Abstract
For many applications, antibodies need to be engineered toward maximum affinity. Strategies are in demand to especially optimize this process toward slower dissociation rates, which correlate with the (un)binding forces. Using single-molecule force spectroscopy, we have characterized three variants of a recombinant antibody single-chain Fv fragment. These variants were taken from different steps of an affinity maturation process. Therefore, they are closely related and differ from each other by a few mutations only. The dissociation rates determined with the atomic force microscope differ by one order of magnitude and agree well with the values obtained from surface plasmon resonance measurements. However, the effective potential width of the binding complexes, which was derived from the dynamic force spectroscopy measurements, was found to be the same for the different mutants. The large potential width of 0.9 nm indicates that both the binding pocket and the peptide deform significantly during the unbinding process.
INTRODUCTION
In recent years, recombinant antibodies have become increasingly important as therapeutic agents (1–4), for proteomics applications, and for diagnostic assays (5). In addition, they might prove useful as building blocks for the self-assembly of nanostructures. Antibodies with high affinities are needed in most of the cases, and the application sets the requirements.
Several different approaches have been developed for the in vitro affinity maturation of recombinant antibody fragments such as single-chain Fv (scFv) or Fab fragments (1–4,6–11). If a number of clones have been selected, they need to be characterized according to their affinity improvement. Often, the determination of the equilibrium dissociation constant KD yields sufficient information, and a ranking of mutants is possible. However, in some cases it is necessary to measure the kinetic rate constants of both the binding and unbinding processes as a way to describe both equilibrium and kinetic behavior in an application. This characterization is of particular importance if the affinity improvement needs to be correlated with the structure of the mutants and the position and type of the acquired mutation(s), e.g., during an affinity maturation process and its structural interpretation.
Several different methods exist for the determination of the equilibrium dissociation constant. It can be measured, e.g., with ELISA (enzyme-linked immunosorbent assay) (12), surface plasmon resonance (SPR) (13), fluorescence titration (14), and fluorescence-activated cell sorting (9). The quantitative measurement of the kinetic constants can be more difficult. Usually, SPR is used for this purpose. To obtain exact values, one has to take care of possible rebinding effects on the surface during the dissociation phase, which can slow down the apparent dissociation rate, koff, artificially. In addition, in the case of slow dissociation rates (koff < 10−5 s−1), an accurate determination of koff is difficult due to the small amount of analyte dissociating. The signal change can then approach the rate of drift of the SPR instrument (15).
Single-molecule force spectroscopy is an alternative method to obtain information about the unbinding process of receptor-ligand interactions. This measurement method has been used for a broad range of different biological systems, including antibody-antigen interactions (14,16–21). Force spectroscopy makes use of the fact that koff is increased if an external force is applied. Measuring the rupture forces of a receptor-ligand interaction for different loading rates (dynamic force spectroscopy) allows extrapolation to the dissociation rate at zero force, which represents the natural koff. Furthermore, force spectroscopy yields additional information about the width of the potential Δx (see below). This information might be useful for interpreting the influence of different mutations on unbinding kinetics. Mutations could lead to changes in the geometry of the binding site or to other conformational rearrangements of the molecule, resulting in an altered unbinding pathway that can be detected as a change in the width of the potential.
In this report we have analyzed three different variants of an scFv fragment with force spectroscopy using an atomic force microscope (AFM). These variants represent a series of clones obtained from different steps of an affinity maturation process by using ribosome display (11,22). All three variants bind the same peptide antigen, which is a random coil in solution. The crystal structure of a closely related variant complexed with the antigen has been determined (11). As the peptide forms an α-helical structure in complex with the antibody fragment, the peptide is considered to undergo a conformational change upon binding and unbinding, giving rise to more complex unbinding pathways compared to the unbinding of small and compact ligands. The study described here focuses on two aspects. First, we address some methodological aspects of the AFM measurements, mainly dealing with data evaluation. The obtained data were used to compare two different methods for analyzing AFM measurements. Both methods are based on the well-established Bell-Evans model (23–25) and allow determination of the koff and Δx values. In addition, the koff values obtained from force spectroscopy measurements were compared with the koff values determined by SPR. Second, as the variants only differ in a few amino acids it is possible to examine the influence of these mutations, acquired during the affinity maturation process, on koff and Δx.
MATERIALS AND METHODS
Cloning, expression, and purification of the antibody fragments
The three scFv variants (C11, C11L34, and 52SR4) were expressed with a C-terminal His tag followed by a cysteine to allow site-specific immobilization of the scFv fragments. The plasmids for periplasmic expression were based on the pAK series (26). The gene for coexpression of the periplasmic chaperone Skp was introduced (27). The original His tag was replaced by a tag of six histidines followed by two glycines and a cysteine. For the expression and purification of the scFv variants, the protocol of Hanes et al. (22) was slightly modified. Briefly, the Escherichia coli strain SB536 was transformed with the plasmids. Cells were grown at 25°C in SB medium (20 g L−1 tryptone, 10 g L−1 yeast extract, 5 g L−1 NaCl, 50 mM K2HPO4) containing 30 μg ml−1 chloramphenicol. Expression was induced with 1 mM isopropyl-β-D-thiogalactopyranoside at an OD600 between 1.0 and 1.5. The cells were harvested by centrifugation 3 h after induction. Cell disruption was achieved by French Press lysis. The scFv fragments were purified using two chromatography steps. After chromatography on a Ni2+-NTA column (Qiagen, Hilden, Germany) using standard protocols, the eluted fraction was directly loaded onto an affinity column with immobilized antigen. The fractions from the affinity column were dialyzed against coupling buffer (50 mM sodium phosphate, pH 7.2, 50 mM NaCl, 10 mM EDTA) and concentrated using Centricon YM-10 (Millipore, Eschborn, Germany). The actual concentration of the purified scFv fragments was determined by measuring the absorbance at 280 nm. The extinction coefficients of the different variants were calculated using the program Vector NTI (Invitrogen, Karlsruhe, Germany). The preparations of the purified proteins were adjusted to a concentration of 0.8 mg ml−1 and stored in aliquots at −80°C.
Preparation of slides and cantilevers for the AFM measurements
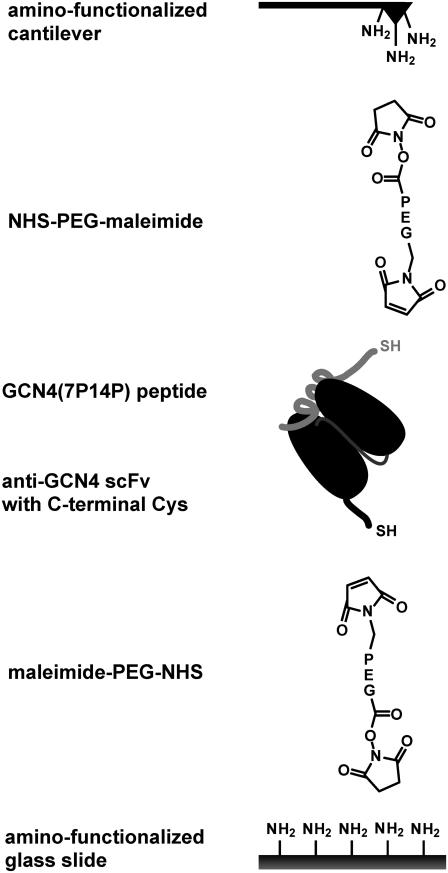
Poly(ethylene) glycol (PEG) was used as a spacer between the biomolecules and the surfaces. Due to its properties, PEG is an ideal spacer for force spectroscopy measurements (14,16,19,21,28–30). It provides protein-resistant surfaces (31), thereby reducing the number of nonspecific binding events. In addition, PEG shows a characteristic force-extension curve, allowing discrimination between specific and nonspecific interactions during data analysis. The scFv fragments possessing a C-terminal Cys were immobilized on an amino-functionalized slide using a heterobifunctional NHS-PEG-maleimide (molecular mass 5000 g/mol; Nektar, Huntsville, AL). The peptide GCN4(7P14P) (RMKQLEPKVEELLPKNYHLENEVARLKKL VGER), which has been used for the generation and affinity-maturation of antibodies, was used for the force spectroscopy measurements (11,22,32). A cysteine residue followed by three glycines was attached to the N-terminus during peptide synthesis (Jerini Peptide Technologies, Berlin, Germany). The Cys was used to couple the peptide to an amino-functionalized cantilever, again using the NHS-PEG-maleimide spacer (Fig. 1).
FIGURE 1.
Experimental setup. The antibody fragments having a C-terminal cysteine were covalently immobilized onto amino-functionalized glass slides using a heterobifunctional PEG spacer. The same coupling chemistry was used for immobilizing the peptide on the cantilever.
The cantilevers (Bio-lever, Olympus, Tokyo, Japan) were cleaned and functionalized as described (21). However, instead of epoxy-functionalized cantilevers, amino-modified surfaces were prepared using 3-aminopropyl-dimethylethoxysilane (ABCR, Karlsruhe, Germany). Commercially available amino-functionalized slides (Slide A, Nexterion, Mainz, Germany) were used. For the next steps, both surfaces (slide and cantilever) were treated in parallel as described (33). Briefly, they were incubated in borate buffer, pH 8.5, to increase the fraction of unprotonated amino groups for coupling to the NHS groups of the PEG. NHS-PEG-maleimide was dissolved at a concentration of 50 mM in borate buffer at pH 8.5 and incubated on the surfaces for 1 h. In parallel, the peptide and one of the scFv fragments were reduced using TCEP beads (Perbio Science, Bonn, Germany) to generate free thiols. After washing both surfaces with ultrapure water, a solution of the peptide (200 μM) was incubated on the cantilever and a solution of the scFv fragment (0.13 mg/ml) was incubated on the slide for 1 h. Finally, both surfaces were rinsed with phosphate-buffered saline (PBS) (10 mM Na phosphate, pH 7.4, 137 mM NaCl, 2.7 mM KCl) to remove noncovalently bound material and stored in PBS until use.
Force spectroscopy
All force measurements were performed with a MFP-1D AFM (Asylum Research, Santa Barbara, CA) at room temperature in PBS. Cantilever spring constants ranged from 6 to 8 pN/nm (B-Bio-Lever) and were measured as described previously (34,35). During one experiment, the approach and retract velocity were held constant, whereas the applied force was adjusted by changing the distance between the cantilever tip and the surface to obtain single binding events. To achieve good statistics, several hundreds of approach-retract cycles were carried out. To obtain measurements over a broad range of different loading rates, several experiments were performed, each at a different retract velocity ranging from 50 nm/s to 10 μm/s.
Data extraction
The obtained data was converted into force-extension curves. From these force-extension curves, the rupture force (the force at which the antibody-antigen complex ruptures), the rupture length, and the corresponding loading rate were determined using the program Igor Pro 5.0 (Wavemetrics, Lake Oswego, OR) and a custom-written set of procedures. The rupture force was determined as described previously (24,25). The loading rate was determined using the two-state freely jointed chain fit to the force-extension curve, according to previous studies (36).
Data analysis
To analyze the data set obtained from one experiment, which was recorded at a constant retract velocity, the rupture forces, rupture lengths, and loading rates were plotted in three histograms. The loading rates were plotted logarithmically. The histograms were analyzed with two methods based on the Bell-Evans model (23–25). The first method refers to the basis of dynamic force spectroscopy and has been applied broadly in the past to analyze force spectroscopy data. The histograms of the force and the loading rate (plotted logarithmically) for each data set, i.e., for each retract velocity, were fitted with a Gaussian distribution to determine the maxima. Finally, these obtained maxima of the Gaussian distributions were plotted in a force versus loading rate diagram. The maximum force (from the Gaussian distribution of the force histogram) represents the most probable force 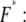
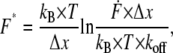 |
(1) |
where kB is the Boltzmann constant, T the temperature, Δx the potential width, koff the natural dissociation rate at zero force, and 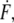 equal to
equal to 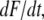 is the loading rate. From a linear fit of the force versus loading rate (pictured logarithmically) plot and Eq. 1, koff and Δx of the antibody-antigen complex can be determined.
is the loading rate. From a linear fit of the force versus loading rate (pictured logarithmically) plot and Eq. 1, koff and Δx of the antibody-antigen complex can be determined.
Whereas the first analysis method requires measurements at different retract velocities, the values for koff and Δx can be obtained from one data set measured at one retract velocity when using the second analysis method. The second method was introduced by Friedsam et al. (25) and takes into account a distribution of spacer lengths of the used PEG. The bond rupture probability density function p(F) was calculated according to Eq. 2 for every spacer length in the measured rupture-length histogram:
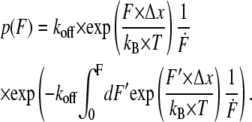 |
(2) |
These p(F) functions were weighted according to their occurrence in the rupture-length histogram and finally added up. This results in a semihypothetical rupture-force histogram based on the two input parameters koff and Δx, which were varied to find the best fit to the measured rupture-force histogram. Additionally, to account for the detection noise, the probability density function, p(F), was convolved with a Gaussian distribution. The standard deviation of the Gaussian distribution equals the typical noise value of the cantilever, which was used in the experiment (37).
The main difference between these two analysis methods is that the first method only uses the maxima of the force and loading-rate distributions for the fit procedure. As a consequence, the spacer length of the PEG spacer is averaged, which therefore results in an averaged loading rate. In contrast, the second method takes into account a certain spacer-length distribution. In addition, with the second method, the force histogram is fitted directly with the probability density function and therefore considers the shape of the histogram. To analyze the experimentally obtained data using the second method, it is extremely important to eliminate nonspecific interactions during data analysis, as they can shift or broaden the force histogram. This would lead to incorrect fit values for koff and Δx.
Proof of specificity
To prove the specificity of the force spectroscopy measurements, experiments were performed either without the antibody fragment or without the peptide. By measuring the antibody fragment, attached to the surface, against a cantilever tip passivated with PEG, >1000 force-extension curves were recorded. Thereby, <1% nonspecific interactions were detected. The measurements without the peptide led to similar results.
SPR measurements
For the measurement of koff (25°C) of the scFv fragments with a Biacore 3000 instrument (BIAcore, Freiburg, Germany), two different assay formats were used: 1), an antigen-immobilized assay for clone C11; and 2), an antibody-immobilized assay for the clones C11L34 and 52SR4. For both formats, a CM5 sensor chip (BIAcore) was modified via amine coupling according to the manufacturer's protocol. For assay format 1, biotinylated peptide GCN4(7P14P) (22) was bound to the amine-coupled neutravidin (Perbio Science, Lausanne, Switzerland) surface to a final signal intensity of 20 RU. Clone C11 was diluted in HBST buffer (20 mM Hepes, pH 7.2, 150 mM NaCl, 0.005% Tween 20) to final concentrations of 20–100 nM and injected on the chip. For format 2, either clone C11L34 or clone 52SR4 was amine-coupled to the surface to a final signal intensity of 100–300 RU. A series of GCN4(7P14P) peptide solutions in HBST buffer in the range 50–0.023 nM, using threefold dilutions, was injected on the chip. After binding, dissociation was followed at a flow rate of 100 μl/min and 50 μl/min for assay formats 1 and 2, respectively. The dissociation phase was fitted globally, using the single-exponential fit function of the program SigmaPlot or Clamp, alternatively.
RESULTS
The antibody fragments used in our study are closely related and have been described previously (11,22). They all bind the same peptide antigen, which has been derived from the transcription factor GCN4. For a better understanding, we briefly summarize the positions (Table 1) and the influence of the mutations. The antibody fragments differ in a few amino acids only. Starting from clone C11, clone C11L34 has one mutation. Compared to clone C11L34, clone 52SR4 has four additional mutations. The mutated amino acids do not interact directly with the antigen, although three of them are located in the complementarity determining regions (CDRs). Mutation L42 (N→S; AHo numbering scheme (38)), which has already been introduced into clone C11L34, may reduce the flexibility of CDR L1 and may allow a more favorable domain orientation. Mutation L107 (A→V) of clone 52SR4 lies in close proximity to mutation L42 and therefore might contribute to this effect. Therefore, these two mutations are thought to influence the loop position and/or geometry and the relative domain orientation and thereby optimize the binding geometry. Most likely, mutation L135 (N→D in clone 52SR4) has a different effect. The exchange of asparagine to aspartic acid introduces a negative charge. This charged residue might be able to establish an electrostatic interaction with the peptide, as the peptide has a positive charge at the corresponding position (K15 in the original peptide). From the structure, it appears that the mutations L13 (T→S in clone 52SR4) and H30 (S→L in clone 52SR4) only have a small contribution to the affinity.
TABLE 1.
Summary of the results obtained for the three different clones
| AFM analysis method 1
|
AFM analysis method 2*
|
SPR
|
||||
|---|---|---|---|---|---|---|
| Clone | Mutations | koff (s−1) | Δx (nm) | koff (s−1) | Δx (nm) | koff (s−1) |
| C11 | None | (3.9 ± 5.7) × 10−3 | 0.88 ± 0.12 | (1.0 ± 0.3) × 10−3 | 0.90 ± 0.02 | 2.9 × 10−3 |
| C11L34 | L42 (N→S) | (4.9 ± 7.2) × 10−4 | 0.90 ± 0.10 | (2.0 ± 1.0) × 10−4 | 1.00 ± 0.10 | 3.0 × 10−4 |
| 52SR4 | L13 (T→S) | (8.2 ± 7.9) × 10−4 | 0.92 ± 0.07 | (5.0 ± 2.0) × 10−4 | 0.86 ± 0.04 | 1.6 × 10−4 |
| L42 (N→S) | ||||||
| L107 (A→V) | ||||||
| L135 (N→D) | ||||||
| H30 (S→L) | ||||||
koff and Δx for the analysis method 2 have been determined for dF/dt between 80 and 90 pN s−1.
To analyze the interaction of the variants with their peptide antigen, force spectroscopy measurements were performed using an atomic force microscope (AFM). To be able to compare the two different analysis methods for the AFM data (see Materials and Methods) and the data from the surface plasmon resonance (SPR) measurements, it was essential to minimize nonspecific interactions and to ensure that only specific and single antibody-peptide interactions were analyzed. As an effective approach to discriminate nonspecific interactions, we chose to attach both the antibody fragment and the peptide via PEG, which is known to provide protein-resistant surfaces. The antibody fragment was coupled to a surface containing covalently attached PEG and the peptide was immobilized onto the cantilever tip in the same way (Fig. 1). An additional advantage of this approach is that PEG acts as an elastic spacer with a known length. When the PEG spacers are stretched, the elastic properties of this molecule lead to a characteristic extension curve, which can be fitted with the two-state freely jointed chain (FJC) fit with the values from the literature (36). Specific interactions were thus selected by considering only those extension curves that show the appropriate length and the characteristic shape of the PEG spacers.
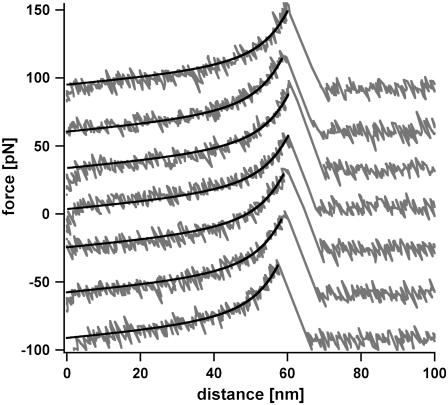
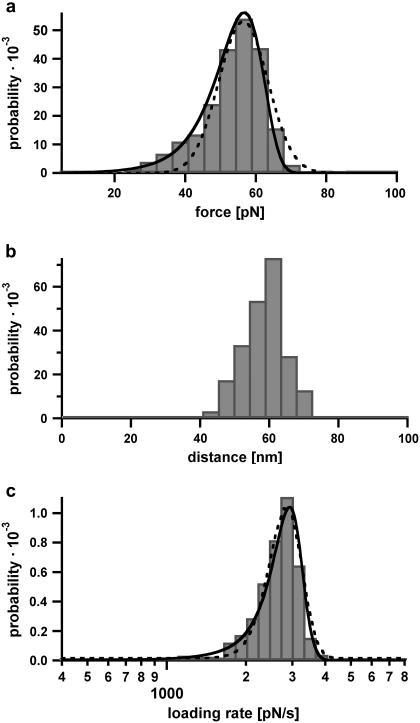
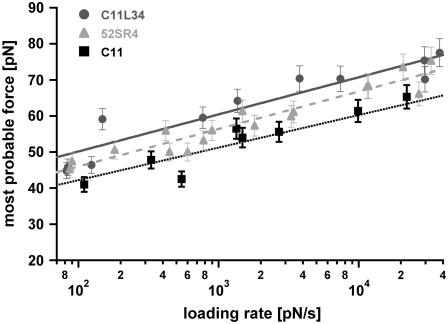
In all experiments, the surface was approached with the tip of the cantilever, allowing the antibody-peptide complex to bind. Subsequently, the cantilever was retracted and the antibody-peptide complex was loaded with an increasing force until the complex finally ruptured and the cantilever relaxed back into its equilibrium position. The force applied to this complex was recorded as a function of the distance between the cantilever tip and the surface. Fig. 2 shows a series of typical force-extension curves representing the interaction between clone C11 and the peptide. To obtain good statistics, several hundred force-extension curves were recorded for all three variants. From these curves, the rupture force, rupture length, and corresponding loading rate were determined. Fig. 3 a shows the rupture-force, Fig. 3 b the rupture-length, and Fig. 3 c the loading-rate distributions for the interaction of clone C11 with the peptide, measured at a retract velocity of 1000 nm/s. The rupture-force histogram in Fig. 3 a was fitted with a Gaussian distribution (dotted curve) and exhibits a most probable force of 55.6 pN. The Gaussian distribution of the histogram of the loading rates (plotted logarithmically) (Fig. 3 c) shows a maximum at 2697 pN s−1. The maxima of the force and the loading-rate distributions were determined for a large range of loading rates, and in the following step were plotted in a force versus loading rate (pictured logarithmically) diagram (first analysis method). The determination of koff and Δx from a linear fit to these data points using Eq. 1 is described in Materials and Methods. The measurements of clone C11 resulted in a koff of (3.9 ± 5.7) × 10−3 s−1 and a Δx of (0.88 ± 0.12) nm. For a complete analysis of the experimental results, all data sets for the three variants were examined by the first analysis method, using Eq. 1 (Fig. 4). The obtained values for koff and Δx for all three variants are listed in Table 1.
FIGURE 2.
Example of seven typical force-extension curves. The force-extension curves show the rupture event of the scFv C11 peptide complex, experimentally recorded at a retract velocity of 1000 nm/s. The elastic behavior of the spacer PEG can be described using the two-state FJC fit (solid curve) with the values from the literature (36). The values for the rupture force, rupture length, and corresponding loading rate were obtained from these force-extension curves.
FIGURE 3.
Example of the obtained rupture-force, rupture-length, and loading-rate distributions. (a) Rupture-force histogram of the scFv C11 peptide complex. The rupture-force histogram contains 859 rupture events and was fitted with a Gaussian curve (dotted curve). Additionally, the obtained rupture-force distribution was compared with the calculated probability density function p(F) (solid curve) with Δx = (0.9 ± 0.03) nm and koff = (2.0 ± 3.0) × 10−3 s−1, as described in Materials and Methods. Within the analysis error, the values for Δx and koff are identical for both analysis methods (experimental data obtained from the first analysis method are shown in Fig. 4). (b) Rupture-length histogram of the scFv C11 peptide complex. (c) Histogram of the loading rates of the scFv C11 peptide complex, plotted logarithmically. This histogram was fitted with a Gaussian curve (dotted curve) and additionally compared with the calculated probability density function 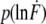 (solid curve).
(solid curve).
FIGURE 4.
Diagram showing the most probable rupture force plotted against the corresponding loading rate (pictured logarithmically) for all three scFv-peptide complexes. The data points were gained from the Gaussian fits of the rupture-force histogram and the histogram of loading rates, plotted logarithmically. The black data points (▪) correspond to the scFv C11 peptide complex. These data points were fitted to a straight line (black dotted curve). From this linear fit, Δx = (0.88 ± 0.12) nm and koff = (3.9 ± 5.7) × 10−3 s−1 were obtained. The dark gray data set (•) was measured for the forced dissociation of the scFv C11L34 peptide complex. From the linear fit (dark gray), Δx = (0.90 ± 0.10) nm and koff = (4.9 ± 7.2) × 10−4 s−1 were obtained. Finally, the scFv 52SR4 peptide complex, plotted with light gray data points (▴) and the linear fit (light gray dashed curve) gave Δx = (0.92 ± 0.07) nm and koff = (8.2 ± 7.9) × 10−4 s−1.
Additionally, the measured rupture-force distributions for all three variants were analyzed using the second analysis method based on the probability density function p(F) (Eq. 2). The respective fit for the presented data set of clone C11 is shown in Fig. 3 (solid curve). For this clone, the second analysis method resulted in a Δx of (0.9 ± 0.03) nm and a koff of (2.0 ± 3.0) × 10−3 s−1. To be able to compare the obtained values for koff and Δx for all the variants, we performed an analysis for similar loading rates in the lower range (dF/dt between 80 and 90 pN s−1). The values for all three variants are also listed in Table 1. A comparison of the three different variants shows that the Δx and koff values, determined using the first and second analysis methods, are identical within the analysis error (first analysis method). The values for koff obtained from the SPR measurements are also summarized in Table 1. Within experimental error, both methods (AFM and SPR) exhibit consistent values for koff for all three variants.
A statistical analysis of the results obtained from the first analysis method using the AFM was performed using Student's t-tests. The potential widths Δx of the three variants have been determined from the corresponding slopes of the linear fits (see Materials and Methods), which are identical with a probability of 96%. Thus, none of the mutations changes the potential width, Δx, significantly. In contrast, the dissociation rates, koff, were determined from the interpolation to zero force. Since the slopes are identical, the significance in the difference in koff can be obtained from the intersections of the linear fits with the ordinate.
These intersections for clones C11 and C11L34 are different with a probability of 66%. This rather low value results from the lack of data points at very low loading rates that are not accessible with the AFM. Since the AFM data from the first and second analysis methods are in full accordance with the SPR results, a comparison of the koff values clearly reveals that clone C11L34 has a slower dissociation rate than clone C11 (5–8-fold). This is the consequence of one single-point mutation at the end of CDR L1 of the VL domain. Clones C11L34 and 52SR4 differ in four amino acids. However, these mutations do not show any significant influence on koff.
DISCUSSION
Receptor-ligand interactions often display a marked deviation from the linear relation between the unbinding force and the logarithm of the force-loading rate, predicted by the Bell-Evans model. Biotin-Avidin (39) and digoxigenin-antibody interactions (21) are prominent examples, where two barriers in series are suggested due to the marked nonlinearity. Alternatively, other models based on the Kramers theory (40,41) are discussed. For the antibody-peptide system investigated here, we found that the simplest level of analysis, which is based on a mechanothermally activated transition in a two-level system, already provided a satisfactory description of the unbinding process: for each mutant, the plot in Fig. 4 revealed a linear relation, showing virtually indistinguishable slopes and a separation of the intersections at zero force.
However, this simple approach has two severe drawbacks: it requires the measurement of a series of data points for a wide spectrum of force-loading rates and it assumes a monodisperse spacer length. This has only a limited validity if polymeric spacers, such as PEG, are used (as in this study). We therefore also employed a second method, which analyzes the shape of the rupture-force histogram and requires only one loading rate. We found that this method provided more accurate results (see Table 1), as it considers the distribution of the spacer lengths (25). For all three variants used in this study, the koff and Δx values agreed well for both analysis methods for slow loading rates. In general, the second analysis method may also be applied to faster loading rates (data not shown). However, for faster loading rates, the experimental noise increases and, therefore, additional correction factors would have to be included for analysis of the data (37). In addition, one should keep in mind that the Bell-Evans model assumes a constant Δx over the entire range of loading rates, which is probably not the case for most receptor-ligand systems. Therefore, using the second analysis method for slow loading rates ensures that the potential width is not changed, as the system is still close to equilibrium.
Last but not least, this second method also reduces the experimental effort significantly, as the potential width and the dissociation rate are obtained from one experiment at one loading rate only, thereby making the method competitive with SPR measurements. This is particularly true in view of the rebinding problems that may hamper SPR analysis for very low koff values. A decreasing koff results in an increasing binding force, thus making this regime favorable for single-molecule force spectroscopy. The main advantage, however, lies in the accessibility of an otherwise not measurable parameter of receptor-ligand interactions: their potential width, Δx.
The most remarkable finding of our study is that the different variants have a more or less identical potential width, which is calculated from the indistinguishable slopes of the linear fits of the different clones. Although, in the Bell-Evans model, the potential width is only a rough measure of the steepness of the binding potential or, in other words, a measure of how far the binding complex can be stretched and deformed until it finally ruptures, this finding leads to the conclusion that neither the geometry of the binding site nor the unbinding pathways were significantly affected by the mutations introduced during the affinity-maturation process. Compared to the potential width of the well characterized antifluorescein scFv fragments (14,42,43), the value for Δx obtained for the system analyzed here is significantly higher. In addition, for the fluorescein system a correlation between Δx and koff was observed, which is not the case for the variants investigated here. When comparing both systems, one has to keep in mind that there are three significant differences. First, in the fluorescein system the mutations are mainly located in the binding sites of the scFv fragments. Second, in the fluorescein system, affinity-matured scFv fragments were taken as a starting point and systematic mutations were made to reduce the number of contacts in the binding site. In the study presented here, a starting clone was improved sequentially by directed evolution. And finally, fluorescein is a very rigid antigen, which cannot adopt multiple conformations. In contrast, the peptide antigen of the system described here is a random coil in solution and has an α-helical structure in complex with the antibody fragment.
Considering these aspects, the observed differences for Δx can be rationalized as follows: during the forced unbinding, one or both binding partners can be deformed in the direction of the applied force. If a certain point is reached, the deformation is so large that the complex dissociates. In the case of the antifluorescein scFvs, the number and quality of the contacts in the binding site differs among the analyzed scFvs. As fluorescein is a rigid antigen, only the scFv fragment itself can respond to the applied force. Therefore, by stretching the scFv fragment with an externally applied force, it is deformed in the direction of the force. Finally, the contacts in the binding site cannot resist the force any longer, and the complex ruptures. Therefore, the correlation between Δx and koff can be explained by the fact that a complex with a higher koff is stabilized by a lower number of contacts and can withstand smaller deformations until the complex dissociates.
However, for the system investigated here, the antigen is extremely flexible. The helical peptide is most likely stretched along its axis, so that a deformation can be induced easily by applying force. In addition, a deformation of the scFv, as observed for the antifluorescein scFvs (42), might occur. That the values for Δx are identical leads to the conclusion that the unbinding process is the same for all variants: before the antibody-peptide complex finally dissociates, the peptide has to be stretched far enough to destabilize the complex. This point of destabilization is identical for the variants. As no additional contacts have been introduced in the binding site during the affinity-maturation process, a stabilization of the complex is only conceivable if the binding site is more rigid and possesses a higher resistance to the applied force. This can be realized with a lower koff value. Indeed, the only relevant mutation that improves koff among the mutants examined here is the mutation in clone C11L34, which is assumed to reduce the flexibility of CDR L1. This interpretation can also be supported by the fact that the flexibility of the binding pocket can be reduced during the affinity maturation process in vivo, as found in other antibody systems (44–46).
This latter finding is an interesting aspect of the affinity maturation process. However, more data from more, different variants would be needed to investigate whether evolution to higher affinity generally results in more rigid binding sites. Furthermore, it would be of great interest to investigate the forced unbinding process of the antibody-peptide system in much greater detail. As the structure of the complex is known, molecular dynamics simulations in combination with additional experiments can provide further insights (42,43,47). For example, it would be interesting to measure whether truncated or mutated peptides show an altered potential width. In addition, the system investigated here is an interesting model system to investigate whether the unbinding pathway is influenced by the direction of the applied force. This can be easily tested by changing the site of attachment of the peptide. A more detailed understanding of the response of biological systems to externally applied forces is of great importance. Even thermodynamically very stable complexes can rupture at low forces, and, conversely, complexes with identical dissociation rates can withstand a broad range of forces depending on the potential width. As more and more systems are discovered that respond to forces in their natural environment, a more detailed knowledge is required of the mechanisms governing how molecules sense and detect forces in biological systems.
Acknowledgments
The authors thank Julia Sedlmair and Holger Hesse for help in recording a large number of data points as well as Gregor Neuert, Hauke Clausen-Schaumann, Matthias Rief, Robert Lugmaier, Ludmila Mendelevitch, Elias Puchner, Thomas Nicolaus, and Gary Morfill for helpful discussions.
This work was supported by the European Union and the Deutsche Forschungsgemeinschaft.
Kerstin Blank's present address is Institut de Science et d'Ingénierie Supramoléculaires (ISIS-ULP), Laboratoire de Biologie Chimique, 8, allée Gaspard Monge, BP 70028, F-67083 Strasbourg cedex, France.
Editor: Jane Clarke.
References
- 1.Yang, W. P., K. Green, S. Pinz-Sweeney, A. T. Briones, D. R. Burton, and C. F. Barbas 3rd. 1995. CDR walking mutagenesis for the affinity maturation of a potent human anti-HIV-1 antibody into the picomolar range. J. Mol. Biol. 254:392–403. [DOI] [PubMed] [Google Scholar]
- 2.Chen, Y., C. Wiesmann, G. Fuh, B. Li, H. W. Christinger, P. McKay, A. M. de Vos, and H. B. Lowman. 1999. Selection and analysis of an optimized anti-VEGF antibody: crystal structure of an affinity-matured Fab in complex with antigen. J. Mol. Biol. 293:865–881. [DOI] [PubMed] [Google Scholar]
- 3.Maynard, J. A., C. B. Maassen, S. H. Leppla, K. Brasky, J. L. Patterson, B. L. Iverson, and G. Georgiou. 2002. Protection against anthrax toxin by recombinant antibody fragments correlates with antigen affinity. Nat. Biotechnol. 20:597–601. [DOI] [PubMed] [Google Scholar]
- 4.Wu, H., D. S. Pfarr, Y. Tang, L. L. An, N. K. Patel, J. D. Watkins, W. D. Huse, P. A. Kiener, and J. F. Young. 2005. Ultra-potent antibodies against respiratory syncytial virus: effects of binding kinetics and binding valence on viral neutralization. J. Mol. Biol. 350:126–144. [DOI] [PubMed] [Google Scholar]
- 5.Kusnezow, W., and J. D. Hoheisel. 2002. Antibody microarrays: promises and problems. Biotechniques. 33(Suppl.):14–23. [PubMed] [Google Scholar]
- 6.Schier, R., J. Bye, G. Apell, A. McCall, G. P. Adams, M. Malmqvist, L. M. Weiner, and J. D. Marks. 1996. Isolation of high-affinity monomeric human Anti-c-erbB-2 single chain Fv using affinity-driven selection. J. Mol. Biol. 255:28–43. [DOI] [PubMed] [Google Scholar]
- 7.Chen, G., I. Dubrawsky, P. Mendez, G. Georgiou, and B. L. Iverson. 1999. In vitro scanning saturation mutagenesis of all the specificity determining residues in an antibody binding site. Protein Eng. 12:349–356. [DOI] [PubMed] [Google Scholar]
- 8.Boder, E. T., K. S. Midelfort, and K. D. Wittrup. 2000. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc. Natl. Acad. Sci. USA. 97:10701–10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van den Beucken, T., H. Pieters, M. Steukers, M. van der Vaart, R. C. Ladner, H. R. Hoogenboom, and S. E. Hufton. 2003. Affinity maturation of Fab antibody fragments by fluorescent-activated cell sorting of yeast-displayed libraries. FEBS Lett. 546:288–294. [DOI] [PubMed] [Google Scholar]
- 10.Graff, C. P., K. Chester, R. Begent, and K. D. Wittrup. 2004. Directed evolution of an anti-carcinoembryonic antigen scFv with a 4-day monovalent dissociation half-time at 37° C. Protein Eng. Des. Sel. 17:293–304. [DOI] [PubMed] [Google Scholar]
- 11.Zahnd, C., S. Spinelli, B. Luginbühl, P. Amstutz, C. Cambillau, and A. Plückthun. 2004. Directed in vitro evolution and crystallographic analysis of a peptide-binding single chain antibody fragment (scFv) with low picomolar affinity. J. Biol. Chem. 279:18870–18877. [DOI] [PubMed] [Google Scholar]
- 12.Friguet, B., A. F. Chaffotte, L. Djavadi-Ohaniance, and M. E. Goldberg. 1985. Measurements of the true affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J. Immunol. Methods. 77:305–319. [DOI] [PubMed] [Google Scholar]
- 13.Nieba, L., A. Krebber, and A. Plückthun. 1996. Competition BIAcore for measuring true affinities: large differences from values determined from binding kinetics. Anal. Biochem. 234:155–165. [DOI] [PubMed] [Google Scholar]
- 14.Schwesinger, F., R. Ros, T. Strunz, D. Anselmetti, H. J. Güntherodt, A. Honegger, L. Jermutus, L. Tiefenauer, and A. Plückthun. 2000. Unbinding forces of single antibody-antigen complexes correlate with their thermal dissociation rates. Proc. Natl. Acad. Sci. USA. 97:9972–9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schier, R., A. McCall, G. P. Adams, K. W. Marshall, H. Merritt, M. Yim, R. S. Crawford, L. M. Weiner, C. Marks, and J. D. Marks. 1996. Isolation of picomolar affinity anti-c-erbB-2 single-chain Fv by molecular evolution of the complementarity determining regions in the center of the antibody binding site. J. Mol. Biol. 263:551–567. [DOI] [PubMed] [Google Scholar]
- 16.Hinterdorfer, P., W. Baumgartner, H. J. Gruber, K. Schilcher, and H. Schindler. 1996. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 93:3477–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dammer, U., M. Hegner, D. Anselmetti, P. Wagner, M. Dreier, W. Huber, and H. J. Güntherodt. 1996. Specific antigen/antibody interactions measured by force microscopy. Biophys. J. 70:2437–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen, S., X. Chen, J. Davies, M. C. Davies, A. C. Dawkes, J. C. Edwards, C. J. Roberts, J. Sefton, S. J. B. Tendler, and P. M. Williams. 1997. Detection of antigen-antibody binding events with the atomic force microscope. Biochemistry. 36:7457–7463. [DOI] [PubMed] [Google Scholar]
- 19.Ros, R., F. Schwesinger, D. Anselmetti, M. Kubon, R. Schafer, A. Plückthun, and L. Tiefenauer. 1998. Antigen binding forces of individually addressed single-chain Fv antibody molecules. Proc. Natl. Acad. Sci. USA. 95:7402–7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moy, V. T., E. L. Florin, and H. E. Gaub. 1994. Intermolecular forces and energies between ligands and receptors. Science. 266:257–259. [DOI] [PubMed] [Google Scholar]
- 21.Neuert, G., C. Albrecht, E. Pamir, and H. E. Gaub. 2006. Dynamic force spectroscopy of the digoxigenin-antibody complex. FEBS Lett. 580:505–509. [DOI] [PubMed] [Google Scholar]
- 22.Hanes, J., L. Jermutus, S. Weber-Bornhauser, H. R. Bosshard, and A. Plückthun. 1998. Ribosome display efficiently selects and evolves high-affinity antibodies in vitro from immune libraries. Proc. Natl. Acad. Sci. USA. 95:14130–14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell, G. I. 1978. Models for the specific adhesion of cells to cells. Science. 200:618–627. [DOI] [PubMed] [Google Scholar]
- 24.Evans, E., and K. Ritchie. 1999. Strength of a weak bond connecting flexible polymer chains. Biophys. J. 76:2439–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedsam, C., A. K. Wehle, F. Kühner, and H. E. Gaub. 2003. Dynamic single-molecule force spectroscopy: bond rupture analysis with variable spacer length. J. Phys. Condens. Matter. 15:S1709–S1723. [Google Scholar]
- 26.Krebber, A., S. Bornhauser, J. Burmester, A. Honegger, J. Willuda, H. R. Bosshard, and A. Plückthun. 1997. Reliable cloning of functional antibody variable domains from hybridomas and spleen cell repertoires employing a reengineered phage display system. J. Immunol. Methods. 201:35–55. [DOI] [PubMed] [Google Scholar]
- 27.Bothmann, H., and A. Plückthun. 1998. Selection for a periplasmic factor improving phage display and functional periplasmic expression. Nat. Biotechnol. 16:376–380. [DOI] [PubMed] [Google Scholar]
- 28.Kienberger, F., V. P. Pastushenko, G. Kada, H. J. Gruber, C. K. Riener, H. Schindler, and P. Hinterdorfer. 2000. Static and dynamic properties of single poly(ethylene glycol) molecules investigated by force spectroscopy. Single Mol. 1:123–128. [Google Scholar]
- 29.Morfill, J., F. Kühner, K. Blank, R. Lugmaier, J. Sedlmair, and H. E. Gaub. 2007. B-S transition in short oligonucleotides. Biophys. J. 93:2400–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kühner, F., J. Morfill, R. A. Neher, K. Blank, and H. E. Gaub. 2007. Force-induced DNA slippage. Biophys. J. 92:2491–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alcantar, N. A., E. S. Aydil, and J. N. Israelachvili. 2000. Polyethylene glycol-coated biocompatible surfaces. J. Biomed. Mater. Res. 51:343–351. [DOI] [PubMed] [Google Scholar]
- 32.Berger, C., S. Weber-Bornhauser, J. Eggenberger, J. Hanes, A. Plückthun, and H. R. Bosshard. 1999. Antigen recognition by conformational selection. FEBS Lett. 450:149–153. [DOI] [PubMed] [Google Scholar]
- 33.Blank, K., J. Morfill, and H. E. Gaub. 2006. Site-specific immobilization of genetically engineered variants of Candida antarctica lipase B. ChemBioChem. 7:1349–1351. [DOI] [PubMed] [Google Scholar]
- 34.Butt, H. J., and M. Jaschke. 1995. Calculation of thermal noise in atomic-force microscopy. Nanotechnology. 6:1–7. [Google Scholar]
- 35.Hugel, T., and M. Seitz. 2001. The study of molecular interactions by AFM force spectroscopy. Macromol. Rapid Commun. 22:989–1016. [Google Scholar]
- 36.Oesterhelt, F., M. Rief, and H. E. Gaub. 1999. Single molecule force spectroscopy by AFM indicates helical structure of poly(ethylene-glycol) in water. New J. Phys. 1:6.1–6.11. [Google Scholar]
- 37.Kühner, F., and H. E. Gaub. 2006. Modelling cantilever-based force spectroscopy with polymers. Polym. 47:2555–2563. [Google Scholar]
- 38.Honegger, A., and A. Plückthun. 2001. Yet another numbering scheme for immunoglobulin variable domains: an automatic modeling and analysis tool. J. Mol. Biol. 309:657–670. [DOI] [PubMed] [Google Scholar]
- 39.Merkel, R., P. Nassoy, A. Leung, K. Ritchie, and E. Evans. 1999. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 397:50–53. [DOI] [PubMed] [Google Scholar]
- 40.Sheng, Y. J., S. Jiang, and H. K. Tsao. 2005. Forced Kramers escape in single-molecule pulling experiments. J. Chem. Phys. 123:91102–91102-4. [DOI] [PubMed] [Google Scholar]
- 41.Schlierf, M., and M. Rief. 2006. Single-molecule unfolding force distributions reveal a funnel-shaped energy landscape. Biophys. J. 90:L33–L35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paci, E., A. Caflisch, A. Plückthun, and M. Karplus. 2001. Forces and energetics of hapten-antibody dissociation: a biased molecular dynamics simulation study. J. Mol. Biol. 314:589–605. [DOI] [PubMed] [Google Scholar]
- 43.Curcio, R., A. Caflisch, and E. Paci. 2005. Change of the unbinding mechanism upon a mutation: a molecular dynamics study of an antibody-hapten complex. Protein Sci. 14:2499–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wedemayer, G. J., P. A. Patten, L. H. Wang, P. G. Schultz, and R. C. Stevens. 1997. Structural insights into the evolution of an antibody combining site. Science. 276:1665–1669. [DOI] [PubMed] [Google Scholar]
- 45.Yin, J., E. C. Mundorff, P. L. Yang, K. U. Wendt, D. Hanway, R. C. Stevens, and P. G. Schultz. 2001. A comparative analysis of the immunological evolution of antibody 28B4. Biochemistry. 40:10764–10773. [DOI] [PubMed] [Google Scholar]
- 46.Jimenez, R., G. Salazar, J. Yin, T. Joo, and F. E. Romesberg. 2004. Protein dynamics and the immunological evolution of molecular recognition. Proc. Natl. Acad. Sci. USA. 101:3803–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heymann, B., and H. Grubmüller. 2001. Molecular dynamics force probe simulations of antibody/antigen unbinding: entropic control and nonadditivity of unbinding forces. Biophys. J. 81:1295–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]