Abstract
Frequent allelic loss at human chromosome 11q23-q24 occurs in a wide variety of cancers, suggesting that this region may harbor a tumor suppressor gene. By constructing a physical map of the LOH11CR2 minimal region of loss on 11q23-q24 associated with lung and breast carcinomas, we were able to clone a hereditary translocation, t(11;12)(q23;q24), in a patient with early-onset breast cancer and family history of cancer. The breakpoint was found within 6 kb of the BCSC-1 candidate tumor suppressor gene located in the LOH11CR2 region whereas additional loss of heterozygosity (LOH) analysis in breast and ovarian tumors, including that of the patient with the t(11;12)(q23;q24), implicated the BCSC-1 locus as the primary target of deletion. Northern analysis of the BCSC-1 mRNA revealed a lack of expression in 33 of 41 (80%) tumor cell lines, and its ectopic expression led to the suppression of colony formation in vitro and tumorigenicity in vivo. These data suggest that BCSC-1 may exert a tumor suppressor activity and is a likely target of the LOH observed on 11q23-q24 in cancer.
Chromosome 11q23-q24 has been identified as a region frequently deleted in a variety of solid tumors including, but not limited to, breast (1–7), lung (8–10), and ovarian cancer ranging from 45–63%, suggesting that a putative tumor suppressor gene exists at this locus. Functional evidence supporting the existence of a tumor suppressor gene(s) is also provided by microcell fusion experiments involving the transfer of partial regions of normal chromosome 11q into a variety of tumorigenic cells lines, including breast, lung, cervical, and ovarian cancers (11). In addition, the introduction of normal regions of chromosome 11q, corresponding to regions of frequent loss of heterozygosity (LOH), had the ability to suppress in vitro growth as well as in vivo growth in nude mice (12, 13). Whereas the distal half of chromosome 11q contains several genes implicated in tumorigenesis such as the ATM gene (14), ALL-1 (15), PPP2R1B (16, 17), and CHEK1 (18), none of these displays a high frequency of gene inactivation in human tumors suggesting that other candidate tumor suppressors exist in the region.
We have previously found that at least two independent regions of LOH at 11q23-q24 are present in breast cancer: a 6- to 7-Mb region between loci D11S2000 and D11S897 and a second region between D11S528 and D11S990 (3). The first region contains the ATM gene associated with the genetic disorder ataxia telangiectasia and an increased risk of cancer (14, 19, 20). A second, more telomeric region, LOH region 2 (LOH11CR2), is defined by the loci D11S528 and D11S990 in breast carcinoma (3) and minimally defined by the loci D11S1345 and D11S1328 in lung adenocarcinoma (9). LOH frequencies at LOH11CR2 ranged from 40–60% in breast and lung cancers, as well as in tumors of the cervix (21), of the ovary (22), and of the thyroid (23), suggesting that genes found within this region could reasonably be considered candidate tumor suppressor genes whose inactivation may be required in many of these tumors.
To identify possible candidate tumor suppressor genes in LOH11CR2, we constructed a physical and sequence map defined by the microsatellite markers D11S1345 and D11S934 containing the LOH11CR2 critical region on chromosome 11q23-q24. In addition, we present the identification of an inherited constitutional translocation, t(11;12)(q24;q23), in a family characterized by early-onset breast cancer. Positional cloning of the translocation breakpoint places it within the LOH11CR2 region and 6 kb upstream of the BCSC-1 gene, also known as LOH11CR2A (24). Whereas previous results failed to implicate this gene as a tumor suppressor, additional data presented here point to a role of the BCSC-1 gene in tumorigenesis and as possibly being the critical tumor suppressor gene located within the LOH11CR2 region at chromosome 11q23-q24.
Materials and Methods
Patient Samples, Tumor Specimens, and Cell Lines. Peripheral blood specimens and archival material were obtained with informed consent. Cytogenetic analysis was carried out by using standard procedures according to the International System for Cytogenetic Nomenclature (ISCN) (25). Patient peripheral blood lymphocytes were isolated by Ficoll gradient (Pharmacia) and used for the isolation of RNA and DNA as well as the establishment of an Epstein–Barr virus-transformed lymphoblastoid cell line by standard methods (26). Somatic cell hybrids of peripheral blood lymphocytes isolated from the proband and the mouse cell line LMTK– (ATCC no. CCL-1.3) were created as described (27). All mouse and human tumor cell lines were obtained from the American Type Culture Collection (ATCC) and maintained in either RPMI medium 1640 containing 10% FBS or DMEM containing 10% FBS (GIBCO), each supplemented with 100 μg/ml gentamicin (BioWhittaker).
LOH Analysis. Matched normal and tumor genomic DNA pairs (28) were analyzed for LOH by the amplification of dinucleotide or tetranuclueotide repeats by using fluorescently end-labeled primers. Primer sequences for each highly polymorphic (>70%) microsatellite marker (Table 1) are available at the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov). Markers were selected due to their location in LOH11CR2 (3, 9) and their existence in a well defined region of loss associated with cervical carcinomas (21). PCR amplifications and analysis were performed according to Cesari et al. (28).
Table 1. Frequency of LOH in cases of sporadic breast and ovarian carcinomas observed for markers at the 11q23–q24 locus.
| No. of cases with LOH/no. of informative cases (% LOH)
|
|||
|---|---|---|---|
| Locus (% HET)*† | Breast | Ovary | Total |
| D11S4094 (71) | 5/14 (36) | 3/11 (27) | 9/25 (36) |
| D11S1353 (74) | 5/14 (36) | 5/16 (31) | 10/30 (33) |
| D11S4144 (75) | 5/19 (26) | 2/16 (13) | 7/35 (20) |
| D11S1328 (64) | 7/16 (44) | 5/13 (38) | 12/29 (41) |
| D11S933 (82) | 5/15 (33) | 1/16 (6) | 6/31 (19) |
| D11S934 (89) | 2/14 (14) | 3/18 (17) | 5/32 (16) |
Marker sequences available at www.ncbi.nlm.nih.gov.
Marker heterozygosities obtained from www.cephb.fr.
Tumorigenicity in Nude Mice. Polyclonal populations of A549, ES-2, and H460 cells transduced with and without the BCSC-1 cDNA were evaluated for tumorigenicity in nude mice. Six-week-old female athymic mice nude (nu/nu) were given s.c. injections of 1–10 × 106 cells in 0.2 ml of PBS. Cells transduced with each construct were injected into three to four mice each and followed individually. Mice were examined two to three times per week for tumor formation at the sites of injection. Tumors were measured with linear calipers, and tumor volumes were calculated (v = ab2/2).
Results
Cloning of a Hereditary, Constitutional t(11;12)(q24;q23) in a Family with Breast and Brain Cancer. An unreported pedigree with a predisposition to cancer was ascertained (Fig. 1A). The proband (III:1) is a Caucasian female whose medical history includes recurrent miscarriages, a constitutional translocation, and cancer of the left breast at age 42. Therapeutic mastectomy showed the histopathology of an infiltrating lobular carcinoma with additional areas of lobular carcinoma in situ. The tumor was histologically determined to be estrogen receptor negative and progesterone receptor positive, and her karyotype revealed the presence of a constitutional t(11;12)(q24;q23) (Fig. 1 A). A history of glioblastoma multiforme was identified in the proband's mother (II:1), with disease onset occurring at age 69, and a maternal aunt (II:4) reportedly developed breast cancer also at the age of 42. II:4 also has the constitutional t(11;12)(q24;q23) and has been in remission. Cytogenetic analysis confirmed a history of the constitutional translocation t(11;12)(q24;q23) in one of the proband's sisters (III:3), who was healthy at age 44, and one of her daughters (IV:2), also healthy at age 14. Although no genetic material was available for testing, pedigree analysis indicates that individual II:1 is an obligate carrier of the t(11;12)(q24;q23) and I:1 or I:2 as the origin of this heritable genetic lesion.
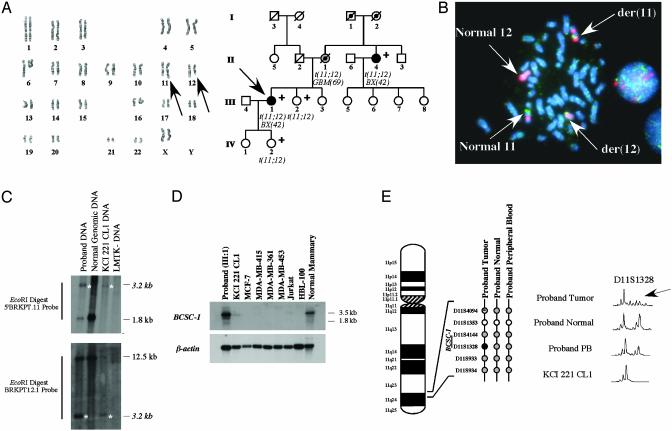
Fig. 1.
Cloning and characterization of the constitutional translocation, t(11;12)(q24;q23), in a family with breast and brain cancer. (A Left) The constitutional t(11, 12)(q24;q23) in an individual with early onset breast cancer (III:1). Karyotype analysis was performed by using standard GTG-banding techniques and the der(11) t(11;12)(q24;q23) and der(12) t(11;12)(q24;q23) are designated by arrows. (Right) Pedigree of a family with early onset breast cancer and brain cancer with onset of 69 years of age. The proband (III:1) is indicated by an arrow whereas individuals available for molecular and cytogenetic testing are indicated with a plus sign. Identified constitutional translocation carriers are designated with t(11;12), and the obligate carriers are marked with a filled black circle. (B) Fluorescence in situ hybridization analysis of the t(11;12)(q24;q23) using the BAC clone 44d21. FITC-labeled BAC44d21 spans the translocation as shown by the presence of signals (green) present on both der(11) t(11;12)(q24;q23) and der(12) t(11;12)(q24;q23). Rhodamine-labeled (red) chromosome 12 whole chromosome probe and chromosome 11 alpha-satellite probe were used as a reference. (C) Southern blot analysis of the t(11;12)(q24;q23). A 3.2-kb aberrant EcoRI fragment (asterisks) was identified by using the 5′BRKPT probe. Confirmation of the sequence obtained from cloning of the 3.2-kb EcoRI fragment was performed by using a chromosome 12-specific probe, BRKPT12.1, derived from the aberrant sequence. (D) Northern analysis of the BCSC-1 gene. (E) LOH analysis of 11q23-q24 in an individual with early onset breast cancer and a constitutional t(11;12)(q24;q23). Six microsatellite markers from the LOH11CR2 region were used to analyze microdissected normal/tumor and peripheral blood lymphocytes (PB) DNA from individual III:1 and the somatic cell hybrid clone KCI 221 CL1 DNA. Noninformative loci are indicated by open circles, retention of heterozygosity by gray circles, and loss of heterozygosity by a filled circle. Results of the marker D11S1328 are shown and loss of the normal chromosome 11 allele is indicated by an arrow.
Because of its association with early-onset breast cancer and glioblastoma, we hypothesized that the t(11;12)(q24;q23) chromosomal translocation breakpoint could be responsible for the disruption or inactivation of a tumor suppressor locus on chromosome 11q23-q24. Fluorescence in situ hybridization analysis using bacterial artificial chromosome (BAC) clones from the D11S1284-D11S933 interval (Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org.) was performed on metaphase chromosomes from a lymphoblastoid cell line derived from the proband. Subsequently, the BAC clone 44d21 (BAC44d21) was found to cover the breakpoint thereby localizing it to within a cloned 220-kb region (Fig. 1B).
Subsequently, Southern blot analysis was used to identify novel genomic fragments amenable for cloning that were the consequence of the t(11;12)(q24;23) fusion localized to within the 5′BRKPT10–5′BRKPT11 interval (See Supporting Materials and Methods, Supporting Data, and Fig. 6, which are published as supporting information on the PNAS web site). The 5′BRKPT11 PCR product was used to probe EcoRI-digested genomic DNA obtained from the proband, which identified an aberrant fragment of ≈3.2 kb (Fig. 1C). The aberrant 3.2-kb EcoRI band was cloned into lambda phage and sequenced (see Fig. 7, which is published as supporting information on the PNAS web site). blast analysis identified homology at the 5′ end to the chromosome 12q BAC RP11-864A19 (GenBank no. AC012083), and a probe generated for chromosome 12 sequence confirmed the isolation of t(11;12)(q24;q23) breakpoint (Fig. 1C). In addition, PCR primers derived from the chromosome 11 and 12 ends of the fragment were used to amplify across the der(12) junction in both the proband and the affected maternal aunt (Fig. 6). Direct sequencing of these PCR products demonstrated that the sequence of the t(11;12)(q24;q23) breakpoint is identical in both individuals (data not shown).
blast comparisons indicated that the translocation did not disrupt any known gene locus either side of the breakpoint and placed the translocation break ≈6 kb centromeric to exon 1A of the BCSC-1 gene located at chromosome 11q23-q24 (Fig. 6).
Whereas PCR analysis followed by direct sequencing of proband and somatic cell hybrid KCI 221 CL1 cDNA did not identify any mutation of the BCSC-1 gene, the proximity with the translocation breakpoint affected BCSC-1 expression. Northern blotting of RNA derived from the lymphoblastoid cell line derived from the proband's peripheral blood lymphocytes revealed the presence of the expected BCSC-1 transcripts (Fig. 1D), but the somatic cell hybrid KCI 221 CL1, containing only human der(12), showed relatively low expression of BCSC-1 transcript, possibly due to cross-reactivity with the endogenous murine bcsc-1 homolog of the parental mouse line LMTK–.
Because tumor suppressor genes are generally biallelically inactivated in tumor cells, we verified whether the LOH11CR2 region was deleted in the tumor of the proband. LOH analysis was performed on DNA from microdissected archival material, peripheral blood, the somatic cell hybrid clone KCI 221 CL1, and the mouse parental line LMTK–. Four of six 11q23-q24 markers were found to be informative in the proband tumor sample (Fig. 1E). LOH was observed only for the D11S1328, a marker located 10 kb from the 3′ end of the BCSC-1 locus, suggesting that it is specifically deleted in tumor cells. Further inspection revealed that the retained allele at D11S1328 is that of der(12), as indicated by its amplification in the somatic cell hybrid clone KCI 221 CL1 (Fig. 1E).
Characterization of the Human BCSC-1 Gene. The BCSC-1 (also known as LOH11CR2A) gene was previously identified as part of an effort to identify putative tumor suppressor genes that were localized to the frequently deleted 11q23 region (24). The cloned cDNA (GenBank no. AF002672) was sequenced and found to contain an ORF of 1,314 bp encoding a single protein with a predicted molecular mass of 48 kDa, with no significant homologies to known proteins. The sequence of the coding region was found to contain 10 exons, with at least two additional exons in the 5′UTR. Northern analysis revealed a single 3.3-kb transcript ubiquitously expressed at low levels. However, our analysis shows the genomic structure of the BCSC-1 gene to be comprised of at least 20 exons across 27 kb and with the most 3′ exon located 10 kb centromeric to the marker D11S1328 (Fig. 2A). Comparison of the AF002672 cDNA sequence to our reference sequence revealed the absence of a single adenine base pair at nucleotide position 915 in the published sequence (data not shown). This result was confirmed by sequencing of the BCSC-1 cDNA in several normal human tissues and normal human genomic DNA samples. With the insertion of an adenine nucleotide, the original ORF is extended to 2,361 bp encoding for a predicted protein of 786 aa and a molecular mass of 86 kDa.
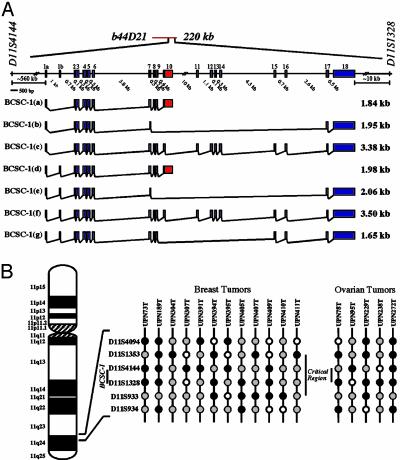
Fig. 2.
Molecular characterization of the BCSC-1 gene at human chromosome 11q23-q24. (A) Transcription map of the human BCSC-1 gene located within the LOH11CR2 critical region on 11q23-q24 (see Supporting Data). (B) LOH analysis of chromosome 11q23-q24 in breast and ovarian cancer. Summary of informative cases with LOH in at least one of the six loci examined. Filled black circles, loss of heterozygosity; filled gray circles, retention of heterozygosity; open circles, not informative (homozygous).
Northern analysis using the 2.3-kb ORF as a probe demonstrated the existence of at least two additional transcripts that are expressed at various levels in all normal human tissues examined (Fig. 3A). EST sequences derived from the BCSC-1 unigene cluster (Hs.152944) and direct sequencing of the IMAGE clone 1461599 also supported the existence of multiple alternative splice variants. Through EST alignment and assembly, cDNA sequencing, and 5′ and 3′ RACE, we were able to determine the structure of the various BCSC-1 splicing variants (see Supporting Data). Our analysis revealed that seven mRNA splice variants (a to g in Fig. 2 A) exist and are compatible with the observed BCSC-1 transcripts detected by Northern blot (Fig. 3).
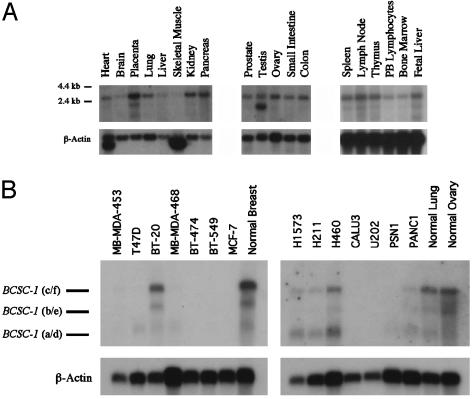
Fig. 3.
Northern blot analysis of the human BCSC-1 gene in normal and cancer-derived tissues. (A) BCSC-1 expression in normal human tissues. A probe containing the entire coding region of the BCSC-1 gene was used to detect its expression in a panel of normal tissues by Northern blotting. (B) BCSC-1 gene expression in human tumor-derived cell lines. Representative Northern blots are shown, and expected transcripts are indicated by each dash. β-Actin was used as a control.
Each of the BCSC-1 splicing variants are predicted to encode for four different proteins where all of the proteins share the same N terminus while terminating at different points along the C terminus (Fig. 8, which is published as supporting information on the PNAS web site). Comparisons of the predicted amino acid sequences failed to reveal any significant matches to known proteins, but conserved protein domains were identified. The first domain is a Vault protein Inter-alpha-Trypsin Inhibitor domain, which is located in the amino terminus of all BCSC-1 isoforms whereas the second, the von Willebrand Factor Type A domain, is present only in the BCSC-1(a/d), BCSC-1(c/f), and BCSC-1(g) variants (Fig. 8).
To confirm the structural features of the human BCSC-1 gene, we cloned the murine homologue. The mouse gene spans a genomic region of 25 kb and is comprised of 20 exons (see Supporting Data and Fig. 9, which is published as supporting information on the PNAS web site). Unlike the human BCSC-1 gene, it is expressed as a single mRNA transcript of ≈4 kb and encodes for a 793-aa protein with a predicted molecular mass of 87 kDa. Similarly to human, the murine bcsc-1 lacks homology to any protein of known function while possessing the two conserved Vault protein Inter-alpha-Trypsin and the von Willebrand Type A domains.
LOH Analysis and Identification of a Common Minimal Region of Loss at 11q23-q24 Encompassing the BCSC-1 Gene Locus. To refine the LOH11CR2 minimal region of loss at 11q23-q24, we examined 20 breast and 20 ovarian matched normal-tumor DNA pairs by using 6 microsatellite markers (Table 1 and Fig. 2B). Markers were selected based on their high heterozygosity (>60%), positions in our physical map of the LOH11CR2 region (Supporting Data), and association with previously reported LOH studies (3, 21, 24). Allelotyping of these samples confirmed the existence of a frequently deleted minimal region of loss centered around the D11S1328 locus in both breast and ovarian tumors. In the combined analysis, the highest LOH frequency observed was 41% for D11S1328 (Table 1).
Tumors with interstitial deletions, that is, tumors with LOH at one or more loci while retaining heterozygosity at flanking loci, were used to identify a shared minimal region of loss. Of the 13 breast and 7 ovarian tumors with partial loss of 11q23-q24, 12 (92%) and 7 (100%), respectively, exhibited deletions including at least one or more loci in the interval D11S1353-D11S4144-D11S1328 (Fig. 2B). Within this interval, a common region of loss containing either LOH of D11S4144 or D11S1328 was minimally defined by four breast (UPN73, UPN391, UPN394, and UPN405) and three ovarian (UPN78, UPN95, and UPN238) samples. Boundaries of this shared deletion are flanked proximally by retention of heterozygosity at D11S1353 in 2 tumors (UPN73 and UPN391) and D11S1933 distally by retention of heterozygosity in 3 tumors (UPN73, UPN95, and UPN238). Our analysis suggests that the D11S4144-D11S1328 region, where BCSC-1 is located, is the more likely to harbor a tumor suppressor gene.
Mutation and Expression Analysis of the BCSC-1 Gene in Cancer Cells. Tumor biopsies available from the breast and ovarian cancer patients (n = 40) used in the LOH analysis were screened for mutations in the BCSC-1 coding sequence by PCR and direct sequencing. Several previously reported polymorphisms (24) were found although no tumor-specific, somatic mutations were observed. BCSC-1 expression was examined in normal tissues and in a panel of 41 human cancer cell lines (Fig. 3B; Table 2, which is published as supporting information on the PNAS web site) by using a 2.3-kb cDNA probe containing the entire ORF of the BCSC-1(c) transcript. Nine of 41 (22%) tumor cell lines exhibited variable expression of each expected transcript by Northern blot analysis whereas each normal tissue mRNA control was positive for expression. Of interest, only 1 of 13 (7%) breast and 0 of 7 (0%) ovarian cell lines were positive for BCSC-1 expression whereas 3 of 6 (50%) lung cancer cell lines were positive for expression (Table 2). These results indicate that the mechanism for BCSC-1 gene inactivation in tumor cells is the loss of gene expression at one allele and deletion of the second.
In Vitro and in Vivo Effects of Ectopic BCSC-1 Expression. To evaluate the BCSC-1 gene's potential as a tumor suppressor gene, we examined the effects of expression on cell growth. Stable transfection of an expression vector containing BCSC-1 or vector alone into MCF-7 and NPA cells, BCSC-1 negative breast and thyroid cancer cell lines, respectively, was performed and assayed for colony formation. After several independent experiments, MCF-7 (n = 3) and NPA (n = 2) cells transfected with pIRESneo2.BCSCHA exhibited significantly fewer G418 resistant colonies than those with only the vector, suggesting that BCSC-1 has some influence on their growth (Fig. 10, which is published as supporting information on the PNAS web site).
To introduce and stably integrate the BCSC-1 gene into a variety of human tumor cell lines with high efficiency, we used a self-inactivating lentiviral system of gene transfer. Modifications were made to a lentivirus vector system (29–32) such that gene expression was driven by a cytomegalovirus promoter upstream of an internal ribosomal entry site (IRES) element and enhanced green fluorescent protein (EGFP) and pseudotyped with the vesicular stomatitis virus glycoprotein (VSVG) receptor. Infection efficiencies of >50% were observed in target human tumor cell lines such as HeLa, A549, H460, ES-2, SK-OV-3, ARO, and NPA whereas low efficiencies of infection (<50%) were obtained for PANC-1 and MDA-MB-453 (data not shown). Typically, BCSC-1 and EGFP gene expression levels became stable 48 h postinfection and continued for several weeks (>12 weeks). In each of these infections, there were no cytotoxic effects or changes in growth characteristics and cell cycle (Fig. 11, which is published as supporting information on the PNAS web site). This finding was consistently observed in cells that both express endogenous BCSC-1 (H460 and ARO) or that lack its expression (A549, ES-2, and NPA) (data not shown). These data suggest that in vitro there are no direct consequences of ectopic BCSC-1 expression on the cell cycle in those human tumor cell lines examined.
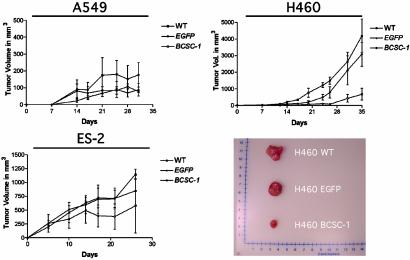
Because in vitro results do not necessarily predict in vivo behavior, we injected 1, 5, and 10 × 106 of each of the H460, ES-2, and A549 tumor-derived cell lines, respectively, infected with either EGFP or BCSC-1 lentiviruses into the s.c. flanks of 6-week-old female nude mice to evaluate the growth suppression ability of ectopic BCSC-1. Tumor cells transduced with virus particles containing BCSC-1 or EGFP alone showed no significant differences in their ability to proliferate over the course of 96 h (Fig. 12, which is published as supporting information on the PNAS web site). High levels of ectopic gene expression were maintained in nearly 100% of the cells examined by flow cytometry and Western analysis (data not shown). After injection, mice were monitored for up to 1 month, and tumor volumes were measured with a metric caliper (Fig. 4). The H460 and ES-2 cell lines infected with BCSC-1 virus appeared to be reduced relative to controls.
Fig. 4.
In vivo effects of ectopic BCSC-1 expression. In vivo tumorigenicity of H460, A549, and ES-2 cells in athymic (nu/nu) mice infected with BCSC-1. Tumor volumes (mm3) of xenografts from uninfected (WT) cells or cells transduced with either EGFP alone or BCSC-1 lentiviral particles.
Discussion
Cancers are associated with frequent deletions of genetic material that select for the loss of genes that prevent cells from acquiring a neoplastic phenotype. Although several tumor suppressor genes have been identified from these areas of deletion, the identities of the vast majority, including those located at human chromosome 11q23-q24, remain elusive. Frequent allelic loss at 11q23-q24 has been observed in a wide spectrum of human neoplasms, including breast, lung, and ovarian carcinomas, suggesting the existence of a gene or genes whose inactivation is involved in the initiation and/or progression of these diseases (1–3, 6–10, 22, 33–39). Despite the identification of a few genes at 11q23-q24 mutated in human cancers, the frequency of their genetic alterations was too low to correspond to the 30–60% detected by LOH analysis of the this region.
To search for a candidate tumor suppressor gene at 11q23-q24, we took advantage of an inherited translocation, t(11;12)(q24;q23), associated with an individual who presented with early-onset breast cancer and a family history of cancer. By physical mapping and cloning of the translocation breakpoint, we could place it ≈6 kb upstream of the BCSC-1 gene. Within 11q23-q24, at least two independent regions of allelic loss have been identified by LOH analysis in breast and lung cancers (3, 9, 24). The more telomeric region, LOH11CR2, spans ≈2 Mb and is minimally defined in lung adenocarcinoma between loci D11S1345 and D11S1328. In addition, the BCSC-1 gene, also known as LOH11CR2A, mapped to the LOH11CR2 region (24).
Whereas initial studies failed to prove, but did not exclude, a role for the BCSC-1 gene in breast, lung, and ovarian cancers (24), we present data suggesting that this gene may have tumor suppressive properties. We have shown that BCSC-1 is down-regulated as a consequence of the t(11;12)(q24;q23) translocation associated with early onset breast cancer. Furthermore, we observed that the tumor material obtained from the patient carrying the translocation displayed a very narrow deletion at the normal, nontranslocated allele, demonstrating a “two-hit” mechanism at the BCSC-1 locus typical for the inactivation of many tumor suppressor genes. In addition, the absence of breast cancer in individual II:1 who developed glioblastoma at age 69, as well as the remaining unaffected carriers, clearly indicates that the penetrance of the breast cancer trait associated with the translocation is incomplete. In addition, by careful LOH analysis, specifically designed to refine the area of minimal deletion within the LOH11CR2 region, we have also shown that the critical area seems to be within the loci D11S4144-D11S1328. This region is precisely where the BCSC-1 gene is located, again demonstrating this gene to be the target of deletion in this region.
Despite identifying a larger ORF of the BCSC-1 gene, we did not observe any inactivating mutations in any human cancer cells. However, we could demonstrate that most of the breast and ovarian cancer cell lines and several lung cancer cell lines examined do not express BCSC-1 relative to their normal tissue counterparts. Northern analysis of several tumor cell lines demonstrated a lack or a reduction of expression in 33 of 41 (80%) analyzed. These results suggest that disruption of BCSC-1 expression may be a necessary component of malignant transformation whereby inactivation of this gene in tumor cells occurs by loss of gene expression of one allele in combination with deletion of the second allele.
With the aim to understand its potential functions, we also characterized in detail the BCSC-1 gene and transcriptional structure. Contrary to the original characterization of this gene (24), we found that it consists of 20 exons instead of 10 and possesses a larger ORF. These structural features were confirmed also for the mouse homologue. We also found that, in a panel of normal human tissues, the human BCSC-1 gene is alternatively spliced, encoding for several proteins with a common N-terminal sequence. The most common transcript seems to be a 3.5-kb mRNA that encodes for a protein of 786 aa encoding for a predicted protein with a molecular mass of 86 kDa. The mouse gene expresses only a single 4-kb transcript that encodes a protein of 793 aa and a molecular mass of 87 kDa. Other variants encode for proteins of predicted molecular mass of 29 and 46 kDa. Despite this new information, all BCSC-1 isoforms continue to lack significant homology with any proteins of known function, and the presence of potential functional domains did not help to clarify any possible functional mechanism of tumor suppression associated with the BCSC-1 gene.
Finally, to establish the BCSC-1 as a bona fide tumor suppressor gene, we examined the biological effect of its ectopic expression in several cell lines derived from human tumors. We found that BCSC-1 could significantly suppress in vitro and in vivo growth, suggesting that it may indeed have tumor suppressor capabilities.
Overall, our observations suggest that the BCSC-1 gene is the frequent target of deletions in a variety of solid tumors leading to the deregulation of its normal expression. Altered expression of this gene and disruption of the molecular pathways it is involved in may contribute directly to or modify tumorigenesis. In addition, these results will provide the basis for future investigations into its role in normal physiological functions and carcinogenesis.
Supplementary Material
Acknowledgments
We thank Roberta Bichi, Jean Letofsky, Maria Cristina Picchio, Koshi Mimori, and Luisa Veronese for technical assistance and insightful advice. This work was supported by National Cancer Institute grants (to A.K.G. and C.M.C.), European Community Contract QLG2-CT-1999-00786, and a grant from Associazione Italiana per la Ricerca sul Cancro (to M.N.).
Abbreviations: LOH, loss of heterozygosity; BAC, bacterial artificial chromosome; UPN, unique patient number; EGFP, enhanced GFP.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY366501–AY366509).
References
- 1.Carter, S. L., Negrini, M., Baffa, R., Gillum, D. R., Rosenberg, A. L., Schwartz, G. F. & Croce, C. M. (1994) Cancer Res. 54, 6270–6274. [PubMed] [Google Scholar]
- 2.Hampton, G. M., Mannermaa, A., Winquist, R., Alavaikko, M., Blanco, G., Taskinen, P. J., Kiviniemi, H., Newsham, I., Cavenee, W. K. & Evans, G. A. (1994) Cancer Res. 54, 4586–4589. [PubMed] [Google Scholar]
- 3.Negrini, M., Rasio, D., Hampton, G. M., Sabbioni, S., Rattan, S., Carter, S. L., Rosenberg, A. L., Schwartz, G. F., Shiloh, Y., Cavenee, W. K., et al. (1995) Cancer Res. 55, 3003–3007. [PubMed] [Google Scholar]
- 4.Koreth, J., Bethwaite, P. B. & McGee, J. O. (1995) J. Pathol. 176, 11–18. [DOI] [PubMed] [Google Scholar]
- 5.Kerangueven, F., Noguchi, T., Coulier, F., Allione, F., Wargniez, V., Simony-Lafontaine, J., Longy, M., Jacquemier, J., Sobol, H., Eisinger, F. & Birnbaum, D. (1997) Cancer Res. 57, 5469–5474. [PubMed] [Google Scholar]
- 6.Laake, K., Odegard, A., Andersen, T. I., Bukholm, I. K., Karesen, R., Nesland, J. M., Ottestad, L., Shiloh, Y. & Borresen-Dale, A. L. (1997) Genes Chromosomes Cancer 18, 175–180. [PubMed] [Google Scholar]
- 7.Launonen, V., Laake, K., Huusko, P., Niederacher, D., Beckmann, M. W., Barkardottir, R. B., Geirsdottir, E. K., Gudmundsson, J., Rio, P., Bignon, Y. J., et al. (1999) Br. J. Cancer 80, 879–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iizuka, M., Sugiyama, Y., Shiraishi, M., Jones, C. & Sekiya, T. (1995) Genes Chromosomes Cancer 13, 40–46. [DOI] [PubMed] [Google Scholar]
- 9.Rasio, D., Negrini, M., Manenti, G., Dragani, T. A. & Croce, C. M. (1995) Cancer Res. 55, 3988–3991. [PubMed] [Google Scholar]
- 10.Wang, S. S., Virmani, A., Gazdar, A. F., Minna, J. D. & Evans, G. A. (1999) Genes Chromosomes Cancer 25, 154–159. [PubMed] [Google Scholar]
- 11.Negrini, M., Sabbioni, S., Possati, L., Rattan, S., Corallini, A., Barbanti-Brodano, G. & Croce, C. M. (1994) Cancer Res. 54, 1331–1336. [PubMed] [Google Scholar]
- 12.Murakami, Y., Nobukuni, T., Tamura, K., Maruyama, T., Sekiya, T., Arai, Y., Gomyou, H., Tanigami, A., Ohki, M., Cabin, D., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 8153–8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koreth, J., Bakkenist, C. J., Larin, Z., Hunt, N. C., James, M. R. & McGee, J. O. (1999) Oncogene 18, 1157–1164. [DOI] [PubMed] [Google Scholar]
- 14.Savitsky, K., Sfez, S., Tagle, D. A., Ziv, Y., Sartiel, A., Collins, F. S., Shiloh, Y. & Rotman, G. (1995) Hum. Mol. Genet. 4, 2025–2032. [DOI] [PubMed] [Google Scholar]
- 15.Baffa, R., Negrini, M., Schichman, S. A., Huebner, K. & Croce, C. M. (1995) Proc. Natl. Acad. Sci. USA 92, 4922–4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, S. S., Esplin, E. D., Li, J. L., Huang, L., Gazdar, A., Minna, J. & Evans, G. A. (1998) Science 282, 284–287. [DOI] [PubMed] [Google Scholar]
- 17.Calin, G. A., di Iasio, M. G., Caprini, E., Vorechovsky, I., Natali, P. G., Sozzi, G., Croce, C. M., Barbanti-Brodano, G., Russo, G. & Negrini, M. (2000) Oncogene 19, 1191–1195. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez, Y., Wong, C., Thoma, R. S., Richman, R., Wu, Z., Piwnica-Worms, H. & Elledge, S. J. (1997) Science 277, 1497–1501. [DOI] [PubMed] [Google Scholar]
- 19.Vorechovsky, I., Luo, L., Lindblom, A., Negrini, M., Webster, A. D., Croce, C. M. & Hammarstrom, L. (1996) Cancer Res. 56, 4130–4133. [PubMed] [Google Scholar]
- 20.Vorechovsky, I., Rasio, D., Luo, L., Monaco, C., Hammarstrom, L., Webster, A. D., Zaloudik, J., Barbanti-Brodani, G., James, M., Russo, G., et al. (1996) Cancer Res. 56, 2726–2732. [PubMed] [Google Scholar]
- 21.Pulido, H. A., Fakruddin, M. J., Chatterjee, A., Esplin, E. D., Beleno, N., Martinez, G., Posso, H., Evans, G. A. & Murty, V. V. (2000) Cancer Res. 60, 6677–6682. [PubMed] [Google Scholar]
- 22.Davis, M., Hitchcock, A., Foulkes, W. D. & Campbell, I. G. (1996) Cancer Res. 56, 741–744. [PubMed] [Google Scholar]
- 23.Oriola, J., Halperin, I., Mallofre, C., Muntane, J., Angel, M. & Rivera-Fillat, F. (2001) Eur. J. Cancer 37, 2470–2474. [DOI] [PubMed] [Google Scholar]
- 24.Monaco, C., Negrini, M., Sozzi, G., Veronese, M. L., Vorechovsky, I., Godwin, A. K. & Croce, C. M. (1997) Genomics 46, 217–222. [DOI] [PubMed] [Google Scholar]
- 25.Mitelman, F., ed. (1995) ISCN: An International System for Human Cytogenetic Nomenclature (Karger, Basel).
- 26.Sambrook, J., Fritsch, E. F., Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab., Plainview, NY).
- 27.Martinis, J. & Croce, C. M. (1978) Proc. Natl. Acad. Sci. USA 75, 2320–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cesari, R., Martin, E. S., Calin, G. A., Pentimalli, F., Bichi, R., McAdams, H., Trapasso, F., Drusco, A., Shimizu, M., Masciullo, V., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 5956–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naldini, L., Blomer, U., Gallay, P., Ory, D., Mulligan, R., Gage, F. H., Verma, I. M. & Trono, D. (1996) Science 272, 263–267. [DOI] [PubMed] [Google Scholar]
- 30.Naldini, L., Blomer, U., Gage, F. H., Trono, D. & Verma, I. M. (1996) Proc. Natl. Acad. Sci. USA 93, 11382–11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Follenzi, A., Ailles, L. E., Bakovic, S., Geuna, M. & Naldini, L. (2000) Nat. Genet. 25, 217–222. [DOI] [PubMed] [Google Scholar]
- 32.Zufferey, R., Dull, T., Mandel, R. J., Bukovsky, A., Quiroz, D., Naldini, L. & Trono, D. (1998) J. Virol. 72, 9873–9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerangueven, F., Eisinger, F., Noguchi, T., Allione, F., Wargniez, V., Eng, C., Padberg, G., Theillet, C., Jacquemier, J., Longy, M., et al. (1997) Oncogene 14, 339–347. [DOI] [PubMed] [Google Scholar]
- 34.Koreth, J., Bakkenist, C. J. & McGee, J. O. (1997) Oncogene 14, 431–437. [DOI] [PubMed] [Google Scholar]
- 35.Foulkes, W. D., Ragoussis, J., Stamp, G. W., Allan, G. J. & Trowsdale, J. (1993) Br. J. Cancer 67, 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gabra, H., Taylor, L., Cohen, B. B., Lessels, A., Eccles, D. M., Leonard, R. C., Smyth, J. F. & Steel, C. M. (1995) Br. J. Cancer 72, 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Launonen, V., Mannermaa, A., Stenback, F., Kosma, V. M., Puistola, U., Huusko, P., Anttila, M., Bloigu, R., Saarikoski, S., Kauppila, A. & Winqvist, R. (2000) Cancer Genet. Cytogenet. 122, 49–54. [DOI] [PubMed] [Google Scholar]
- 38.Launonen, V., Stenback, F., Puistola, U., Bloigu, R., Huusko, P., Kytola, S., Kauppila, A. & Winqvist, R. (1998) Gynecol. Oncol. 71, 299–304. [DOI] [PubMed] [Google Scholar]
- 39.Watson, R. H., Neville, P. J., Roy, W. J., Jr., Hitchcock, A. & Campbell, I. G. (1998) Oncogene 17, 207–212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.