Abstract
Biological supramolecular systems are commonly built up by the self-assembly of identical protein subunits to produce symmetrical oligomers with cyclical, icosahedral, or helical symmetry that play roles in processes ranging from allosteric control and molecular transport to motor action. The large size of these systems often makes them difficult to structurally characterize using experimental techniques. We have developed a computational protocol to predict the structure of symmetrical protein assemblies based on the structure of a single subunit. The method carries out simultaneous optimization of backbone, side chain, and rigid-body degrees of freedom, while restricting the search space to symmetrical conformations. Using this protocol, we can reconstruct, starting from the structure of a single subunit, the structure of cyclic oligomers and the icosahedral virus capsid of satellite panicum virus using a rigid backbone approximation. We predict the oligomeric state of EscJ from the type III secretion system both in its proposed cyclical and crystallized helical form. Finally, we show that the method can recapitulate the structure of an amyloid-like fibril formed by the peptide NNQQNY from the yeast prion protein Sup35 starting from the amino acid sequence alone and searching the complete space of backbone, side chain, and rigid-body degrees of freedom.
Keywords: Monte Carlo method, protein structure prediction, symmetry
Symmetry is a recurrent theme in nature, from macroscopic objects like animals and plants to microscopic protein assemblies. A number of different point group and helical symmetries are found in naturally occurring protein assemblies (1). The most common type of symmetry is cyclic (Cn symmetry) where the oligomeric structure can be described by a rotation around a single rotation axis of one subunit. Cyclic symmetry generates ring structures found in pores, chambers and molecular motors generating rotational motion. Another common point-group symmetry is the dihedral group (Dn symmetry), which combines one rotational symmetry axis with perpendicular axes of twofold symmetry. D2 symmetry is particularly suited for allosteric control because it involves extensive contact surfaces between subunits (1). Icosahedral symmetry produces roughly spherical assemblies that are often used for storage and transport, as in virus capsids (1). Helical symmetries are produced by rotation and translation along a single symmetry axis and have been observed in microtubules, flagella and actin filaments (1). Amyloid fibers displaying helical symmetry are associated with a number of diseases, such as Creutzfeldt–Jacob's disease and Alzheimer's disease, and are formed by a large number of proteins (2).
The size of larger symmetrical assemblies can make it challenging to obtain high-resolution structures of these biologically important systems. Molecular modeling provides an attractive route to studying symmetrical protein assemblies to provide structural models and answer mechanistic questions. By enforcing symmetry the number of degrees of freedom can be reduced, making calculations on otherwise quite large systems tractable. To date, there have only been a few attempts to predict the structure of larger protein assemblies. Eisenstein et al. (3) assembled the helical protein coat of tobacco mosaic virus by starting from a set of docked dimers. A similar approach was later used to dock structures with Cn and Dn symmetry (4, 5). Comeau and Camacho (6) developed a protocol to predict symmetry type (Cn and Dn symmetry) and the structure of oligomers given an oligomerization state by assembling sets of docked dimers into alternative symmetric assemblies. Schneidman-Duhovny et al. (7) developed a protocol for prediction of cyclic symmetry and Huang and Mayo (8) implemented a method for docking of C2 dimers for use in protein design. In these methods, the side chain and backbone degrees were not sampled.
In this work, we present a general computational framework for prediction of the structure of symmetrical protein assemblies implemented in the computer program ROSETTA (9). Symmetry is imposed in backbone, side chain, and rigid body degrees of freedom. The conformational search space is reduced by sampling only symmetric degrees of freedom and the sizes of the systems are effectively limited by only explicitly simulating a subset of the interacting monomers. Using this method, we can accurately predict the structure of protein assemblies with cyclic, helical, and icosahedral symmetries from the structure of a single subunit while keeping the backbone torsion angles fixed, and recapitulate the structure of an amyloid-like fibril formed by the peptide NNQQNY from the yeast prion protein Sup35 (10) starting from amino acid sequence alone by searching the complete set of backbone, side chain, and rigid body degrees of freedom.
Results
Overview of Method.
We implemented a protocol for modeling of symmetrical protein assemblies within the computer program ROSETTA (9). In this work, we keep bond lengths and bond angles fixed, and assume perfect symmetry of the subunits, and hence the degrees of freedom are the backbone and side chain torsion angles of a single subunit and the parameters describing the rigid body transforms relating the subunits. The simulation starts from a random symmetrical configuration. The conformational search process is divided into low- and high-resolution phases. In the low-resolution search, the backbone and rigid-body degrees of freedom are optimized by using a reduced representation of the complex in which each amino acid in the protein is described by the position of the four backbone heavyatoms and a single “pseudoatom” representing the side chain (referred to as a centroid).
In the more time-intensive, high-resolution search, side chains are added to each protein copy using a Monte Carlo simulated annealing algorithm together with a backbone-dependent rotamer library (11). Then the backbone, side chain, and rigid body degrees of freedom are simultaneously optimized by using a Monte Carlo-plus-minimization (MCM) protocol in which each move consists of three steps: (i) random perturbation of the rigid-body and backbone degrees of freedom; (ii) optimization of side chain conformations by either full combinatorial repacking or by cycling through alternative rotamers for each side chain in a randomized order and selecting the lowest energy conformation (referred to as rotamer trials); and (iii) gradient-based minimization of the backbone, side chain, and rigid body degrees of freedom. Moves are accepted or rejected according to the standard Metropolis criterion; typical simulations involve ≈100 MCM attempted moves. The lowest-energy structures produced in a large number of independent trajectories are clustered and the lowest-energy member of the largest cluster is chosen. Typically, the global search is followed by a local search where the free energy landscape is further explored in the vicinity of the conformational space of the lowest energy models.
Implementation of Symmetry.
A symmetrical system is unchanged under a symmetry transformation. These transformations can be rotation, translation, inversion, and mirror operations. Due to the chiral nature of amino acids, oligomeric proteins exhibit only rotational and translational symmetry. Given the coordinates of a single subunit together with a set of symmetry transformations consistent with the desired symmetry, the position of all subunits in an oligomer can be computed. This simple description of a symmetric system leads to difficulties in gradient computations (see below). To avoid these difficulties, we take advantage of a recently implemented a general kinematic framework for optimization of molecular systems with rigid-body and torsional degrees of freedom (12). We extended this tree-based framework to support symmetric systems by including in the molecular description a set of local reference frames, one frame associated with each subunit.
These local reference frames are related by symmetry transforms (which may vary during the simulation); additional rigid-body transforms link each subunit to its associated reference frame. The latter transforms are identical for all subunits; equivalently, each subunit has identical coordinates when viewed in its associated reference frame. For example, in a cyclic system each reference coordinate system can be chosen with the z axis along the rotation axis, the x axis pointing toward the rotation parallel to the axis, and with y perpendicular to the plane spanned by x and z. A translation along x in one reference system will preserve symmetry if an identical translation is applied to the other subunits. In this representation, it is straightforward to preserve symmetry during gradient-based minimization and rigid-body perturbations. In addition, the partial derivative of the energy function with respect to a symmetric degree of freedom (rigid-body or torsional) can be calculated by multiplying the corresponding derivative for a single subunit by a factor of ns, where ns is the number of subunits in the system. As an example, consider the partial derivative of the energy E of a cyclic system with respect to the symmetric x-coordinate introduced above
 |
where ∇E is the gradient of the full (nonsymmetric) system and x̂i is the unit vector corresponding to translation of subunit i along the x axis of its local frame.
Symmetry of side chain degrees of freedom is implemented by modification of the combinatorial packing (described in ref. 13) and rotamer trials algorithms. Insertions of rotamers are symmetrized, leading to the insertion of identical rotamers at all symmetry-related positions and the energy of insertion is evaluated for all positions at once.
Cyclic Symmetry.
A cyclic system has four rigid body degrees of freedom; the subunits have three rotational and a translational degree of freedom (the radius). For larger oligomers the system can be fully described by a smaller subsystem. Systems with more than three subunits are simulated with three subunits to avoid edge effects. The global search starts with random orientations of the subunits, which are brought into contact by a symmetric translation toward the n-fold rotation axis. A total of 3 × 103 independent models are typically generated in the global search, which is followed by a local refinement (generating ≈1 × 103 models) to explore the local energy landscape. In the test calculations described here, the backbone torsion angles were kept fixed and the search was done over the side chain and rigid-body degrees of freedom.
We tested the symmetrical assembly protocol on a range of randomly selected symmetrical oligomers from the Protein Data Bank (14) containing noncrystallographic symmetry. The set includes a homodimer (dihydrofolate; ref. 15), two trimers [acyl carrier protein (16) and Chorismate mutase (17)], one pentamer (lumizine synthetase; ref. 18), one heptamer (archael sm protein; ref. 19) and an oligomer with unknown oligomerization state from the type III secretion system. These experimental structures do not obey strict symmetry (the backbone rmsd between subunits in the oligomers range between 0.2 and 1.0 Å) but deviations are relatively small, with rotation angles between subunits differing 0.2–4.9% relative to the values expected for perfect symmetry.
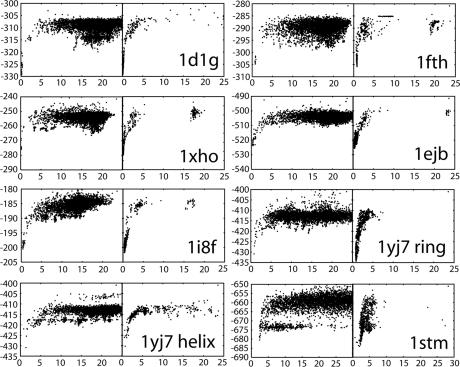
Plots of energy vs. root mean square deviation (rmsd, calculated over all common Cα for the simulated subsystem) relative to the native structure for the models generated in the global search are shown in Fig. 1, and lowest energy models after further refinement are compared with the native structures in Fig. 2. The result for the pentameric structure of lumizine synthetase (1ejb) serves as a representative of the results obtained for all of the systems. The global search (Fig. 1 Left) produced models with a large spread in energy and rmsd and with a significant fraction of models with low backbone rmsd relative to the native structure. The lowest-energy model was subjected to a local refinement, in which the rigid body orientation is randomly perturbed around the starting conformation and the MCM protocol is repeated to sample the local energy landscape (Fig. 1 Right). The energy funnel is steep and narrow, with a width of ≈3 Å. The lowest energy model is 0.3 Å away from the experimentally determined structure (Fig. 2) calculated over the full oligomer. A detailed analysis of the binding interface of the best scoring model shows that the side chain conformations for a large fraction of residues are correctly predicted. The fraction of interface residues with native-like side chain conformations is 71% for this model (Fig. 3). The results for the other studied systems are very similar to the results for lumizine synthetase and are summarized in Table 1.
Fig. 1.
Energy versus rmsd distribution after global sampling (Left) and local refinement (Right). x axis, rmsd over the studied subsystem versus the crystal structure; y axis, Rosetta fullatom energy.
Fig. 2.
Comparison of the lowest energy models after refinement to the complete native structures. Native structures are in red and models are in blue.
Fig. 3.
Side chain prediction of selected residues at a subunit interface of 1ejb. The backbone and side chains for the crystal structure subunits A and B are shown in yellow and green, respectively. Side chains for subunits A and B for the lowest-energy models are shown in magenta and blue, respectively.
Table 1.
Results for studied systems
| System | Symmetry | rmsd,Å* | Fraction correct side-chains, %† |
|---|---|---|---|
| Dihydrofolate reductase (1d1g) | C2 | 0.2 | 91. |
| Acyl carrier protein (1fth) | C3 | 1.1 | 91 |
| Chorismate mutase (1xho) | C3 | 0.3 | 77 |
| Lumazine synthase (1ejb) | C5 | 0.3 | 71 |
| Archael sm protein (1i8f) | C7 | 1.3 | 75 |
| YscJ/PrgK like protein (1yj7) | C24‡ | 0.9 | 68 |
| YscJ/PrgK like protein (1yj7) | Helix‡ | 0.7 | 78 |
| Satellite panicum | |||
| Mosaic virus (1 stm) | Icosahedral§ | 2.1 | 68 |
*rmsd for lowest energy model calculated over all common Cα for the full oligomer.
†A side chain is defined as being correctly predicted if both χ1 and χ2 values are <40° away from the native values. A side chain is defined as being in a protein-protein interface if its Cβ is within 8.0 Å of any Cβ in the other subunit.
‡rmsd calculated over three subunits.
§rmsd calculated over six subunits.
Modeling of the Type III Secretion System (TTSS) Component EscJ.
The docking protocol was also used to predict the structure of a component of the TTSS. TTSSs are multicomponent macromolecules found in many Gram-negative pathogens that mediate secretion and translocation of bacterial proteins into the cytoplasm of eukaryotic cells (20, 21). The core of the TTSSs has been shown by electron microscopy to resemble a needle and is referred to as the needle complex. At the base of the needle several proteins form ring-shaped structures. The structure of one of these base proteins, EscJ from enteropathogenic Escherichia coli, has been solved (22). In the crystal unit cell, protein subunits form a supramolecular helix. Biochemical and electron microscopy data have indicated that EscJ forms a 22 ± 1.7 monomer ring in the biological setting. By projecting the helix onto a plane, a model of the circular form of EscJ could be constructed (with 24 subunits in the ring) (22).
We used the symmetrical docking protocol to predict the structure of the cyclical form of EscJ from the crystal structure of EscJ. The structure of the ring was simulated with oligomerization states ranging from 21 to 25 monomers. Lower energy models were found for each case. The 24 membered ring, having lowest energy, was chosen for further studies. The energy vs. rmsd plots display a sharp energy funnel with a large drop in energy relative to the crystallized form of the protein (Fig. 1). The similarity of the lowest energy model (Fig. 2) with the crystal structure (0.9 Å calculated over three subunits) suggests that the model is a reasonable representation of the cyclical form of EscJ.
Helical Symmetry.
Helical systems have six rigid body degrees of freedom. The subunits have three rotational degrees of freedom and a translational degree of freedom, which is the distance from the center of a subunit to the n-fold rotation axis (the radius); one degree of freedom specifies the rotation angle between subunits (α) and the pitch of the helix is set by the sixth degree of freedom. The pitch can have both positive and negative values corresponding to right- or left-handed helices and is constrained because neighbors along the helix axis cannot clash, although they are free to interact. In the models, we assume that the interactions between consecutive subunits are the primary driving force for helix formation and focus on these interactions to reduce the computational complexity. Three consecutive monomers were used to model the system. In the test calculations described here we kept the backbone torsion angles fixed and the search was done over the side chain and rigid body degrees of freedom.
We tested the method by attempting to reproduce the helical form of EscJ in the crystal structure starting with a single monomer. All degrees of freedom were fully randomized except α, which was initialized in the range corresponding to 20–26 monomers per helix turn (which is consistent with experimental information; ref. 22) but is allowed to move outside this range during the simulation. The global search produces a handful of lower energy models. All of these have a pitch corresponding to a left-handed helix except the lowest energy model, which is close to 0 (0.8 Å). This model was subjected to multiple independent refinement calculations followed by filtering to pick out models without lateral clashes. The lowest energy model after this procedure has an rmsd versus the crystal form over three subunits of 0.7 Å (Table 1). The pitch is −2.6 Å, close to the experimental values of −2.8 Å, and the handedness is correctly predicted. The rotation angle between subunits is 13.3° in the model compared with 15° for the crystal form. A reconstruction of the helix can be seen in Fig. 4.
Fig. 4.
Reconstruction of the helical model of 1yj7 and the capsid model of 1stm. Subunits around a fivefold axis or four consecutive monomers are shown in blue, red, magenta, green, and cyan for the virus (Upper) and helix (Lower), respectively. (Left) Lowest energy models. (Right) Crystal structures.
Icosahedral Symmetry.
Icosahedra contain 20 triangular faces and two-, three-, and fivefold symmetry axes. The icosahedral symmetries of virus capsids are classified by a triangulation number. The simplest icosahedral viruses have a triangulation number of 1 (T1), where all subunits have identical interactions with neighboring subunits. Each of the 20 triangular faces of T1 viruses consists of three subunits resulting in a macromolecule with 60 subunits. The icosahedral system has six degrees of freedom. These correspond to three rotational degrees of freedom for the subunit, a translational degree of freedom normal to the triangular face that determines the size of the icosahedron, a translational degree of freedom corresponding to the distance from the subunit to the threefold symmetry axis (a radius), and rotational degree of freedom that rotates the threefold symmetry-related partners around their threefold axis. The two last degrees of freedom are used to define the position of the subunits on the face of the polyhedron.
By carrying out searches over all six rigid-body degrees of freedom and the side chain degrees of freedom as in the previous examples we attempt to reconstruct the T1 virus capsid of Satellite panicum mosaic virus from the structure of its subunit capsid protein (23). A subsystem of six subunits was simulated to avoid edge effects, so that one subunit is completely encapsulated by neighboring interfaces. Before entering the high-resolution phase, models are filtered based on the number of intersubunit contacts, which removes ≈25% of the population. In the energy vs. rmsd plot for the global search an energy funnel can be distinguished (Fig. 1). The lowest energy model is 2.4 Å away from the native structure calculated over six subunits. After the local refinement, the lowest energy model is 2.1 Å away from the native structure (Fig. 2). Sixty-eight percent of the interface residues in this model are correctly predicted. The refinement process produces a number of models with lower rmsd values, but these have slightly higher energies. The lowest rmsd model is only 0.7 Å away from the native structure, but has significantly higher energy. The full model of the reconstructed virus can be seen in Fig. 4.
Modeling an Amyloid-Like Fibril.
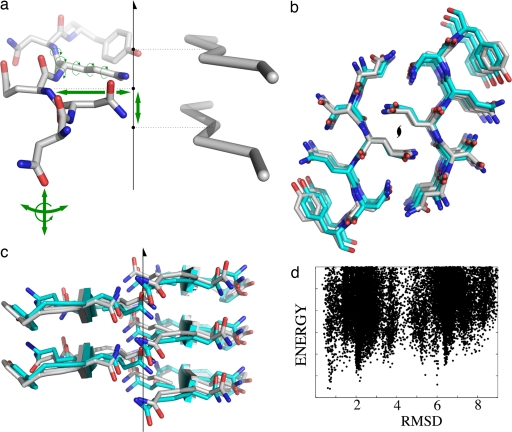
The recent high-resolution structure of a microcrystal formed by the peptide NNQQNY from the yeast prion protein Sup35 has been proposed as a model for the cross-beta core of amyloid fibrils (10). In this structure, a single copy of the six-residue peptide is replicated by a twofold screw symmetry to form two parallel β-sheets that pack tightly together to form a dry interface described as a steric zipper. A distinctive feature of this steric zipper is that it is formed by polar side chains, asparagine and glutamine, which satisfy their hydrogen-bonding requirements by forming stacks of hydrogen bonds parallel to the fibril (symmetry) axis. We set out to recapitulate the structure of this steric zipper using knowledge of the symmetry type and of the presence of backbone hydrogen bonds parallel to the fibril axis (the cross-beta structure, a well established characteristic of amyloid fibrils; ref. 2). The degrees of freedom in this system are the peptide backbone and side chain torsion angles and five rigid-body degrees of freedom (three rotations of the peptide, distance from the peptide to the symmetry axis, and rise along the axis between peptides). Details on the simulation are given in Materials and Methods; briefly, a low-resolution model is built by choosing a random starting orientation for the peptide and sampling backbone torsion angles by fragment insertion. This model is refined by a high-resolution, all-atom simulation in which all degrees of freedom (Fig. 5a) are simultaneously optimized. A plot of energy-vs.-rmsd for a five-peptide slice of the system is shown in Fig. 5d; in Fig. 5 b and c, the lowest-energy model is superimposed on the native structure (0.59 Å Cα rmsd, 0.70 Å over the core side chains). This figure illustrates that we are, starting only from the sequence of the peptide, able to recapitulate the steric zipper to high resolution, suggesting that computational modeling may be a powerful complement to experimental techniques in elucidating the structures of other amyloid-like systems.
Fig. 5.
Amyloid fibril modeling. (a) The green arrows indicate the degrees of freedom sampled during the conformational search: side chain and backbone torsional degrees of freedom; three rotations of the peptide; distance from the peptide to the twofold screw axis; spacing along the axis between peptides. (b and c) Superposition of the lowest-energy model (gray) and the crystal structure of the NNQQNY steric zipper (cyan), showing good agreement over the core side chains. (d) Scatter plot of all-atom rmsd to the crystal structure (x axis) versus energy for the fibril modeling simulations.
Discussion
We have developed a method to predict the structure of symmetrical protein assemblies. The method uses simultaneous optimization of backbone, side chain, and rigid body degrees of freedom in which the search space is restricted to symmetrical conformations. The computational complexity is further reduced by simulating only smaller subsystems of the symmetrical assembly. The method has been applied to systems with cyclical, helical, and icosahedral symmetry but is not restricted to these systems and can be extended to model any type of symmetrical system where all subunits are chemically equivalent. The results show that highly accurate models can be produced with this protocol.
Our approach assumes symmetrical arrangements of protein subunits. With a few notable exceptions, homo-oligomers assemble into symmetrical arrangements despite the fact that there are vastly greater nonsymmetrical possibilities. Symmetry breaking, when present, is usually fairly local. Asymmetry in side chain conformations is sometimes found close to a symmetry axis where for example efficient hydrogen bond formation requires local symmetry breaking (as is the case for leucine zippers; ref. 24). Symmetry breaking is also well established in larger virus capsids (1). As there is usually considerable symmetry present even when there is local symmetry breaking, a reasonable general approach would be to fully constrain symmetry during initial model generation to reduce the size of the space being sampled, and then allow local symmetry breaking, for example by eliminating the symmetrization of the side chain conformations, in later refinement steps. In most of the calculations we have assumed knowledge of the oligomerization state of the system. This information can often be experimentally determined but may also be inferred from simulations with different oligomerization states, as shown in ref. 6.
Comparison with Previous Methods.
Several groups have developed methods to predict the structure of symmetrical protein assemblies using three-dimensional grid-based fast Fourier transform (FFT) docking, a method which optimizes the shape complementarity between binding partners, to produce dimeric complexes. Top scoring dimer orientations are then used to assemble the full symmetrical system, which is scored using an energy function (3–6). Schneidman-Duhovny et al. (7) developed a different method that use a sparse representation of the molecular surface and geometrical hashing techniques to predict Cn symmetries. All these methods have been successful in predicting Cn and Dn symmetries in bound–bound docking experiments. Our approach has the advantage that arbitrary symmetries can be modeled, both side chain and backbone flexibility can be explicitly modeled, and that all degrees of freedom in the oligomeric system are simultaneously optimized. The main disadvantage of our method is the high computational cost associated with high-resolution modeling.
Backbone Flexibility.
In general, backbone conformational changes are expected in real world applications of our symmetrical modeling protocol, either because the starting structure is a comparative model or to allow for conformational change upon oligomerization. Thus, the examples in this paper that utilizes crystal structures coordinates must be viewed as “best case” scenarios. The use of low-resolution experimental data as constraints in the simulations can drastically reduce the conformational search space and compensate for the computational cost associated with full backbone flexibility. These constraints can come from various sources, e.g., alanine scanning (25), chemical cross-linking (26), and hydrogen-deuterium exchange (27) coupled with mass spectrometry. Perhaps the most useful type of intermediate- to low-resolution data are provided by cryo-electron microscopy, which is often used to structurally characterize large multiprotein assemblies (28). Although, in the general case, cryo-EM or other low-resolution data will be highly desirable for building confident models using the methods described in this paper, the striking recapitulation of the crystal structure of the amyloid fiber forming peptide illustrates that, in systems with relatively few degrees of freedom, accurate models can be built from sequence information alone.
Materials and Methods
The symmetrical modeling protocol was implemented in ROSETTA and combines new methods for the treatment of symmetry with methods previously developed for protein-protein docking (29, 30) and ab initio protein structure prediction (31). ROSETTA uses real space Monte-Carlo Minimization to find the lowest energy conformation of binding partners. The protocol consists of a low-resolution search protocol where the side chains are represented by a centroid pseudoatom placed at the average position found in a representative set of structures from the Protein Data Bank. The low-resolution energy function uses residue-scale interaction potential derived from the analysis of high-resolution protein structures (29, 32, 33). In a subsequent high-resolution stage, the energy is calculated by using an all-atom energy function dominated by a Lennard–Jones potential, an orientation-dependent hydrogen bond potential, and an implicit solvation model (11). The time to generate a single model ranged from 3 to 13 min on a 1.6-MHz Athlon AMD processor with 1 Gb of memory.
Symmetrical Placement of Subunits.
The protocol starts with a randomization of the rotational degrees of freedom of one subunit. The first subunit is placed in its local coordinate frame. The coordinate frames of the other subunits are constructed by symmetry transformation of the first subunit's coordinate frame within a static coordinate frame. The other subunits are placed within their coordinate frame with the same internal coordinates as the first subunit. The exact details of this process depend on the symmetry of the system.
For cyclic symmetry the origin of the first coordinate frame is placed at a certain distance (equal to the radius) along the x axis in the static frame with the x axis of the coordinate frame pointing toward the origin of the static frame and the z axis of the first coordinate frame parallel to n-fold rotation axis. The coordinate frames for the other subunits are created by n − 1 rotations around the z axes of the static frame. The first step of the low-resolution search is a “slide into contact,” where the subunits are translated along the x axes of their coordinate frames until they meet in glancing contact. For systems with more than three subunits, three adjacent subunits are chosen and only the energy for the central subunit is calculated to avoid edge effects.
For helical symmetry, the first coordinate frame is placed as described in previous paragraph and the origins of the other coordinate frames are constructed by a rotation of α degrees (where 360/α is the number of subunits per turn) around and translation p (equal to the pitch of the helix) along the z axis of the static coordinate frame. At the start of the simulation α and p are randomized in the range 13.8–18° (corresponding to 20–26 subunits per turn) and 0.5–60 or -0.5-(-60) Å, respectively. Subunits are “slide-into-contact” as in the cyclical case but with the addition that an adjustment of p may be necessary in some cases to get a contact. To make it computationally tractable, a system of three adjacent subunits is chosen where only the central subunit is scored to avoid edge effects. For the energy refinement step an extension of the “slide-in-contact” method was also used where an additional “slide-in-contact” performed by reducing α from a larger value until contact occurs.
The fibril model has a twofold screw symmetry. The reference coordinate systems are chosen to lie along the symmetry axis, with z axes parallel to the symmetry axis and a 180° rotation about the z axis from one coordinate system to the next. The low-resolution simulation begins with the choice of a random starting configuration. To guarantee that backbone hydrogen bonds are present along the fibril axis, we choose at the start of each simulation a parallel β-strand pairing at random from a protein of known structure. The geometry of this pairing is used to determine the i → i + 2 rigid body transformation between subunits (subunits i and i + 1 are on opposite sides of the symmetry axis; backbone hydrogen bonds are present between subunits i and i + 2). The two remaining rigid body degrees of freedom (distance from the subunit to the symmetry axis, and internal rotation of the subunit about its z axis) are chosen randomly from suitable uniform distributions. Backbone torsion angles are initialized to extended values and a low-resolution fragment insertion simulation is used to build a backbone compatible with the starting rigid-body configuration. The resulting low-resolution model is further refined by an all-atom simulation as described above, with the added feature that backbone torsion-angle moves are included, and all degrees of freedom of the system (rigid-body, backbone, and side chain) are minimized simultaneously.
An icosahedron contains 12 vertices and 20 triangular faces. For a T1 symmetrical system, each face contains three subunits coupled by a threefold symmetry axis. In the icosahedral setup, the vertices are created at s*(0,±1, ±φ), s*(±1, ±φ,0), s*(±φ,0,±1) where s controls the size of the icosahedron and φ = (1+√5)/2 is the golden ratio. Reference frames are placed at the center of each face with the z axis normal to the face, the x axis pointing toward one vertex describing the triangular face, and the y axis perpendicular to x and z axis. The origin of the coordinate frames of the first subunit of a face is placed along the x axis of the reference frames (the distance to the center is the radius). In the coordinate frames of the first subunits of the faces, the x axis points to the center of the face and the z axis is parallel to the z axis of the reference frame. The coordinate frames of the two other subunits in a face are constructed by 120° rotations of the first coordinate frame around the z axis of the reference frame. At the start of the simulation the size of the icosahedron (s), which is controlled by a translation along the z axes of reference frames, and the radius is set to a large values so that different subunits do no contact each other. The first step of the low-resolution search is a “slide-into-contact” where the subunits are translated along the x axes of their coordinate frames followed by a second slide-into-contact where the size of the icosahedron is reduced by sliding along the z axis of the reference frame. The subunits are initially placed on lines from the vertices to the center of the faces. Then all of the subunits related by threefold symmetry are rotated together around the z axis of the reference frame by a random angle in the range ±30°. A reduced system of six subunits is used to simulate the icosahedral symmetry corresponding to the three subunits related by a threefold axis together with three subunits from two other faces. Only the subunit that is surrounded by the largest number of neighboring subunits is chosen for scoring to avoid edge effects.
Symmetrization of Side Chain Degrees of Freedom.
Rosetta uses two side chain rotamer optimization methods: simulated annealing (“packing”) (11) and greedy one at a time optimization (“rotamer trials”). Packing or rotamer trials are used for side chain optimization within MCM after the random pertubation but before the gradient-based minimization. Packing is used instead of rotamer trials every eight cycles of MCM. Both the packing algorithm (13) and rotamer trials were modified to allow for symmetrical rotamer placement.
Software.
All figures were made with gnuplot and pymol (DeLano Scientific). The rosetta source code is available without charge for academic users from http://depts.washington.edu/ventures/UW_Technology/Express_Licenses/Rosetta.
Acknowledgments
We thank Ora Schueler-Furman for stimulating conversations about the origin of symmetry and Keith Laidig for flawless maintenance of computer resources and Sam Miller and Natalie Strynadka for introducing us to the type III secretion system. This work was supported by a National Institutes of Health grant. A postdoctoral fellowship to I.A. from the Knut and Alice Wallenberg foundation is gratefully acknowledged.
Abbreviation
- MCM
Monte Carlo plus minimization.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Goodsell DS, Olson AJ. Annu Rev Biophys Biomol Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 2.Stefani M, Dobson CM. J Mol Med. 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 3.Eisenstein M, Shariv I, Koren G, Friesem AA, Katchalski-Katzir E. J Mol Biol. 1997;266:135–143. doi: 10.1006/jmbi.1996.0773. [DOI] [PubMed] [Google Scholar]
- 4.Berchanski A, Eisenstein M. Proteins. 2003;53:817–829. doi: 10.1002/prot.10480. [DOI] [PubMed] [Google Scholar]
- 5.Berchanski A, Segal D, Eisenstein M. Proteins. 2005;60:202–206. doi: 10.1002/prot.20558. [DOI] [PubMed] [Google Scholar]
- 6.Comeau SR, Camacho CJ. J Struct Biol. 2005;150:233–244. doi: 10.1016/j.jsb.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. Proteins. 2005;60:224–231. doi: 10.1002/prot.20562. [DOI] [PubMed] [Google Scholar]
- 8.Huang PS, Love JJ, Mayo SL. J Comput Chem. 2005;26:1222–1232. doi: 10.1002/jcc.20252. [DOI] [PubMed] [Google Scholar]
- 9.Rohl CA, Strauss CE, Misura KM, Baker D. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 10.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhlman B, Baker D. Proc Natl Acad Sci USA. 2000;97:10383–8. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradley P, Baker D. Proteins. 2006;65:922–929. doi: 10.1002/prot.21133. [DOI] [PubMed] [Google Scholar]
- 13.Thompson MJ, Sievers SA, Karanicolas J, Ivanova MI, Baker D, Eisenberg D. Proc Natl Acad Sci USA. 2006;103:4074–4078. doi: 10.1073/pnas.0511295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, et al. Acta Crystallogr D. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 15.Dams T, Auerbach G, Bader G, Jacob U, Ploom T, Huber R, Jaenicke R. J Mol Biol. 2000;297:659–672. doi: 10.1006/jmbi.2000.3570. [DOI] [PubMed] [Google Scholar]
- 16.Chirgadze NY, Briggs SL, McAllister KA, Fischl AS, Zhao G. EMBO J. 2000;19:5281–5287. doi: 10.1093/emboj/19.20.5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu H, Yang C, Chen L, Kataeva IA, Tempel W, Lee D, Habel JE, Nguyen D, Pflugrath JW, Ferrara JD, et al. Acta Crystallogr D. 2005;61:960–966. doi: 10.1107/S0907444905010644. [DOI] [PubMed] [Google Scholar]
- 18.Meining W, Mortl S, Fischer M, Cushman M, Bacher A, Ladenstein R. J Mol Biol. 2000;299:181–197. doi: 10.1006/jmbi.2000.3742. [DOI] [PubMed] [Google Scholar]
- 19.Mura C, Cascio D, Sawaya MR, Eisenberg DS. Proc Natl Acad Sci USA. 2001;98:5532–5537. doi: 10.1073/pnas.091102298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson S, Deane JE, Lea SM. Curr Opin Struct Biol. 2005;15:700–707. doi: 10.1016/j.sbi.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Yip CK, Strynadka NC. Trends Biochem Sci. 2006;31:223–230. doi: 10.1016/j.tibs.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Yip CK, Kimbrough TG, Felise HB, Vuckovic M, Thomas NA, Pfuetzner RA, Frey EA, Finlay BB, Miller SI, Strynadka NC. Nature. 2005;435:702–707. doi: 10.1038/nature03554. [DOI] [PubMed] [Google Scholar]
- 23.Ban N, McPherson A. Nat Struct Biol. 1995;2:882–890. doi: 10.1038/nsb1095-882. [DOI] [PubMed] [Google Scholar]
- 24.O'Shea EK, Klemm JD, Kim PS, Alber T. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 25.DeLano WL. Curr Opin Struct Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 26.Back JW, de Jong L, Muijsers AO, de Koster CG. J Mol Biol. 2003;331:303–313. doi: 10.1016/s0022-2836(03)00721-6. [DOI] [PubMed] [Google Scholar]
- 27.Lanman J, Prevelige PE., Jr Curr Opin Struct Biol. 2004;14:181–188. doi: 10.1016/j.sbi.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 28.Jiang W, Ludtke SJ. Curr Opin Struct Biol. 2005;15:571–577. doi: 10.1016/j.sbi.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. J Mol Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 30.Schueler-Furman O, Wang C, Baker D. Proteins. 2005;60:187–194. doi: 10.1002/prot.20556. [DOI] [PubMed] [Google Scholar]
- 31.Bradley P, Misura KM, Baker D. Science. 2005;309:1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 32.Simons KT, Kooperberg C, Huang E, Baker D. J Mol Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 33.Simons KT, Ruczinski I, Kooperberg C, Fox BA, Bystroff C, Baker D. Proteins. 1999;34:82–95. doi: 10.1002/(sici)1097-0134(19990101)34:1<82::aid-prot7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]