Abstract
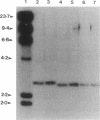
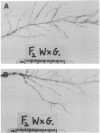
Insertion mutagenesis identified two negatively acting gene loci which restrict the ability of Rhizobium leguminosarum bv. trifolii TA1 to infect the homologous host Trifolium subterraneum cv. Woogenellup. One locus was confirmed by DNA sequence analysis as the nodM gene, while the other locus, designated csn-1 (cultivar-specific nodulation), is not located on the symbiosis plasmid. The presence of these cultivar specificity loci could be suppressed by the introduction of the nodT gene from ANU843, a related R. leguminosarum bv. trifolii strain. Other nod genes, present in R. leguminosarum bv. viciae (including nodX) and R. meliloti, were capable of complementing R. leguminosarum bv. trifolii TA1 for nodulation on cultivar Woogenellup. Nodulation studies conducted with F2 seedlings from a cross between cultivar Geraldton and cultivar Woogenellup indicated that a single recessive gene, designated rwt1, is responsible for the Nod- association between strain TA1 and cultivar Woogenellup. Parallels can be drawn between this association and gene-for-gene systems common in interactions between plants and biotrophic pathogens.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aguilar O. M., Kapp D., Pühler A. Characterization of a Rhizobium meliloti fixation gene (fixF) located near the common nodulation region. J Bacteriol. 1985 Oct;164(1):245–254. doi: 10.1128/jb.164.1.245-254.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelbaum E. R., McLoughlin T. J., O'Connell M., Chartrain N. Expression of symbiotic genes of Rhizobium japonicum USDA 191 in other rhizobia. J Bacteriol. 1985 Jul;163(1):385–388. doi: 10.1128/jb.163.1.385-388.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canter Cremers H., Spaink H. P., Wijfjes A. H., Pees E., Wijffelman C. A., Okker R. J., Lugtenberg B. J. Additional nodulation genes on the Sym plasmid of Rhizobium leguminosarum biovar viciae. Plant Mol Biol. 1989 Aug;13(2):163–174. doi: 10.1007/BF00016135. [DOI] [PubMed] [Google Scholar]
- Cervantes E., Sharma S. B., Maillet F., Vasse J., Truchet G., Rosenberg C. The Rhizobium meliloti host range nodQ gene encodes a protein which shares homology with translation elongation and initiation factors. Mol Microbiol. 1989 Jun;3(6):745–755. doi: 10.1111/j.1365-2958.1989.tb00223.x. [DOI] [PubMed] [Google Scholar]
- Cregan P. B., Keyser H. H., Sadowsky M. J. Host Plant Effects on Nodulation and Competitiveness of the Bradyrhizobium japonicum Serotype Strains Constituting Serocluster 123. Appl Environ Microbiol. 1989 Oct;55(10):2532–2536. doi: 10.1128/aem.55.10.2532-2536.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E. O., Evans I. J., Johnston A. W. Identification of nodX, a gene that allows Rhizobium leguminosarum biovar viciae strain TOM to nodulate Afghanistan peas. Mol Gen Genet. 1988 Jun;212(3):531–535. doi: 10.1007/BF00330860. [DOI] [PubMed] [Google Scholar]
- Debellé F., Maillet F., Vasse J., Rosenberg C., de Billy F., Truchet G., Dénarié J., Ausubel F. M. Interference between Rhizobium meliloti and Rhizobium trifolii nodulation genes: genetic basis of R. meliloti dominance. J Bacteriol. 1988 Dec;170(12):5718–5727. doi: 10.1128/jb.170.12.5718-5727.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debellé F., Rosenberg C., Vasse J., Maillet F., Martinez E., Dénarié J., Truchet G. Assignment of symbiotic developmental phenotypes to common and specific nodulation (nod) genetic loci of Rhizobium meliloti. J Bacteriol. 1986 Dec;168(3):1075–1086. doi: 10.1128/jb.168.3.1075-1086.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic M. A., Zurkowski W., Rolfe B. G. Plasmids and stability of symbiotic properties of Rhizobium trifolii. J Bacteriol. 1982 Aug;151(2):560–568. doi: 10.1128/jb.151.2.560-568.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic M. A., Zurkowski W., Shine J., Rolfe B. G. Sym plasmid transfer to various symbiotic mutants of Rhizobium trifolii, R. leguminosarum, and R. meliloti. J Bacteriol. 1983 Dec;156(3):1035–1045. doi: 10.1128/jb.156.3.1035-1045.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Gray J. X., Djordjevic M. A., Rolfe B. G. Two genes that regulate exopolysaccharide production in Rhizobium sp. strain NGR234: DNA sequences and resultant phenotypes. J Bacteriol. 1990 Jan;172(1):193–203. doi: 10.1128/jb.172.1.193-203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges R. W., Jacob A. E. Transposition of ampicillin resistance from RP4 to other replicons. Mol Gen Genet. 1974;132(1):31–40. doi: 10.1007/BF00268228. [DOI] [PubMed] [Google Scholar]
- Horvath B., Kondorosi E., John M., Schmidt J., Török I., Györgypal Z., Barabas I., Wieneke U., Schell J., Kondorosi A. Organization, structure and symbiotic function of Rhizobium meliloti nodulation genes determining host specificity for alfalfa. Cell. 1986 Aug 1;46(3):335–343. doi: 10.1016/0092-8674(86)90654-9. [DOI] [PubMed] [Google Scholar]
- Jacobs T. W., Egelhoff T. T., Long S. R. Physical and genetic map of a Rhizobium meliloti nodulation gene region and nucleotide sequence of nodC. J Bacteriol. 1985 May;162(2):469–476. doi: 10.1128/jb.162.2.469-476.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge P., Roche P., Faucher C., Maillet F., Truchet G., Promé J. C., Dénarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990 Apr 19;344(6268):781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- McIver J., Djordjevic M. A., Weinman J. J., Bender G. L., Rolfe B. G. Extension of host range of Rhizobium leguminosarum bv. trifolii caused by point mutations in nodD that result in alterations in regulatory function and recognition of inducer molecules. Mol Plant Microbe Interact. 1989 May-Jun;2(3):97–106. doi: 10.1094/mpmi-2-097. [DOI] [PubMed] [Google Scholar]
- Rolfe B. G., Gresshoff P. M., Shine J., Vincent J. M. Interaction Between a Non-Nodulating and an Ineffective Mutant of Rhizobium trifolli Resulting in Effective (Nitrogen-Fixing) Nodulation. Appl Environ Microbiol. 1980 Feb;39(2):449–452. doi: 10.1128/aem.39.2.449-452.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowsky M. J., Tully R. E., Cregan P. B., Keyser H. H. Genetic Diversity in Bradyrhizobium japonicum Serogroup 123 and Its Relation to Genotype-Specific Nodulation of Soybean. Appl Environ Microbiol. 1987 Nov;53(11):2624–2630. doi: 10.1128/aem.53.11.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-Mob transposon. Mol Gen Genet. 1984;196(3):413–420. doi: 10.1007/BF00436188. [DOI] [PubMed] [Google Scholar]
- Staskawicz B., Dahlbeck D., Keen N., Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987 Dec;169(12):5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surin B. P., Downie J. A. Characterization of the Rhizobium leguminosarum genes nodLMN involved in efficient host-specific nodulation. Mol Microbiol. 1988 Mar;2(2):173–183. doi: 10.1111/j.1365-2958.1988.tb00019.x. [DOI] [PubMed] [Google Scholar]
- Surin B. P., Watson J. M., Hamilton W. D., Economou A., Downie J. A. Molecular characterization of the nodulation gene, nodT, from two biovars of Rhizobium leguminosarum. Mol Microbiol. 1990 Feb;4(2):245–252. doi: 10.1111/j.1365-2958.1990.tb00591.x. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truchet G., Debellé F., Vasse J., Terzaghi B., Garnerone A. M., Rosenberg C., Batut J., Maillet F., Dénarié J. Identification of a Rhizobium meliloti pSym2011 region controlling the host specificity of root hair curling and nodulation. J Bacteriol. 1985 Dec;164(3):1200–1210. doi: 10.1128/jb.164.3.1200-1210.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfeld P. L., Seeburg P. H., Shine J. The human pro-opiomelanocortin gene: organization, sequence, and interspersion with repetitive DNA. DNA. 1982;1(2):133–143. doi: 10.1089/dna.1.1982.1.133. [DOI] [PubMed] [Google Scholar]