Abstract
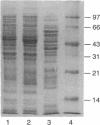
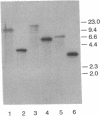
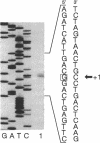
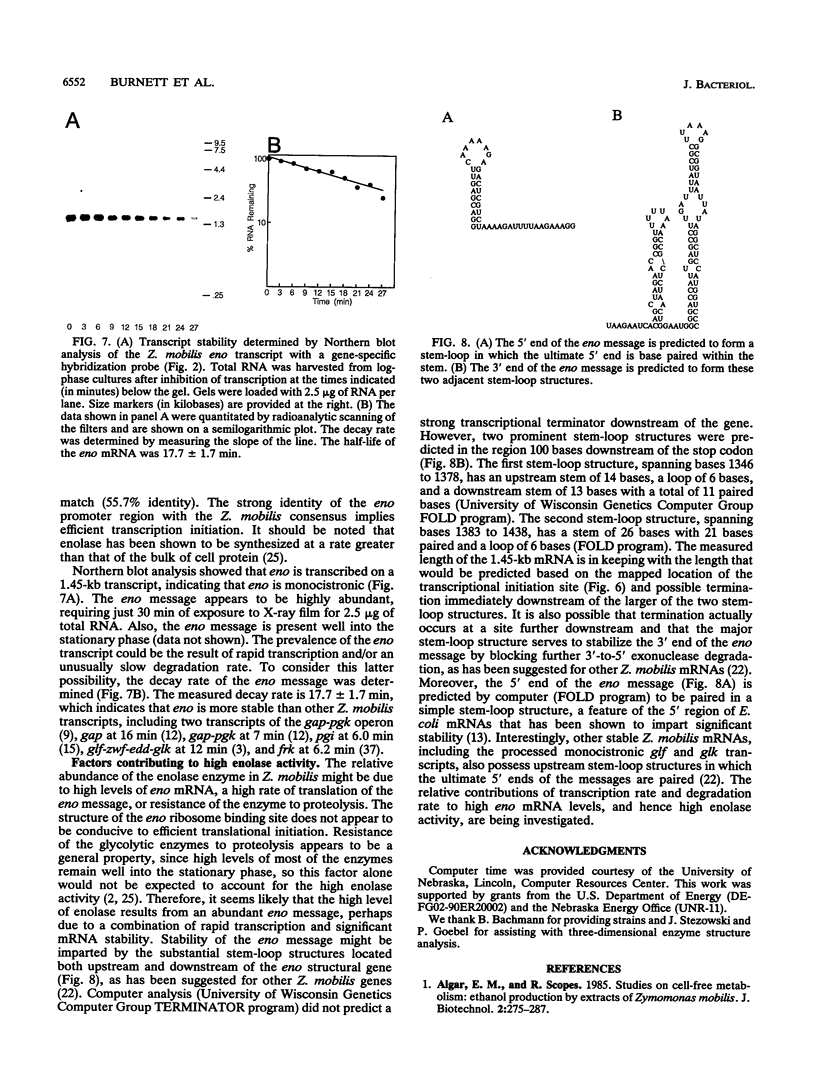
The Zymomonas mobilis gene encoding enolase was cloned by genetic complementation of an Escherichia coli eno mutant. An enzyme assay and sodium dodecyl sulfate-polyacrylamide gel electrophoresis confirmed the overexpression of enolase in E. coli clones carrying the Z. mobilis eno gene. The eno gene is present in a single copy of the Z. mobilis genome. Nucleotide sequence analysis of the eno region revealed an open reading frame of 1,293 bp that encodes a protein of 428 amino acids with a predicted molecular weight of 45,813. Comparison of the sequence of Z. mobilis enolase with primary amino acid sequences for other enolases indicates that the enzyme is highly conserved. Unlike all of the previously studied glycolytic genes from Z. mobilis that possess canonical ribosome binding sites, the eno gene is preceded by a modest Shine-Dalgarno sequence. The transcription initiation site was mapped by primer extension and found to be located within a 115-bp sequence that is 55.7% identical to a highly conserved consensus sequence found within the regulatory regions of highly expressed Z. mobilis genes. Northern RNA blot analysis revealed that eno is encoded on a 1.45-kb transcript. The half-life of the eno mRNA was determined to be 17.7 +/- 1.7 min, indicating that it is unusually stable. The abundance of the eno message is proposed to account for enolase being the most prevalent protein in Z. mobilis.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- An H., Scopes R. K., Rodriguez M., Keshav K. F., Ingram L. O. Gel electrophoretic analysis of Zymomonas mobilis glycolytic and fermentative enzymes: identification of alcohol dehydrogenase II as a stress protein. J Bacteriol. 1991 Oct;173(19):5975–5982. doi: 10.1128/jb.173.19.5975-5982.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnell W. O., Liu J., Hesman T. L., O'Neill M. C., Conway T. The Zymomonas mobilis glf, zwf, edd, and glk genes form an operon: localization of the promoter and identification of a conserved sequence in the regulatory region. J Bacteriol. 1992 May;174(9):2816–2823. doi: 10.1128/jb.174.9.2816-2823.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnell W. O., Yi K. C., Conway T. Sequence and genetic organization of a Zymomonas mobilis gene cluster that encodes several enzymes of glucose metabolism. J Bacteriol. 1990 Dec;172(12):7227–7240. doi: 10.1128/jb.172.12.7227-7240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow K. D., Collins J. G., Norton R. S., Rogers P. L., Smith G. M. 31P nuclear magnetic resonance studies of the fermentation of glucose to ethanol by Zymomonas mobilis. J Biol Chem. 1984 May 10;259(9):5711–5716. [PubMed] [Google Scholar]
- Bishop J. G., Corces V. G. The nucleotide sequence of a Drosophila melanogaster enolase gene. Nucleic Acids Res. 1990 Jan 11;18(1):191–191. doi: 10.1093/nar/18.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Byun M. O., Ingram L. O. Expression Vector for Zymomonas mobilis. Appl Environ Microbiol. 1987 Feb;53(2):235–241. doi: 10.1128/aem.53.2.235-241.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Fliege R., Jones-Kilpatrick D., Liu J., Barnell W. O., Egan S. E. Cloning, characterization and expression of the Zymononas mobilis eda gene that encodes 2-keto-3-deoxy-6-phosphogluconate aldolase of the Entner-Doudoroff pathway. Mol Microbiol. 1991 Dec;5(12):2901–2911. doi: 10.1111/j.1365-2958.1991.tb01850.x. [DOI] [PubMed] [Google Scholar]
- Conway T., Ingram L. O. Phosphoglycerate kinase gene from Zymomonas mobilis: cloning, sequencing, and localization within the gap operon. J Bacteriol. 1988 Apr;170(4):1926–1933. doi: 10.1128/jb.170.4.1926-1933.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Ingram L. O. Similarity of Escherichia coli propanediol oxidoreductase (fucO product) and an unusual alcohol dehydrogenase from Zymomonas mobilis and Saccharomyces cerevisiae. J Bacteriol. 1989 Jul;171(7):3754–3759. doi: 10.1128/jb.171.7.3754-3759.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy C. K., Mejia J. P., Conway T., Ingram L. O. Differential expression of gap and pgk genes within the gap operon of Zymomonas mobilis. J Bacteriol. 1989 Dec;171(12):6549–6554. doi: 10.1128/jb.171.12.6549-6554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emory S. A., Bouvet P., Belasco J. G. A 5'-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992 Jan;6(1):135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- Giallongo A., Feo S., Moore R., Croce C. M., Showe L. C. Molecular cloning and nucleotide sequence of a full-length cDNA for human alpha enolase. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6741–6745. doi: 10.1073/pnas.83.18.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesman T. L., Barnell W. O., Conway T. Cloning, characterization, and nucleotide sequence analysis of a Zymomonas mobilis phosphoglucose isomerase gene that is subject to carbon source-dependent regulation. J Bacteriol. 1991 May;173(10):3215–3223. doi: 10.1128/jb.173.10.3215-3223.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman J. D., Fraenkel D. G. Glyceraldehyde 3-phosphate dehydrogenase mutants of Escherichia coli. J Bacteriol. 1975 Jun;122(3):1175–1179. doi: 10.1128/jb.122.3.1175-1179.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland M. J., Holland J. P., Thill G. P., Jackson K. A. The primary structures of two yeast enolase genes. Homology between the 5' noncoding flanking regions of yeast enolase and glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1981 Feb 10;256(3):1385–1395. [PubMed] [Google Scholar]
- Irani M. H., Maitra P. K. Properties of Escherichia coli mutants deficient in enzymes of glycolysis. J Bacteriol. 1977 Nov;132(2):398–410. doi: 10.1128/jb.132.2.398-410.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal S. K., Johnson S., Conway T., Kelley P. M. Characterization of a maize cDNA that complements an enolase-deficient mutant of Escherichia coli. Plant Mol Biol. 1991 May;16(5):787–795. doi: 10.1007/BF00015071. [DOI] [PubMed] [Google Scholar]
- Lebioda L., Stec B., Brewer J. M. The structure of yeast enolase at 2.25-A resolution. An 8-fold beta + alpha-barrel with a novel beta beta alpha alpha (beta alpha)6 topology. J Biol Chem. 1989 Mar 5;264(7):3685–3693. doi: 10.2210/pdb2enl/pdb. [DOI] [PubMed] [Google Scholar]
- Lessie T. G., Phibbs P. V., Jr Alternative pathways of carbohydrate utilization in pseudomonads. Annu Rev Microbiol. 1984;38:359–388. doi: 10.1146/annurev.mi.38.100184.002043. [DOI] [PubMed] [Google Scholar]
- Liu J., Barnell W. O., Conway T. The polycistronic mRNA of the Zymomonas mobilis glf-zwf-edd-glk operon is subject to complex transcript processing. J Bacteriol. 1992 May;174(9):2824–2833. doi: 10.1128/jb.174.9.2824-2833.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman Y. A., Conway T., Bonetti S. J., Ingram L. O. Glycolytic flux in Zymomonas mobilis: enzyme and metabolite levels during batch fermentation. J Bacteriol. 1987 Aug;169(8):3726–3736. doi: 10.1128/jb.169.8.3726-3736.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A., Scopes R. K., Griffiths-Smith K. Isolation and properties of the glycolytic enzymes from Zymomonas mobilis. The five enzymes from glyceraldehyde-3-phosphate dehydrogenase through to pyruvate kinase. Biochem J. 1986 Aug 15;238(1):275–281. doi: 10.1042/bj2380275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segil N., Shrutkowski A., Dworkin M. B., Dworkin-Rastl E. Enolase isoenzymes in adult and developing Xenopus laevis and characterization of a cloned enolase sequence. Biochem J. 1988 Apr 1;251(1):31–39. doi: 10.1042/bj2510031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring T. G., Wold F. The purification and characterization of Escherichia coli enolase. J Biol Chem. 1971 Nov 25;246(22):6797–6802. [PubMed] [Google Scholar]
- Thomson J., Gerstenberger P. D., Goldberg D. E., Gociar E., Orozco de Silva A., Fraenkel D. G. ColE1 hybrid plasmids for Escherichia coli genes of glycolysis and the hexose monophosphate shunt. J Bacteriol. 1979 Jan;137(1):502–506. doi: 10.1128/jb.137.1.502-506.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M., Makaroff C. A., Zalkin H. Nucleotide sequence of Escherichia coli pyrG encoding CTP synthetase. J Biol Chem. 1986 Apr 25;261(12):5568–5574. [PubMed] [Google Scholar]
- Zembrzuski B., Chilco P., Liu X. L., Liu J., Conway T., Scopes R. Cloning, sequencing, and expression of the Zymomonas mobilis fructokinase gene and structural comparison of the enzyme with other hexose kinases. J Bacteriol. 1992 Jun;174(11):3455–3460. doi: 10.1128/jb.174.11.3455-3460.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]