Abstract
We have investigated the effects of inhibiting the expression of cofilin to understand its role in protrusion dynamics in metastatic tumor cells, in particular. We show that the suppression of cofilin expression in MTLn3 cells (an apolar randomly moving amoeboid metastatic tumor cell) caused them to extend protrusions from only one pole, elongate, and move rectilinearly. This remarkable transformation was correlated with slower extension of fewer, more stable lamellipodia leading to a reduced turning frequency. Hence, the loss of cofilin caused an amoeboid tumor cell to assume a mesenchymal-type mode of movement. These phenotypes were correlated with the loss of uniform chemotactic sensitivity of the cell surface to EGF stimulation, demonstrating that to chemotax efficiently, a cell must be able to respond to chemotactic stimulation at any region on its surface. The changes in cell shape, directional migration, and turning frequency were related to the re-localization of Arp2/3 complex to one pole of the cell upon suppression of cofilin expression.
Introduction
Changes in the regulation and expression of key molecules of the actin cytoskeleton contribute in a major way to differences between metastatic and non-metastatic cancer cells (Condeelis et al., 2005). Cofilin, and its regulatory proteins, are differentially expressed and regulated in metastatic cancer cells and the activity status of the cofilin pathway is directly correlated with metastasis in mammary tumors (Wang et al., 2004, 2006, 2007a,b). Cofilin is a small ubiquitous protein (∼19 kD) that is able to bind both monomeric (G) and filamentous (F) actin (Maciver and Hussey, 2002; Ono, 2007). By severing actin filaments, cofilin increases the number of filament ends for polymerization and depolymerization (Ichetovkin et al., 2002; Andrianantoandro and Pollard, 2006; Yamaguchi and Condeelis, 2007). Cofilin can be regulated through different upstream effectors; LIM 1 and 2, and TES 1 and 2 kinases phosphorylate cofilin on the serine 3 residue, thus rendering it inactive (Mizuno et al., 1994; Okano et al., 1995; Toshima et al., 2001); whereas type 1, 2A, 2B, slingshot, and chronophin phosphatases dephosphorylate cofilin (Meberg et al., 1998; Ambach et al., 2000; Niwa et al., 2002; Zhan et al., 2003; Gohla et al., 2005). In addition, cofilin is inhibited when bound to phosphatydalinositol-4,5-bisphosphate (PIP2) (Yonezawa et al., 1990, 1991). In vivo studies suggest that in metastatic tumor cells, PLC-mediated hydrolysis of PIP2 can release cofilin from this complex, thereby activating it (Kassis et al., 1999; Mouneimne et al., 2004).
Cofilin is a key player in regulating the dynamics of the actin cytoskeleton of migrating cells both in vivo and in vitro. Cultured cells with low cofilin levels show defects in actin polymerization and depolymerization (Mouneimne et al., 2004, 2006; Hotulainen et al., 2005). The direct stimulation of cofilin by uncaging in vivo demonstrates that the local activation of cofilin causes free barbed end formation, and can define the site of actin polymerization, protrusion, and cell direction (Ghosh et al., 2004). Consistently, PLC and cofilin are required for the initiation of protrusions toward an EGF gradient (Mouneimne et al., 2004, 2006). All of these considerations suggest that cofilin is a key regulator of directional migration, chemotaxis, and cell polarity.
The ability of cofilin to affect actin dynamics specifically at the leading edge is amplified by two factors. First, cofilin activity is spatially restricted at the leading edge because this is the only region in crawling mammalian cells where filaments are not saturated with tropomyosin; filaments containing tropomyosin are resistant to cofilin severing, thereby concentrating the activity of cofilin to the leading edge (DesMarais et al., 2002; Gupton et al., 2005). Second, cofilin severs actin filaments to generate free barbed ends which elongate forming newly polymerized filaments that are preferred by Arp2/3 complex for dendritic nucleation. Cofilin stimulates Arp2/3 activity by producing these filament ends (Ichetovkin et al., 2002). This synergistic amplification of Arp2/3 complex's dendritic nucleation activity by cofilin has been observed both in vitro (Ichetovkin et al., 2002) and in vivo (DesMarais et al., 2004). This synergy may explain the location and the bias of the dendritic nucleation activity of Arp2/3 complex toward the barbed ends of newly polymerized filaments (Carlier et al., 1997; Bailly et al., 1999; Ichetovkin et al., 2002).
In this study, we investigated the effects of inhibiting the expression of cofilin in mammary tumor cells. Our goal was to determine the cofilin knockdown phenotype in enough detail to understand the role of cofilin in protrusion dynamics, including the effects of cofilin on stability and directionality of lamellipodial protrusions in metastatic cells in particular.
Results
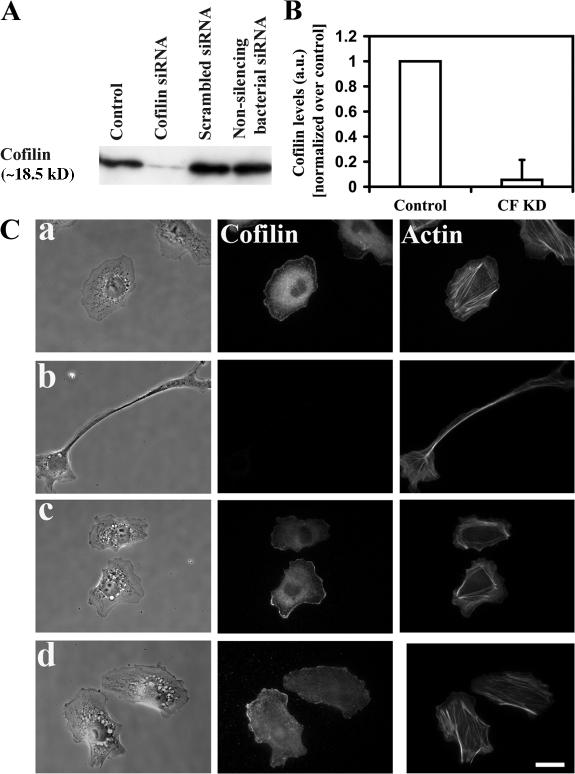
Cofilin is the dominant isoform compared with the actin depolymerizing factor (ADF) in several cells (Hotulainen et al., 2005), including MTLn3 cells (Wang et al., 2004, 2006). To suppress cofilin expression we used siRNA that specifically targets cofilin, but not ADF. Using this sequence, Western blot analysis showed that the cofilin levels were lowered by an average of 95% in MTLn3 cells, as compared with control (oligofectamine only) treated cells at 36 h after transfection (Fig. 1 A); reversion to normal levels was observed by 96 h (Mouneimne et al., 2004). Because the antibody used for Western blotting in these experiments recognizes both cofilin and ADF, the small amount of protein remaining in cells after the siRNA treatment is consistent with the small amount of ADF relative to cofilin. We tested two other siRNA sequences as negative controls: a scrambled version of the cofilin siRNA, and a non-silencing bacterial siRNA, which does not have target sequences in MTLn3 cells. Using Western blotting and immunofluorescence, both siRNAs did not cause any change in the cofilin expression levels, whereas the cofilin siRNA significantly reduced cofilin expression (Fig. 1).
Figure 1.
Suppression of cofilin using siRNA. (A) Western blotting of whole cell lysates of cells treated with the different siRNAs, blotted for cofilin. The western was repeated four times, and the corresponding quantification is shown in B. (C) representative figures of (a) control cells (treated with oligofectamine only), (b) cells treated with cofilin siRNA, (c) cells treated with scrambled siRNA, and (d) cells treated with non-silencing bacterial siRNA. Bar, 10 μm.
Control, scrambled, and bacterial siRNA treated cells (Fig. 1 C, a, c, and d, respectively) showed similar morphology to that of untreated cells, whereas cofilin siRNA treated cells (referred to hereafter as CF KD) exhibited an elongated morphology (Fig. 1 C, b). Using FACS analysis, we also tested the viability of the control and CF KD cells and found that at 36 h after siRNA transfection, the majority of cells were viable (unpublished data). Therefore, all further experiments reported in the paper were performed 36 h after siRNA transfection, when maximum suppression of cofilin expression was observed (Fig. 1 B). It is important to note that suppression of cofilin did not affect the expression of other factors that are important in the regulation of the actin cytoskeleton, such as WAVE 2 and N-WASP expression (Fig. S1, C and D; available at http://www.jcb.org/cgi/content/full/jcb.200707009/DC1).
Cofilin siRNA knockdown cells are characterized by an elongated shape and an increase in F-actin–containing structures
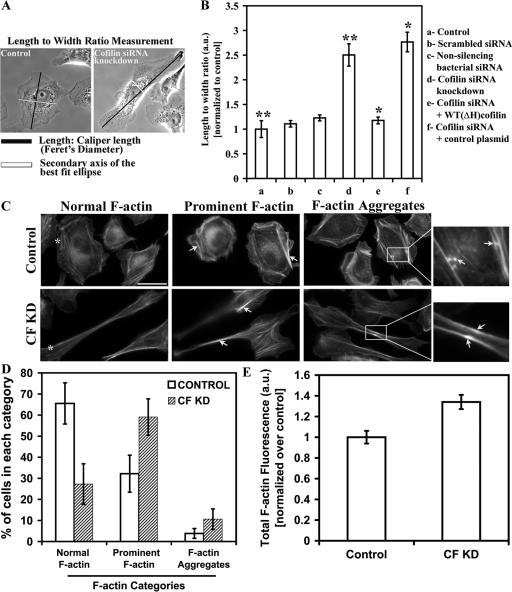
To investigate the effect of CF KD on the morphology of MTLn3 cells, we examined the ratio of length to width (L/W) of control and siRNA-treated cells. CF KD cells exhibited a two- to threefold increase in L/W ratio (Fig. 2 B) compared to control cells. The scrambled and non-silencing bacterial siRNA showed similar L/W ratio as control cells. Next, we transfected the CF KD cells with a human cofilin plasmid that is not targeted by our siRNA sequence, or a control plasmid, and scored their L/W ratio. The cells transfected with the human cofilin rescue plasmid had WT-like morphology, whereas the cells transfected with the control plasmid showed high L/W ratio characteristic of CF KD cells, demonstrating that the elongated phenotype was due to cofilin suppression. Furthermore, shape analysis performed with the DIAS software demonstrated that the maximum length and perimeter of CF KD cells were almost twice that of control cells, while roundness was significantly reduced. These changes were rescued by expression of the human cofilin plasmid (Table I).
Figure 2.
Cofilin siRNA knockdown cells are elongated and show changes in F-actin structures. (A) Method of length/width (L/W) measurement. Length was scored as Feret's diameter, which is the longest distance between any two points along the boundary, black line; width is the secondary axis of the best fit ellipse of the selected cell, white line. (B) quantification of the L/W ratio of (a) control cells transfected with either (b) scrambled or (c) non-silencing bacterial siRNAs, (d) CF KD cells, (e) cofilin siRNA and human cofilin plasmid cotransfected cells, and (f) cofilin siRNA and control plasmid cotransfected cells. Number of cells scored: control (136), scrambled (67), non-silencing bacterial (51), CF KD cells (123), CF KD + rescue plasmid (51), CF KD + control plasmid (38) cells. *, P < 0.001; **, P < 0.001. (C) control and CF KD cells were subdivided into three categories depending on their actin structures as stained with rhodamine-phalloidin. Cells in each group, were compared with the examples shown in panel, and were subdivided according to the following criteria: cells exhibiting normal F-actin (asterisk; cells exhibiting prominent F-actin, showing an increase in the thickness and number of stress fibers as compared with cells with normal F-actin (arrows); and cells exhibiting F-actin aggregates (arrows in magnified inset mark F-actin aggregates). Bar, 10 μm. (D) cells were double-blind scored as explained in C, and presented as percentage of total. Number of cells scored: control (87), CF KD (67). (E) Total F-actin levels of control (n = 69) and CF KD (n = 67) cells. Results shown here and throughout the paper are the average of cells collected from at least three experiments.
Table I. Expression of the human cofilin plasmid can rescue the phentotypic changes seen in CF KD cells.
| Maximum lengtha
(μm) |
Perimeter (μm/min) |
Roundednessb
(%) |
Area (μm2) |
|
|---|---|---|---|---|
| Control | 52.16 ± 0.97 | 148.28 ± 3.2 | 73.68 ± 1.32 | 1,285.35 ± 46.71 |
| Cofilin siRNA knockdown | 99.33 ± 6.2 | 257.02 ± 15.13 | 32.87 ± 3.18 | 1,272.26 ± 37.13 |
| Cofilin siRNA knockdown plus WT (ΔH) cofilin | 50.04 ± 2.16 | 137.18 ± 5.13 | 74.80 ± 2.14 | 1,111.87 ± 67.44 |
DIAS analysis was carried to quantify shape parameters for control, CF KD cells, and CF KD cells + WT(ΔH) cofilin plasmid.
Maximum length is the distance between the farthest points on the cell boundary.
Roundedness is a measure of how efficiently a given amount of perimeter encloses an area (a circle has 100% roundedness; a straight line has 0%). Number of cells scored: control (35), CF KD (31) and cofilin CF KD + WT (ΔH) (19). Numbers are significantly different (P < 0.0001) between control and CF KD groups, and between CF KD and CF KD+WT(ΔH) cofilin groups, for all parameters except for the area measurement.
Previous studies have shown that overexpression of constitutively active LIM-kinase changes F-actin organization in MTLn3 cells, causing the formation of more prominent F-actin fibers and F-actin aggregates (Zebda et al., 2000). To determine if the CF KD cells exhibit a similar phenotype, cells were fixed, stained using rhodamine-phalloidin, and divided into three categories as described previously (Zebda et al., 2000) according to the structure of their actin cytoskeleton. Cells were double-blind scored, where control cells were compared with control cells, and CF KD cells were compared with other CF KD cells (representative cells from each group exhibiting the different F-actin categories are shown in Fig. 2 C). CF KD cells had more prominent F-actin fibers and more F-actin aggregates (Fig. 2 C, white arrows) compared to control cells (Fig. 2, C and D). These results are consistent with the observations reported by Zebda et al. (2000), thus showing that lowering either cofilin expression or activity yields the same result. Recent reports have also shown that siRNA suppression of cofilin in NIH3T3 and mouse neuroblastoma cells led to an accumulation of F-actin, increase in thickness of stress fibers, and decrease in actin stress fiber turnover as measured by FRAP (Hotulainen et al., 2005). Our analysis showed that CF KD caused a significant (P = 0.0024) increase in the total F-actin levels, presumably resulting from the loss of actin depolymerization by cofilin and hence accumulation of F-actin (Fig. 2 E).
Cofilin knockdown increases the directionality and decreases the turning frequency of MTLn3 cells
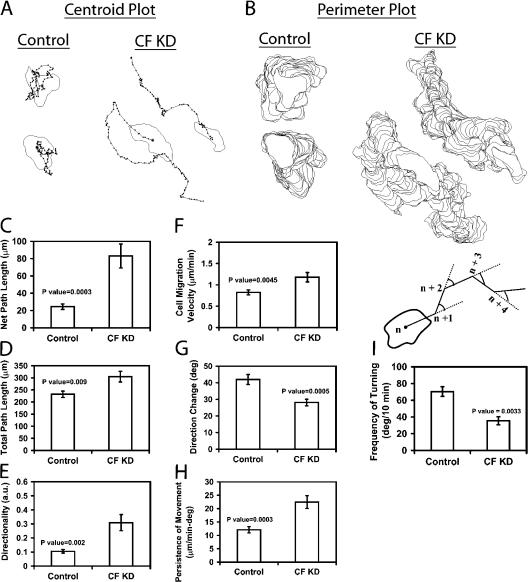
Because of the importance of cofilin in actin-based protrusions, we studied the effects of CF KD on MTLn3 cell motility. Using time-lapse microscopy, control and CF KD cells were imaged and the corresponding videos were analyzed using DIAS (Videos 1–3, available at http://www.jcb.org/cgi/content/full/jcb.200707009/DC1). Centroid plot analysis, which shows the position of the centroids of cells traced at consecutive time intervals, showed that CF KD cells followed a more linear path compared to the random walking path, typical of MTLn3 cells and control cells (Fig. 3 A). Additionally, stacked perimeter plots revealed that CF KD cells translocated at least five to six cell body lengths in 45 min in comparison to control cells, which traversed only one to two cell body lengths. Consistent with this analysis, CF KD cells also showed significant differences in both total and net path lengths (Fig. 3, C and D). Consequently, directionality, which is the ratio of net to total path length, was also significantly higher in CF KD cells (Fig. 3 E). Expression of the WT(ΔH) cofilin plasmid rescued this phenotype by reducing directionality to control levels (Fig. 2, A–D; available at http://www.jcb.org/cgi/content/full/jcb.200707009/DC1). These results strongly suggest that MTLn3 cells with lower cofilin levels are more directional than the control cells.
Figure 3.
Cofilin knockdown cells exhibit directional migration. (A) Centroid plots of representative control and CF KD cells show the trajectory of cells over time (4 h), analyzed using DIAS. Black dots represent path of cells over time (centroid). (B) Perimeter plots of cells in A shown as a stack of the shapes of the cells as they move over time. (C–H) Quantification of the motility parameters. Net path length is the net distance that the cells traversed from the first to the last frame; total path length is the total distance traversed by cells over time; directionality is the ratio of net to total path length, and persistence is the instantaneous velocity divided by direction change. Cartoon: quantification method of the turning frequency (degree of turning) of cells. Solid lines represent the path followed by cells between frames (every 10 min), and the broken lines represent the imaginary straight path, the cells would have followed if they did not turn. The angle of turning calculated is the angle between the followed path and the imaginary straight one. For C–H, number of cells scored: control (17), CF KD (16). (I) is the average of (11) control and (12) CF KD cells.
To pursue this, the direction change and persistence (speed/direction change) parameters were determined using DIAS. Results demonstrated that CF KD cells have lower direction change and are simultaneously more persistent in their movement than control cells (Fig. 3, G and H). The frequency of turning was calculated as the angle between the trajectories that the cells were projected to follow between consecutive frames if they moved in a straight path, and the actual path they followed during the time lapse (Fig. 3, cartoon). Analysis showed that CF KD cells had a significantly lower turning frequency than control cells (Fig. 3 I). Consistent with their higher directionality, CF KD cells also had higher cell migration velocities than control cells because they tended to more efficiently cover distance due to less frequent turns (Fig. 3 F). Overall, similar results were obtained whether cell movements were imaged for 45 min, 4 h, or 12 h, indicating that subtle changes in cell shape do not contribute to the analysis of directionality, persistence, and turning frequency (unpublished data).
In support of the CF KD phenotype in MTLn3 cells, analysis of LIMK overexpressing MTLn3 (F) cells showed that these cells have an elongated morphology and enhanced directionality similar to CF KD cells (Fig. S3, A–E; available at http://www.jcb.org/cgi/content/full/jcb.200707009/DC1). F cells also showed a directional motility behavior similar to the CF KD cells (Fig. S3, B, D, and d) when compared with control cells (Fig. S3, C and c) and cells expressing the kinase-dead domain of LIMK (KS) (Fig. S3, E and e). This illustrates that inactivating cofilin has the same general phenotype as reducing its expression.
Cofilin knockdown phenotype in mesenchymal-like non metastatic tumor cells is different from that in metastatic tumor cells
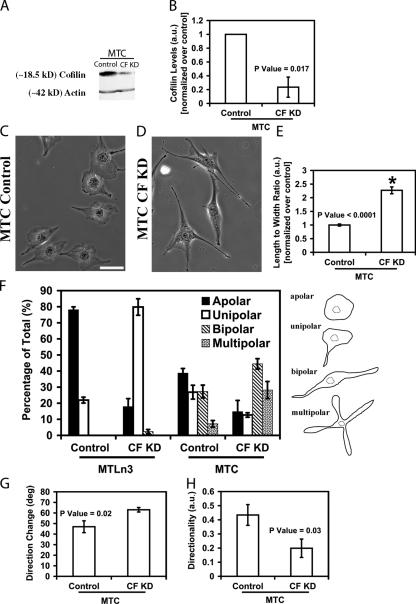
Using the same siRNA, we were able to knockdown cofilin levels ∼80% in MTC, nonmetastatic mammary adenocarcinoma cells (Fig. 4, A and B). MTC cells have a mesenchymal (fibroblast-like) type of motility exhibiting a well-developed unipolar leading edge with a large amount of F-actin and a characteristic rectilinear movement (Shestakova et al., 1999). Suppression of cofilin in MTC cells led to the formation of multipolar lamellipodia (Fig. 4, C–F) and caused a decrease in the directionality of migration consistent with the appearance of multiple lamellipodia (Fig. 4, F–H). These results suggest that cofilin is involved in the maintenance of directionality of migration in mesenchymal-like non-metastatic tumor cells, like that previously described for fibroblasts (Dawe et al., 2003), as compared with decreasing directionality by increasing turning frequency in metastatic amoeboid type tumor cells as described above for MTLn3 cells (Fig. 3, C–I).
Figure 4.
Cofilin knockdown changes the motility behavior of MTC cells. (A) Western blot of whole cell lysate samples taken from MTC cells treated either with oligofectamine (control), or cofilin siRNA (CF KD). (B) quantification of the levels of cofilin in MTC cells treated with cofilin siRNA. (C and D) phase images of control and CF KD MTC cells. Bar, 10 μm. (E) L/W of control and CF KD MTC cells; number of cells scored: control (197) and CF KD (178). F, scoring of the degree of polarity of MTLn3 and MTC cells. Cells were scored as shown in the cartoon on the right, and presented as percentage of total cells scored (n = 350 cells/group). (G and H) quantification of the motility parameters (using DIAS). G, direction change (deg); H, directionality; 8 control and CF KD cells were scored. A–F were analyzed at 36 h after siRNA transfection.
CF KD increases the persistence and stability, and decreases both the frequency and rate, of protrusions in MTLn3 cells
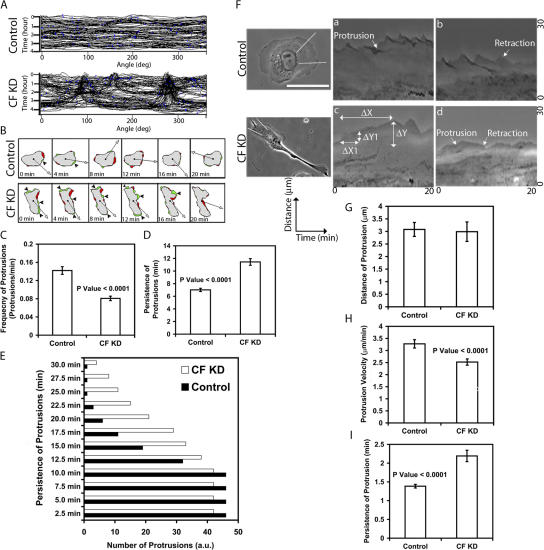
To understand the contributions of changes in protrusion to the phenotype observed in metastatic tumor cells following CF KD, we measured the protrusion dynamics in control and CF KD cells. Using shape and kymograph analysis we found that the protrusions in CF KD cells were concentrated in just a few areas of the cell membrane, whereas in control cells protrusions were observed with equal frequency around the entire perimeter of the cell (Fig. 5 A). Using DIAS, we followed protrusions (green) and retractions (red) in time-lapse videos (Fig. 5 B, arrows) and determined the frequency and persistence of these events. CF KD cells had more persistent but less frequent protrusions than control cells (Fig. 5, C and D). Additionally, the number of protrusions lasting for 10 min was similar in both groups; whereas CF KD cells tend to have more protrusions lasting for longer time intervals (Fig. 5 E).
Figure 5.
Importance of cofilin to the dynamics of protrusions in MTLn3 cells. (A) shape analysis of representative control, and CF KD cells, taken from a time lapse recorded over 4 h (y-axis), shown as unwrapped stacked shapes of cells. Unwrapping was done by angle conversion (x-axis) (see Materials and methods). (B) Tabular display of representative control and CF KD cells. Centroid of cells (black), protrusions (green), retractions (red), and direction of movement (arrow). Consecutive frames are shown as out takes from videos recorded over 4 h (time between frames shown is 4 min). Frequency (C), persistence (D), and lifetime (E) of protrusions were calculated by analyzing the protrusions (green) of cells like ones in B. C, frequency of protrusions (protrusions/min); D average persistence of these protrusions; E, quantification of the number of protrusions lasting for different time intervals. Number of protrusions analyzed in C: control (258) and CF KD cells, (327). For D and E, (13) cells were analyzed. F, phase images of control and CF KD cells. Bar, 10 μm. Two representative kymographs of control (a and b) and CF KD (c and d) are shown. ΔX = persistence of protrusions, ΔY = Distance of protrusions, and ΔY1/ΔX1 = protrusion velocity (rate). (G–I) Quantification of the dynamics of protrusions as measured by kymographs. Number of cells analyzed in G: Control (19 cells, 106 measurements), and CF KD (26 cells, 107 measurements). For H and I, number of cells analyzed: control (21 cells, 105 measurements), and CF KD (26 cells, 126 measurements).
Using kymography, we found that CF KD cells have protrusions of approximately the same size as control cells (Fig. 5 G) but that are more persistent (Fig. 5 I) and with lower velocities (Fig. 5, F and H). Expression of the WT(ΔH) cofilin plasmid in CF KD cells was able to rescue these changes seen in CF KD cells (Fig. S2, E–G; available at http://www.jcb.org/cgi/content/full/jcb.200707009/DC1). Combined, these results suggest that cofilin is important in inducing the formation of globally distributed protrusions with higher velocity in MTLn3 cells, and in helping to turn over the protrusions once formed.
Cofilin knockdown in MTLn3 cells inhibits chemotaxis by desensitizing the cell perimeter to directional stimulation
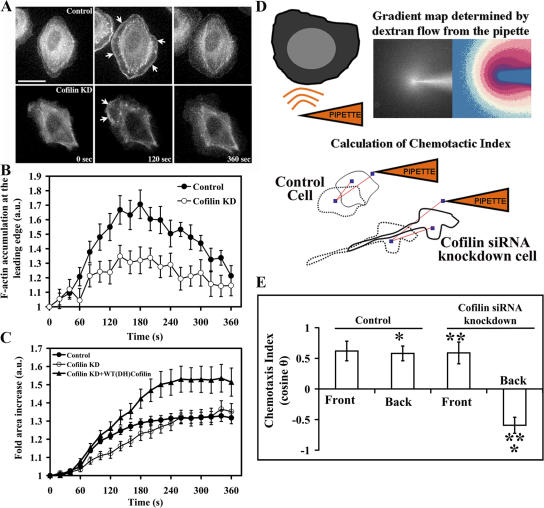
Amoeboid chemotaxis is a hallmark of metastatic mammary tumor cells like MTLn3 (Wyckoff et al., 2007). Thus, we studied the EGF-induced motility and chemotaxis of CF KD MTLn3 cells because EGF is a well-documented chemotactic stimulus in these cells (Mouneimne et al., 2004, 2006). Control and CF KD cells were starved for 3–4 h, and then were bath stimulated with 5 nM EGF. F-actin accumulation at the leading edge (Fig. 6, A and B), and the rate of protrusion were suppressed in CF KD cells (Fig. 6 C; Fig. S4, and Videos 4–6; available at http://www.jcb.org/cgi/content/full/jcb.200707009/DC1). The initial rate of lamellipod protrusion was rescued in CF KD cells that were cotransfected with the human cofilin plasmid (Fig. 6 C; Fig. S4 and Videos 4–6). These results are consistent with the lower turning rate of CF KD cells because lamellipod extension is a first step in the assignment of cell direction (Mouneimne et al., 2004) and fewer lamellipods per time might decrease turning frequency.
Figure 6.
Cofilin knockdown suppresses EGF-induced F-actin assembly and protrusion at the leading edge and the ability of the cells to chemotaxis. (A) representative control and CF KD GFP-actin expressing MTLn3 cells, over a range of 6 min; 0 min (no stimulation), 120 s and 360 s (after EGF stimulation). Arrows refer to global response along the membrane in the control panel compared with localized response mainly at the leading edge in the CF KD cells. Bar, 10 μm. (B) Quantification of F-actin levels at the leading edge after EGF stimulation. (C) Quantification of membrane protrusion, of control, CF KD cells, and CF KD cells + WT(ΔH) cofilin plasmid, after EGF stimulation (standardized over time 0). Number of cells analyzed: B, 15 cells; C, at least 21 cells/group. (D) Cartoon showing the pipette assay technique, and method of calculation of the chemotactic index. Chemotactic index is calculated as the cosine of the angle that is formed between the vector connecting the first and last centroid of the stimulated cells, and the vector between the first centroid and the tip of the pipette. E, quantification of the chemotactic indices of at least 11 cells/ category (control-front, control-back, CF KD-front, and CF KD-back). Cells were either stimulated at the front leading edge or at the back as determined by their shape and the presence of the pipette. *, P < 0.05; **, P < 0.01.
We used tropomyosin staining to determine if loss of cofilin perturbs or abolishes the lamellipodial actin network without affecting the lamellar network. Previous reports using quantitative fluorescent speckle microscopy in migrating epithelial cells found that there are two distinct F-actin regions; the lamellipodium which consists of a dynamic F-actin network and has high concentrations of Arp2/3 complex and cofilin, and the lamella which has myosin II and tropomyosin (Gupton et al., 2005). Tropomyosin has been shown to be a useful marker for distinguishing the lamellipod and lamella compartments (DesMarais et al., 2002). In this context, we analyzed the localization of tropomyosin, and found that CF KD did not abolish the lamellipod compartment (Fig. S4, B and C), consistent with previous work indicating that cofilin alone is not responsible for lamellipod formation (Mouneimne et al., 2004).
The ability of local activation of cofilin to initiate protrusion as described previously (Ghosh et al., 2004), and the requirement of cofilin for cell turning as described here, suggest that cofilin activity is required for protrusion which in turn is required for turning, i.e., setting cell direction. In other words, to chemotax efficiently a cell must be able to respond to chemotactic stimulation at any region on its surface and to turn in response to changes in direction of the chemotactic stimulus. To investigate this, cells were divided into front and back, as defined by the position of the pipette. In control cells this was random because there was no fixed front and back, whereas in CF KD cells the front of the cell was taken as the leading edge of the elongated cells (Fig. 6 D). Control cells exhibited similar sensitivity on all sides of the cell toward a pipette delivering a gradient of EGF (Fig. 6 E). However, CF KD cells were only able to protrude toward the pipette when it was at the front of the cell, but retracted away from the pipette when stimulated at the back (Fig. 6 E). These results indicate that cofilin is required for the ability of a cell to protrude uniformly around its periphery when stimulated with EGF.
The elongated phenotype of CF KD cells was intriguing in its resemblance to the elongated cell shape resulting from the enrichment of myosin II in the uropods of streaming Dictyostelium cells and migrating neutrophils (Zhang et al., 2002). Polarized myosin II localization to the uropod has been hypothesized to specify the rear of the cell and antagonize protrusion activity there. To investigate if myosin II is localized to the rear of CF KD cells thereby causing the elongation and unipolar protrusion phenotype, we stained CF KD MTLn3 cells for myosin IIA and IIB isoforms. As shown in Fig. S5 (available at http://www.jcb.org/cgi/content/full/jcb.200707009/DC1), myosin II is uniformly distributed in control and slightly concentrated in the front of CF KD cells. No accumulation of myosin II was detected in the rear of these elongated cells, and there was no difference between total myosin II levels between control and CF KD cells. Thus, the phenotype of CF KD cells could not be explained by myosin II accumulation at the rear of the cells.
Cofilin knockdown causes relocalization of Arp2/3 to one side of MTLn3 cells
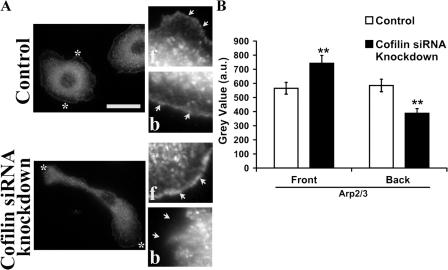
Because cofilin generates newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex in vitro (Ichetovkin et al., 2002), we investigated whether CF KD produced any change in the localization of the Arp2/3 complex in the cell. Interestingly, our results showed that CF KD led to the asymmetric localization of the Arp2/3 complex to the front of the elongated cell (Fig. 7, A and B). These results also showed that in control cells the level of Arp2/3 complex is the same in randomly chosen front and back regions.
Figure 7.
Cofilin knockdown causes relocalization of Arp2/3 to the front of the elongated MTLn3 cells. (A) Arp2/3 staining of control, and CF KD cells. Insets show the front (f) and the back (b) at higher magnification; asterisks represent the regions that are shown in the magnified images and white arrows refer to the leading edge of the cell. Bar, 10 μm. (B) 1-pixel-line intensity analysis of Arp2/3 complex staining in (30) control and (30) CF KD cells; **, P < 0.001. The front and back, were defined by drawing a line through the geometrical center of the cell. The regions were assigned randomly for control cells due lack of polarized shape, while for CF KD cells, the front was assigned to the leading edge of the cell, and the back was assigned to the tail of the cell (the end being geometrically opposite to the leading edge of the cell).
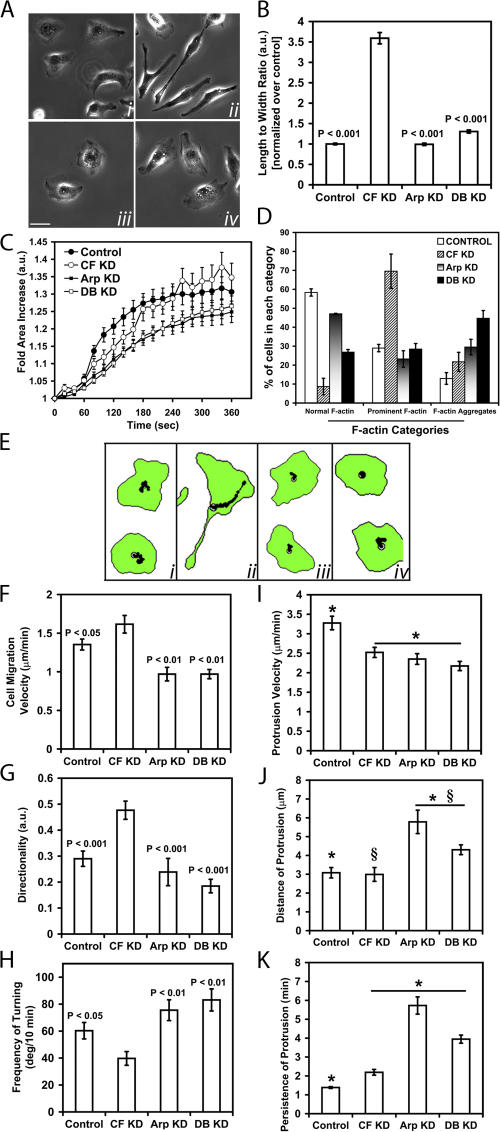
To determine if the asymmetric distribution of Arp2/3 complex in CF KD cells contributed to their elongated shape and directional migration, we suppressed the expression of Arp2/3 complex by siRNA knock down of the p34 subunit. This is sufficient to decrease the expression of other subunits of the complex (Fig. S1, F–H) and the activity of the complex (Kempiak et al., 2003). Although p34 knockdown (referred to hereafter as Arp KD) did not affect the shape and directionality of control cells, Arp KD reverted the elongated shape and directionality of CF KD cells to control levels (Fig. 8, A, B, and E–H).
Figure 8.
Arp2/3 complex is required for the shape and direction of migration of metastatic MTLn3 cells. (A) Representative images of control (i), CF KD (ii), Arp KD (iii), and DB KD (iv). Bar, 10 μm. (B) Quantification of the L/W of the groups in A. Number of cells scored: control (151), CF KD (214), Arp KD (62), and DB KD (203). (C) Quantification of membrane protrusion after EGF stimulation. Number of cells analyzed: control (24), CF KD (27), Arp KD (21), DB KD (23). (D) F-actin analysis, done as shown in Fig. 2. Quantification of the motility parameters: (E) centroid plots; (F) cell migration velocity and (G) directionality (using DIAS). For F and G, number of cells scored: control (35), CF KD cells (32), Arp KD (6), and DB KD (14). (H) Quantification of the turning frequency; number of cells scored: control (30), CF KD (30), Arp KD (20) and DB KD (20). (I–K) Quantification of the dynamics of protrusions as measured by kymography; number of cells analyzed in I: Control (19 cells, 106 measurements), CF KD (26 cells, 107 measurements), Arp KD (15 cells, 81 measurements), and DB KD (12 cells, 109 measurements). For J and K, number of cells analyzed: control (21 cells, 105 measurements), CF KD (26 cells, 126 measurements), and (12 cells, 123 measurements) for both Arp and DB KD groups. In I and K, the groups at the underlined asterisk are each significantly different from the control group. In J, Arp and DB KD are both different from both control and CF KD.
Additionally, we studied the rate of protrusion, stimulated by 5 nM EGF, of Arp KD and DB KD cells (CF KD in which p34 is also knocked down) and found that both types of treatments generated cells that show significant decreases in their protrusion in response to EGF (Fig. 8 C). However, Arp KD and DB KD cells were still able to protrude. The observation of protrusion activity in Arp KD and DB KD cells is consistent with previous work showing that Arp3-siRNA treated cells are still able to generate protrusions and migrate (Di Nardo et al., 2005) and the ability of p34 knockdown cells to protrude lamellipodia in MTLn3 cells in particular (unpublished data).
DIAS analysis of movies of DB KD cells (in serum) also showed reversion to the normal random-walking motility behavior and turning frequency of MTLn3 cells similar to that seen in control cells (Fig. 8, E–H) suggesting that Arp2/3 complex contributes to the directionality and decreased turning frequency seen in CF KD cells. Arp KD and DB KD cells also showed lowered cell migration velocities indicating that optimum cell migration requires the Arp2/3 complex (Fig. 8 G). Interestingly, Arp KD and DB KD cells did not show an increase in prominent F-actin structures like that seen in CF KD cells but had a significant increase in F-actin aggregates compared with control cells, suggesting that F-actin dynamics are altered in Arp KD cells (Fig. 8 D).
Using kymography, we found that the absence of Arp2/3 complex produced a slight decrease in protrusion velocity compared with CF KD cells (Fig. 8 I), whereas protrusions from both Arp KD and DBKD cells were more persistent (Fig. 8 K) and larger (Fig. 8 J) than in control or CF KD cells. This latter result is consistent with increased protrusion size seen in MTLn3 cells resulting from activation of mDia proteins after inhibition of Arp2/3 complex activity (unpublished data).
Effects of activated Rac on the cofilin knock down phenotype
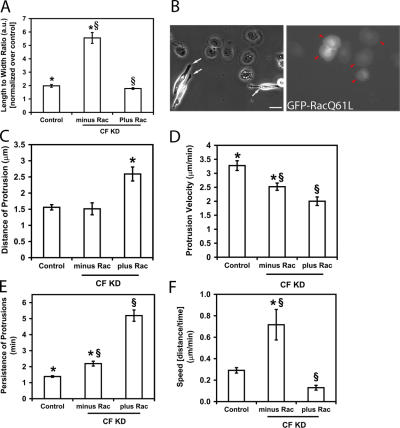
Regulation of the actin cytoskeleton by the Rho family of GTPases is an important factor in the regulation of cell protrusion dynamics (Nobes and Hall, 1995). By pull-down assays in CF KD cells, we found that there was no change in the activity of the Rho GTPases (unpublished data). Previous work on MTLn3 cells has shown that constitutively active Rac results in global lamellipod protrusion (El-Sibai et al., 2007), and that Rac is not required for the initiation of protrusions, but is for lamellipodial stability (Yip et al., 2007). Thus, we investigated if constitutively active Rac is able to affect the shape and motility phenotypes observed in CF KD cells. We found that the transfection of CF KD cells with a constitutively active Rac plasmid (GFP-RacQ61L) rescued their elongated phenotype (Fig. 9, A and B). Cells expressing the plasmid had lower length to width ratio (Fig. 9 B, red arrowheads) than CF KD cells that were not expressing the plasmid (Fig. 9 B, white arrowheads) resulting from the return to apolar protrusion activity. CF KD cells expressing the Rac mutant also showed reversion of the increase in speed seen in CF KD cells consistent with the renewed ability to protrude in all directions. GFP-RacQ61L expression increased the size and persistence of protrusions beyond that resulting from CF KD (Fig. 9, C and E) consistent with previous findings that Rac stabilizes protrusions in MTLn3 cells (Yip et al., 2007). In addition, the affects of activated Rac on the behavior of CF KD cells is consistent with a role for Arp2/3 complex in the motility phenotypes of CF KD cells.
Figure 9.
Activated Rac rescues the elongated phenotype of the cofilin siRNA knockdown cells. (A) Constitutively active GFP-RacQ61L was able to rescue the elongated phenotype of the CF KD cells. (A) L/W ratio of control and CF KD cells transfected with GFP-RacQ61L. CF KD Cells transfected with control GFP plasmid showed elongated phenotype compared with control cells treated with the same plasmid. (B) CF KD cells transfected with GFP-RacQ61L (red arrowheads in right panel), have normal phenotype as opposed to CF KD cells that are not transfected with the plasmid (white arrows in left panel). Bar, 10 μm. For A, at least 70 cells/group were analyzed. (C–E) Quantification of the dynamics of protrusions as measured by kymography (analyzed for cells in serum, as explained in Fig. 5 legend). (C) Distance of protrusion, (D) protrusion rate, and (E) persistence of protrusion.
Discussion
In an effort to understand the role of cofilin in protrusion dynamics, including the effects of cofilin on the stability and directionality of lamellipodial protrusion in metastatic cells in particular, we used siRNA to suppress the expression of cofilin. We found that cofilin suppression leads to the inhibition of actin polymerization, rate of protrusion and an increase in F-actin and its aggregation inside cells. These results are consistent with previous cofilin siRNA studies in other cell types (Hotulainen et al., 2005), and in MTLn3 cells (Zebda et al., 2000; Mouneimne et al., 2004). Further analysis showed that lamellipodial protrusions extend slowly but are more stable in the absence of cofilin. The Arp2/3 complex in cofilin suppressed cells was localized toward one end of the cell, consistent with their ability to protrude and move in only one direction. Knockdown of the Arp2/3 complex and expression of constitutively active Rac in CF KD cells suggest that Arp2/3 complex contributes to the CF KD phenotype
Cofilin, cell morphology, and directional migration in tumor cells
Our results support the involvement of cofilin in the initiation of lamellipodial protrusion and stability. Our results that cofilin suppression decreases the frequency and rate of protrusion but increases stability are consistent with the finding in Drosophila S2 cells, plated on concanavalin A, that suppression of cofilin increases lamellipodial stability (Iwasa and Mullins, 2007). However, unlike the requirement in S2 cells for the Arp2/3 complex (Rogers et al., 2003) but not cofilin in lamellipodial protrusion (Iwasa and Mullins, 2007), MTLn3 cells, like embryonic fibroblasts (Di Nardo et al., 2005), do not have an absolute requirement for Arp2/3 complex for lamellipodial protrusion, and have a requirement for cofilin for the efficient initiation of lamellipodial protrusion. These results emphasize that generalizations about protein function in cell motility from the study of a single cell type are not possible given the many alternative pathways that contribute to cell migration and the different physiological functions of different cell types that contribute different molecular backgrounds as starting points in the analysis. For example, suppression of cofilin in MTC cells, a non metastatic mesenchymal-type tumor cell with a high polarity of movement (Shestakova et al., 1999), led to the formation of multipolar lamellipodia and a decrease in the directionality of migration consistent with the appearance of multiple lamellipodia whereas the opposite was observed in MTLn3 cells. These results suggest that cofilin is involved in the maintenance of both the polar directionality of migration in mesenchymal-like non metastatic tumor cells, as in MTC cells and as previously described for polarized fibroblasts (Dawe et al., 2003), and also in the maintenance of the apolar high turning frequency type of migration characteristic of metastatic amoeboid MTLn3 tumor cells. Clearly the result of cofilin suppression is different in these two related cell types.
In terms of physiological background, MTLn3 and MTC cells are a metastatic and non metastatic cell pair, respectively, derived from the same tumor. MTLn3 cells are an example of amoeboid chemotactic tumor cells, whereas MTC cells are representative of mesenchymal tumor cells (Sahai and Marshall, 2003; Wolf et al., 2003). Major differences in mRNA targeting and dependence of migration in 3D ECM microenvironments on cell surface protease activity have been detected where MTLn3 cells fail to target mRNAs and exhibit protease independent migration while MTC cells target mRNA and require protease activity for their migration (Shestakova et al., 1999; Wyckoff et al., 2006). Failure to target mRNA by MTLn3 cells is correlated with random amoeboid movement and enhanced chemotaxis to EGF and both phenotypes can be suppressed by rescuing mRNA targeting by expression of ZBP1, which causes unipolar directional migration and inhibition of chemotaxis (Wang et al., 2004). ZBP1 is responsible for binding and targeting of β-actin mRNA and other functionally related mRNAs such as the Arp2/3 complex subunits to one location in the cell to define the leading edge (Huttelmaier et al., 2005; Mingle et al., 2005). Our finding here that the suppression of cofilin expression transforms the amoeboid behavior of metastatic MTLn3 cells into the mesenchymal behavior of non metastatic MTC cells suggests that cofilin is a good target for therapeutics that are directed toward the most aggressive tumor cells in mammary tumors, i.e., amoeboid tumor cells like MTLn3 (Condeelis and Segall, 2003; Sidani et al., 2006). This is supported by observations that the activity status of the cofilin pathway is directly correlated with metastatic potential in mammary tumors (Mouneimne et al., 2006; Wang et al., 2007a). Cofilin may be involved in mRNA targeting and/or defining the location of the Arp2/3 complex. Further work on how cofilin may affect mRNA targeting and cell surface protease activity, to affect in different ways the shape and migration phenotypes of MTLn3 and MTC cells, may be helpful in the design of such therapeutics.
A synergy between cofilin and Arp2/3 complex
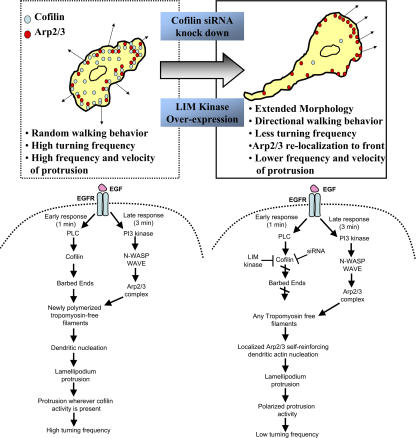
Previous reports have shown that there is a synergistic interaction between the Arp2/3 complex and cofilin which can contribute to lamellipodial extension (DesMarais et al., 2004). Our results indicate that CF KD causes a significant change in the localization of the Arp2/3 complex which becomes asymmetrically localized to one end of the cell. The relocalization of the Arp2/3 complex is involved in the directional motility behavior of the CF KD cells because the DB KD cells showed reversion to the apolar random walking motility characteristic of MTLn3 cells. This observation supports the synergy between cofilin and Arp2/3 complex reported previously in vitro where cofilin is observed as a first step to initiate actin polymerization, and then Arp2/3 complex-mediated dendritic nucleation follows on the cofilin-generated filaments as a later step (Ichetovkin et al., 2002). This is consistent with the observation in vivo that cofilin enters the leading edge earlier than the Arp2/3 complex immediately before the start of lamellipod extension (DesMarais et al., 2004). In the absence of cofilin, the Arp2/3 complex is confined to the sides of actin filaments that are not saturated with tropomyosin because tropomyosin inhibits the binding of Arp2/3 complex to F-actin (Blanchoin et al., 2001). This is predicted to generate a self-reinforcing accumulation of Arp2/3 activity in one location wherever tropomyosin-free mother filaments are initially located (Ichetovkin et al., 2002) unless there is some other factor like cofilin that can generate tropomyosin-free filaments throughout the periphery of the cell for use by Arp2/3 complex (Fig. 10).
Figure 10.
Cofilin sets the directionality of cancer cells. In comparing untreated (left) and cofilin suppressed (right) cells, current work and previous reports (Ghosh et al., 2004), have shown that cofilin is important for the motility for mammary cancer cells. This study shows that decreasing cofilin levels using siRNA or by LIM kinase overexpression, changes the cell “random walking” behavior resulting from frequent turning into a directional motility resulting from infrequent turning. The change in phenotype seen is related to the ability of cofilin to determine the location of Arp2/3 complex by supplying new filaments for dendritic nucleation (left). Cofilin suppression causes the localization of Arp2/3 complex to one region of the cell which becomes the front of the cell causing linear walking. This relocalization is determined by the availibilty of tropomyosin-free filaments in cofilin suppressed cells (right). Lower cofilin levels also caused changes in membrane protrusion dynamics; CF KD cells had lower frequency and velocity of protrusion than control cells.
These considerations may explain the sequence of actin polymerization transients observed in metastatic tumor cells such as MTLn3 (Mouneimne et al., 2004) after EGF stimulation. MTLn3 cells exhibit early and late barbed end transients in response to EGF stimulation where the early transient at 1 min is PLC and cofilin dependent, and the late transient at 3 min is PI3-kinase and Arp2/3 complex dependent (Chan et al., 2000; Mouneimne et al., 2004). These results are consistent with a role for cofilin in initiating actin polymerization that subsequently determines the location of Arp2/3 complex for polymerization and protrusion.
Consistent with the involvement of Arp2/3 complex in the CF KD phenotype was the finding that the expression of activated Rac was able to rescue the elongated phenotype of CF KD cells allowing the cells to protrude uniformly around the periphery resulting in the return to apolar protrusion morphology. This presumably resulted from the activation of the Arp2/3 complex by activated Rac thereby bypassing the need for cofilin in potentiating Arp2/3 activity around the cell periphery. Consistent with this hypothesis was the finding that the expression of activated Rac in CF KD cells also increased the size and stability of protrusions beyond that obtained by knocking down cofilin alone.
Materials and methods
Cell lines and plasmids
MTLn3 and MTC cells (rat mammary adenocarcinoma cell line) were maintained in α-MEM (GIBCO BRL) with 5% FBS, as described previously (Segall et al., 1996; Bailly et al., 1998). For the Rac rescue experiment, 18 h after administration of the cofilin siRNA, cells were transfected with the GFP-RacQ61L plasmid (a gift from Dr. Klaus Hahn; University of North Carolina, Chapel Hill, NC) for 18 h before cells were analyzed (36 h in total after siRNA transfection).
siRNA and transfection procedure
The non-silencing siRNA (AATTCTCCGAACGTGTCACGT), designed against a bacterial target sequence and the cofilin siRNA designed against (AAGGTGTTCAATGACATGAAA), were purchased from QIAGEN. The scrambled siRNA (AAGGTGTCTAATGACATAAAG), purchased from Ambion, was generated by scrambling the cofilin siRNA and blasted to assure the absence of any possible target sequence. Cells were transfected with the siRNA in the presence of oligofectamine (Invitrogen). The transfection was terminated after 4 h by using α-MEM containing 15% FBS. For the DB KD, both siRNAs were added simultaneously to the cells. The p34 siRNA was designed against the following sequence (AAGGAACTTCAGGCACACGGA). All experiments in the paper were performed 36 h after siRNA transfection.
Western blot analysis and antibodies
Whole cell lysates were prepared as described previously (Mouneimne et al., 2004). Anti-p34 antibody (Upstate Biotechnology), anti-Arp3 antibody (Santa Cruz Biotechnology), and anti-actin antibody were purchased from Sigma-Aldrich. Anti-cofilin (chicken IgY) antibody was raised against purified recombinant full-length rat cofilin. For N-WASP and WAVE2, specific monoclonal Ig rabbit antibodies were used (generous donation from Dr. Tadaomi Takenwa's Lab; Tokyo University, Tokyo, Japan). GTPases were pulled down from MTLn3 lysates, and their activity was detected using Western blotting as described (El-Sibai et al., 2007).
Microscopy, kymography, and immunofluorescence quantification
Pictures were taken using either a 20× NA 0.75 or 60× NA 1.4 infinity-corrected optics on a microscope (model IX170; Olympus) supplemented with a computer-driven cooled CCD camera and operated by IPLab Spectrum software (VayTek). For EGF stimulation, time-lapse series were recorded at a rate of 1 frame/10 s, and analyzed in ImageJ. For time lapse, cells were initially transfected on 100-mm culture dishes, split after 12 h onto glass-bottom dishes (MatTek Corporation). After 24 h, cells were then washed twice with Leibovitz's L15 medium (GIBCO BRL), and imaged in L15 (with 5% FBS) media overlaid with mineral oil on a 37°C stage, for 45 min or 12 h (at a rate of 2 frames/min). Analysis of more than 4 h was not feasible because during the time lapse, imaged cells divided or crawled out of the field of imaging. Data collected from movies captured for 45 min or 4 h, gave similar results, and conclusions. For the MTC cell line, cells were plated on collagen-coated glass bottom dishes for at least 24 h before imaging. For kymography, lines were used to generate kymographs (at least 3 lines were drawn/cell) from time-lapse series taken over 20 min, and acquired at 10 or 30 s intervals using 20 or 60× objectives.
For tropomyosin analysis, images were analyzed using a macro designed to measure cell edge fluorescence intensity developed by the Analytical Imaging Facility (AIF) of the Albert Einstein College of Medicine. Cells were traced and the macro automated the collection of pixel intensity in a perimeter of the cell starting at 1.98 μm outside the cell and extending to 4.18 μm inside the cell in 0.22-μm steps (background was automatically subtracted from the measured mean fluorescence intensity. For myosin II experiment, cells were stained with myosin II A and B antibodies (gift from Dr. Anne Bresnick).
F-actin analysis
Cells were fixed and stained using rhodamine-phalloidin. Actin analysis was performed as described previously (Zebda et al., 2000). In brief, cells were double-blind scored and subdivided into three categories: cells exhibiting normal F-actin, prominent F-actin structures (thicker stress fibers); and F-actin aggregates (cells containing F-actin aggregates). Control cells were analyzed in comparison to representative control cells, and CF KD cells were scored in comparison to representative CF KD cells.
Dynamic image analysis system (DIAS) parameter analysis
Time lapse video microscopy images were processed using Image J software. Cells were either traced in ImageJ, or using DIAS. Cell shape (Table I) and motility parameters (Figs. 3–5 ) were then calculated using DIAS, as described in (Wessels et al., 1998), and plotted in Excel. For Fig. 5, unwrapping was done by conversion from polar to rectangular coordinates (centroid of the cell being the origin). The unwrapped points are plotted with (X1,Y1) coordinates, where X is the point of the boundary of the shape and Y is the angle of the vector connecting the centroid to the boundary point.
Length to width ratio measurement
The length/width ratio (L/W) was calculated using ImageJ, as follows: length was scored as the Feret's diameter of the cell [the longest distance between any two points along the boundary] (Fig. 2 A, black line); width was scored as the secondary axis of the best fit ellipse of the circled cells (Fig. 2 A, white line).
Micropipette and the EGF upshift assays
The micropipette assay was performed as described previously (Mouneimne et al., 2004). Time-lapse series were taken using 20× objectives on a microscope and analyzed using Image J. Cells were divided into two areas, front and the back of the stimulated cells, defined in reference to shape of the elongated-CF KD cells. The front was designated as the leading edge of the elongated cell, while the back refers to the elongated tail of the cell. In case of control cells, which lack distinctive elongated front and back polarity, polarized cells were chosen to be consistent with the shape of the CF KD cells and also compared with control cells chosen at random. The EGF upshift assay was performed as described previously (Mouneimne et al., 2004). In brief, cells were starved in L15 medium (GIBCO BRL) supplemented with 0.35% BSA for 3 to 4 h. Cells were then stimulated with a bath application of 5 nM EGF (Invitrogen), treated at 37°C.
Online supplemental material
Fig. S1: analysis of the cofilin siRNA shows high efficiency in suppressing cofilin levels with specificity. Fig. S2: the directionality and persistence of CF KD cells can be rescued by reexpression of Human cofilin plasmid. Fig S3: perturbing cofilin levels by overexpression of LIMK in MTLn3 changes their motility behavior. Fig S4: cofilin knockdown suppresses EGF-induced protrusion, but does not affect the presence of the lamellipodium. Fig. S5: the distribution of myosin II isoforms in cofilin knockdown cells. Video 1: control MTLn3 cells exhibit random walking behavior. Video 2: cofilin siRNA knockdown cells show a directional motility behavior. Video 3: expression of the WT(ΔH) cofilin plasmid rescues the directional walking behavior characteristic of the cofilin siRNA knockdown cells. Video 4: upon EGF stimulation, control MTLn3 cells exhibit lamellipod extension and accumulation of F-actin at the leading edge of the lamellipod. Video 5: cofilin siRNA knockdown suppresses EGF-induced F-actin assembly and protrusion in MTLn3 cells. Video 6: expression of the WT(ΔH) cofilin plasmid rescues the suppression of the EGF-induced F-actin assembly and protrusion observed in the cofilin knockdown cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200707009/DC1.
Supplemental Material
Acknowledgments
The authors would like to thank late Dr. Bruce Terman for all his efforts and help in the design of the human cofilin plasmid. Thanks also to the members of the Gruss-Lipper Biophotonics Center at the Albert Einstein College of Medicine for their help, in particular: Mr. Michael Cammer, Mr. Dustin Grezick, and Ms. Rosana Leonard. Thanks also extended to members of the Condeelis Lab, for their valuable comments throughout the writing of the manuscript; in particular, Dr. Jacco van Rheenan, Dr. Antonia Patsialou, Ms. Marianela Arias, Ms. Wies van Roosmalen, and Mr. Mathew Oser for their help in the double-blind scoring. We would like to thank Dr. Wassim Abou Kheir for help with the p34 antibody and Dr. Amber Wells for her valuable discussion about the myosin II analysis and staining.
This work was funded by NIHGM38511 to J. Condeelis and CA100324 to J. Backer, and in part by HD-18577 to D. Soll. The authors also acknowledge the use of the W.M. Keck Dynamic Image Analysis Facility at the University of Iowa.
D. Wessels and G. Mouneimne contributed equally to this paper.
Abbreviations used in this paper: Arp2/3 complex, actin-related protein complex; CF KD, cofilin siRNA knockdown; DIAS, dynamic image analysis software; ZBP, zip code binding protein.
References
- Ambach, A., J. Saunus, M. Konstandin, S. Wesselborg, S.C. Meuer, and Y. Samstag. 2000. The serine phosphatases PP1 and PP2A associate with and activate the actin-binding protein cofilin in human T lymphocytes. Eur. J. Immunol. 30:3422–3431. [DOI] [PubMed] [Google Scholar]
- Andrianantoandro, E., and T.D. Pollard. 2006. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell. 24:13–23. [DOI] [PubMed] [Google Scholar]
- Bailly, M., L. Yan, G.M. Whitesides, J.S. Condeelis, and J.E. Segall. 1998. Regulation of protrusion shape and adhesion to the substratum during chemotactic responses of mammalian carcinoma cells. Exp. Cell Res. 241:285–299. [DOI] [PubMed] [Google Scholar]
- Bailly, M., F. Macaluso, M. Cammer, A. Chan, J.E. Segall, and J.S. Condeelis. 1999. Relationship between Arp2/3 complex and the barbed ends of actin filaments at the leading edge of carcinoma cells after epidermal growth factor stimulation. J. Cell Biol. 145:331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin, L., T.D. Pollard, and S.E. Hitchcock-DeGregori. 2001. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr. Biol. 11:1300–1304. [DOI] [PubMed] [Google Scholar]
- Carlier, M.F., V. Laurent, J. Santolini, R. Melki, D. Didry, G.X. Xia, Y. Hong, N.H. Chua, and D. Pantaloni. 1997. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136:1307–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, A.Y., M. Bailly, N. Zebda, J.E. Segall, and J.S. Condeelis. 2000. Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J. Cell Biol. 148:531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis, J., and J.E. Segall. 2003. Intravital imaging of cell movement in tumours. Nat. Rev. Cancer. 3:921–930. [DOI] [PubMed] [Google Scholar]
- Condeelis, J., R.H. Singer, and J.E. Segall. 2005. The great escape: when cancer cells hijack the genes for chemotaxis and motility. Annu. Rev. Cell Dev. Biol. 21:695–718. [DOI] [PubMed] [Google Scholar]
- Dawe, H.R., L.S. Minamide, J.R. Bamburg, and L.P. Cramer. 2003. ADF/cofilin controls cell polarity during fibroblast migration. Curr Biol. 13:252–257. [DOI] [PubMed] [Google Scholar]
- DesMarais, V., I. Ichetovkin, J. Condeelis, and S.E. Hitchcock-DeGregori. 2002. Spatial regulation of actin dynamics: a tropomyosin-free, actin-rich compartment at the leading edge. J. Cell Sci. 115:4649–4660. [DOI] [PubMed] [Google Scholar]
- DesMarais, V., F. Macaluso, J. Condeelis, and M. Bailly. 2004. Synergistic interaction between the Arp2/3 complex and cofilin drives stimulated lamellipod extension. J. Cell Sci. 117:3499–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nardo, A., G. Cicchetti, H. Falet, J.H. Hartwig, T.P. Stossel, and D.J. Kwiatkowski. 2005. Arp2/3 complex-deficient mouse fibroblasts are viable and have normal leading-edge actin structure and function. Proc. Natl. Acad. Sci. USA. 102:16263–16268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sibai, M., P. Nalbant, H. Pang, R.J. Flinn, C. Sarmiento, F. Macaluso, M. Cammer, J.S. Condeelis, K.M. Hahn, and J.M. Backer. 2007. Cdc42 is required for EGF-stimulated protrusion and motility in MTLn3 carcinoma cells. J. Cell Sci. 120:3465–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, M., X. Song, G. Mouneimne, M. Sidani, D.S. Lawrence, and J.S. Condeelis. 2004. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 304:743–746. [DOI] [PubMed] [Google Scholar]
- Gohla, A., J. Birkenfeld, and G.M. Bokoch. 2005. Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat. Cell Biol. 7:21–29. [DOI] [PubMed] [Google Scholar]
- Gupton, S.L., K.L. Anderson, T.P. Kole, R.S. Fischer, A. Ponti, S.E. Hitchcock-DeGregori, G. Danuser, V.M. Fowler, D. Wirtz, D. Hanein, and C.M. Waterman-Storer. 2005. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. J. Cell Biol. 168:619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen, P., E. Paunola, M.K. Vartiainen, and P. Lappalainen. 2005. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol. Biol. Cell. 16:649–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier, S., D. Zenklusen, M. Lederer, J. Dictenberg, M. Lorenz, X. Meng, G.J. Bassell, J. Condeelis, and R.H. Singer. 2005. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 438:512–515. [DOI] [PubMed] [Google Scholar]
- Ichetovkin, I., W. Grant, and J. Condeelis. 2002. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 12:79–84. [DOI] [PubMed] [Google Scholar]
- Iwasa, J.H., and R.D. Mullins. 2007. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr. Biol. 17:395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis, J., J. Moellinger, H. Lo, N.M. Greenberg, H.G. Kim, and A. Wells. 1999. A role for phospholipase C-gamma-mediated signaling in tumor cell invasion. Clin. Cancer Res. 5:2251–2260. [PubMed] [Google Scholar]
- Kempiak, S.J., S.C. Yip, J.M. Backer, and J.E. Segall. 2003. Local signaling by the EGF receptor. J. Cell Biol. 162:781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver, S.K., and P.J. Hussey. 2002. The ADF/cofilin family: actin-remodeling proteins. Genome Biol. 3:reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meberg, P.J., S. Ono, L.S. Minamide, M. Takahashi, and J.R. Bamburg. 1998. Actin depolymerizing factor and cofilin phosphorylation dynamics: response to signals that regulate neurite extension. Cell Motil. Cytoskeleton. 39:172–190. [DOI] [PubMed] [Google Scholar]
- Mingle, L.A., N.N. Okuhama, J. Shi, R.H. Singer, J. Condeelis, and G. Liu. 2005. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J. Cell Sci. 118:2425–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, K., I. Okano, K. Ohashi, K. Nunoue, K. Kuma, T. Miyata, and T. Nakamura. 1994. Identification of a human cDNA encoding a novel protein kinase with two repeats of the LIM/double zinc finger motif. Oncogene. 9:1605–1612. [PubMed] [Google Scholar]
- Mouneimne, G., L. Soon, V. DesMarais, M. Sidani, X. Song, S.C. Yip, M. Ghosh, R. Eddy, J.M. Backer, and J. Condeelis. 2004. Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation. J. Cell Biol. 166:697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimne, G., V. Desmarais, M. Sidani, E. Scemes, W. Wang, X. Song, R. Eddy, and J. Condeelis. 2006. Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr. Biol. 16:2193–2205. [DOI] [PubMed] [Google Scholar]
- Niwa, R., K. Nagata-Ohashi, M. Takeichi, K. Mizuno, and T. Uemura. 2002. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 108:233–246. [DOI] [PubMed] [Google Scholar]
- Nobes, C.D., and A. Hall. 1995. Rho, rac and cdc42 GTPases: regulators of actin structures, cell adhesion and motility. Biochem. Soc. Trans. 23:456–459. [DOI] [PubMed] [Google Scholar]
- Okano, I., J. Hiraoka, H. Otera, K. Nunoue, K. Ohashi, S. Iwashita, M. Hirai, and K. Mizuno. 1995. Identification and characterization of a novel family of serine/threonine kinases containing two N-terminal LIM motifs. J. Biol. Chem. 270:31321–31330. [DOI] [PubMed] [Google Scholar]
- Ono, S. 2007. Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int. Rev. Cytol. 258:1–82. [DOI] [PubMed] [Google Scholar]
- Rogers, S.L., U. Wiedemann, N. Stuurman, and R.D. Vale. 2003. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162:1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai, E., and C.J. Marshall. 2003. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 5:711–719. [DOI] [PubMed] [Google Scholar]
- Segall, J.E., S. Tyerech, L. Boselli, S. Masseling, J. Helft, A. Chan, J. Jones, and J. Condeelis. 1996. EGF stimulates lamellipod extension in metastatic mammary adenocarcinoma cells by an actin-dependent mechanism. Clin. Exp. Metastasis. 14:61–72. [DOI] [PubMed] [Google Scholar]
- Shestakova, E.A., J. Wyckoff, J. Jones, R.H. Singer, and J. Condeelis. 1999. Correlation of beta-actin messenger RNA localization with metastatic potential in rat adenocarcinoma cell lines. Cancer Res. 59:1202–1205. [PubMed] [Google Scholar]
- Sidani, M., J. Wyckoff, C. Xue, J.E. Segall, and J. Condeelis. 2006. Probing the microenvironment of mammary tumors using multiphoton microscopy. J. Mammary Gland Biol. Neoplasia. 11:151–163. [DOI] [PubMed] [Google Scholar]
- Toshima, J., J.Y. Toshima, K. Takeuchi, R. Mori, and K. Mizuno. 2001. Cofilin phosphorylation and actin reorganization activities of testicular protein kinase 2 and its predominant expression in testicular Sertoli cells. J. Biol. Chem. 276:31449–31458. [DOI] [PubMed] [Google Scholar]
- Wang, W., S. Goswami, K. Lapidus, A.L. Wells, J.B. Wyckoff, E. Sahai, R.H. Singer, J.E. Segall, and J.S. Condeelis. 2004. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 64:8585–8594. [DOI] [PubMed] [Google Scholar]
- Wang, W., G. Mouneimne, M. Sidani, J. Wyckoff, X. Chen, A. Makris, S. Goswami, A.R. Bresnick, and J.S. Condeelis. 2006. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J. Cell Biol. 173:395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., R. Eddy, and J. Condeelis. 2007. a. The cofilin pathway in breast cancer invasion and metastasis. Nat. Rev. Cancer. 7:429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., J.B. Wyckoff, S. Goswami, Y. Wang, M. Sidani, J.E. Segall, and J.S. Condeelis. 2007. b. Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Res. 67:3505–3511. [DOI] [PubMed] [Google Scholar]
- Wessels, D., E. Voss, N. Von Bergen, R. Burns, J. Stites, and D.R. Soll. 1998. A computer-assisted system for reconstructing and interpreting the dynamic three-dimensional relationships of the outer surface, nucleus and pseudopods of crawling cells. Cell Motil. Cytoskeleton. 41:225–246. [DOI] [PubMed] [Google Scholar]
- Wolf, K., I. Mazo, H. Leung, K. Engelke, U.H. von Andrian, E.I. Deryugina, A.Y. Strongin, E.B. Brocker, and P. Friedl. 2003. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 160:267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff, J.B., S.E. Pinner, S. Gschmeissner, J.S. Condeelis, and E. Sahai. 2006. ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr. Biol. 16:1515–1523. [DOI] [PubMed] [Google Scholar]
- Wyckoff, J.B., Y. Wang, E.Y. Lin, J.F. Li, S. Goswami, E.R. Stanley, J.E. Segall, J.W. Pollard, and J. Condeelis. 2007. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 67:2649–2656. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, H., and J. Condeelis. 2007. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim. Biophys. Acta. 1773:642–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip, S.C., M. El-Sibai, S.J. Coniglio, G. Mouneimne, R.J. Eddy, B.E. Drees, P.O. Neilsen, S. Goswami, M. Symons, J.S. Condeelis, and J.M. Backer. 2007. The distinct roles of Ras and Rac in PI 3-kinase-dependent protrusion during EGF-stimulated cell migration. J. Cell Sci. 120:3138–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa, N., E. Nishida, K. Iida, I. Yahara, and H. Sakai. 1990. Inhibition of the interactions of cofilin, destrin, and deoxyribonuclease I with actin by phosphoinositides. J. Biol. Chem. 265:8382–8386. [PubMed] [Google Scholar]
- Yonezawa, N., Y. Homma, I. Yahara, H. Sakai, and E. Nishida. 1991. A short sequence responsible for both phosphoinositide binding and actin binding activities of cofilin. J. Biol. Chem. 266:17218–17221. [PubMed] [Google Scholar]
- Zebda, N., O. Bernard, M. Bailly, S. Welti, D.S. Lawrence, and J.S. Condeelis. 2000. Phosphorylation of ADF/cofilin abolishes EGF-induced actin nucleation at the leading edge and subsequent lamellipod extension. J. Cell Biol. 151:1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, Q., J.R. Bamburg, and J.A. Badwey. 2003. Products of phosphoinositide specific phospholipase C can trigger dephosphorylation of cofilin in chemoattractant stimulated neutrophils. Cell Motil. Cytoskeleton. 54:1–15. [DOI] [PubMed] [Google Scholar]
- Zhang, H., D. Wessels, P. Fey, K. Daniels, R.L. Chisholm, and D.R. Soll. 2002. Phosphorylation of the myosin regulatory light chain plays a role in motility and polarity during Dictyostelium chemotaxis. J. Cell Sci. 115:1733–1747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.