scholar]
- Seed B. An LFA-3 cDNA encodes a phospholipid-linked membrane protein homologous to its receptor CD2. 1987 Oct 29-Nov 4Nature. 329(6142):840–842. doi: 10.1038/329840a0. [DOI] [PubMed] [Google Scholar]
- Strausbauch P. H., Fischer E. H. Chemical and physical properties of Escherichia coli glutamate decarboxylase. Biochemistry. 1970 Jan 20;9(2):226–233. doi: 10.1021/bi00804a006. [DOI] [PubMed] [Google Scholar]
- Strausbauch P. H., Fischer E. H. Structure of the binding site of pyridoxal 5'-phosphate to Escherichia coli glutamate decarboxylase. Biochemistry. 1970 Jan 20;9(2):233–238. doi: 10.1021/bi00804a007. [DOI] [PubMed] [Google Scholar]
- Tikhonenko A. S., Sukhareva B. S., Braunstein A. E. Electron-microscopic investigation of Escherichia coli glutamate decarboxylase. Biochim Biophys Acta. 1968 Oct 8;167(2):476–479. doi: 10.1016/0005-2744(68)90232-5. [DOI] [PubMed] [Google Scholar]
- Van Vliet F., Cunin R., Jacobs A., Piette J., Gigot D., Lauwereys M., Piérard A., Glansdorff N. Evolutionary divergence of genes for ornithine and aspartate carbamoyl-transferases--complete sequence and mode of regulation of the Escherichia coli argF gene; comparison of argF with argI and pyrB. Nucleic Acids Res. 1984 Aug 10;12(15):6277–6289. doi: 10.1093/nar/12.15.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T., Sugisaki H., Takanami M., Kaziro Y. The nucleotide sequence of the cloned tufA gene of Escherichia coli. Gene. 1980 Dec;12(1-2):25–31. doi: 10.1016/0378-1119(80)90012-8. [DOI] [PubMed] [Google Scholar]
- Youngs T. L., Tunnicliff G. Substrate analogues and divalent cations as inhibitors of glutamate decarboxylase from Escherichia coli. Biochem Int. 1991 Mar;23(5):915–922. [PubMed] [Google Scholar]
- ZINDER N. D., LEDERBERG J. Genetic exchange in Salmonella. J Bacteriol. 1952 Nov;64(5):679–699. doi: 10.1128/jb.64.5.679-699.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]