Abstract
The B10.Q/J strain of mice was serendipitously discovered to be highly susceptible to infection by the intracellular protozoan parasite, Toxoplasma gondii but markedly resistant to induction of autoimmune arthritis. We have previously shown that the B10.Q/J phenotype is controlled by a single recessive locus and is associated with lymphocyte hyporesponsiveness to IL-12. Using genetic approaches, we have now localized the B10.Q/J locus to chromosome 9 and established its identity as Tyk2, a Janus kinase essential for IL-12 and IFN-α/β cytokine signaling. The B10.Q/J Tyk2 gene contained a single missense mutation resulting in a nonconservative amino acid substitution (E775K) in an invariant motif of the pseudokinase (Janus kinase homology 2) domain. This mutation appeared to result in the absence of the B10.Q/J-encoded Tyk2 protein, despite presence of Tyk2-specific transcripts. Phenotypically, B10.Q/J cells were indistinguishable from Tyk2-deficient cells, showing impaired signaling and biologic responses to IL-12, IL-23, and type I IFNs. The analogous E782K mutant of human Tyk2 failed to restore IFN-α responsiveness in Tyk2 null 11,1 cells. Our results indicate a crucial role for Tyk2 in T helper 1-mediated protective and pathogenic immune responses. An additional implication of our findings is that naturally occurring mutations in the Tyk2 gene may underlie altered susceptibilities to infectious or autoimmune diseases in human and animal populations.
Studies with IL-12-deficient mice have demonstrated an essential role for endogenous IL-12 in promoting the generation of IFN-γ-secreting effector lymphocytes required for resistance to intracellular pathogens, such as Toxoplasma (1–3), Mycobacteria (4), and Leishmania (5). Similarly, mice deficient in Stat4, a signal transducer and activator of transcription (Stat) involved in IL-12 signal transduction display, impaired development of TH1 lymphocytes and susceptibility to these intracellular pathogens (6, 7). Patients with hereditary deficiencies in IL-12Rβ1 expression and function present with severe atypical Mycobacteria and Salmonella infections (8, 9). Interestingly, defective IL-12 signaling, evident as impaired phosphorylation of Stat1, -3, and -5, is also associated with susceptibility to atypical mycobacterial and staphylococcal infections in a patient, despite apparently normal expression of IL-12 receptor and Stat4 phosphorylation (10). Thus, in both mice and humans, IL-12 function is critical for resistance to intracellular pathogens.
In contrast to its protective role against infectious pathogens, IL-12 contributes to the pathogenesis of organ-specific autoimmune diseases (mediated by TH1 type lymphocytes reactive to self-antigens) (11). In the DBA/1 model of collagen-induced arthritis induced by active immunization with Type II collagen, IL-12 deficiency results in a profound reduction in severity and incidence of joint disease (12). In addition, a critical role of IL-12 in pathogenesis of insulin-dependent diabetes mellitus (13), inflammatory bowel disease (14), and experimental allergic encephalomyelitis (15) has also been demonstrated. Taken together, IL-12 can be viewed as a “double-edged sword,” playing a requisite protective role against infection yet promoting the tissue-destructive effector functions of self-reactive lymphocytes.
The ability of IL-12 to regulate resistance to autoimmunity and susceptibility to infectious challenges depends critically on an intact IL-12 signaling pathway. Thus, genetic defect(s) along this pathway will have both beneficial and deleterious effects, manifesting as resistance to autoimmune diseases and susceptibility to infectious pathogens. We have recently reported a genetic defect in the B10.Q-H2q/SgJ (B10.Q/J) strain of mice maintained at The Jackson Laboratory, which, unlike B10.Q strain mice from Taconic Farms, are, for unknown reason(s), resistant to of collagen-induced arthritis and extremely susceptible to infection with protozoan parasite, Toxoplasma gondii (3, 16). This B10.Q/J phenotype was associated with hyporesponsiveness to IL-12, resulting in poor IFN-γ responses to autoantigen priming or parasite challenge. Preliminary genetic analysis, by using T. gondii susceptibility as readout, demonstrated that the B10.Q/J phenotype is controlled by a major autosomal recessive locus (3). We have now successfully linked the B10.Q/J locus to mouse chromosome 9 and identified the genetic lesion underlying the opposing effects this locus exerts on autoimmune and infectious disease development.
Materials and Methods
Animals. B10.Q-H2q/Sgj (B10.Q/J) mice were obtained from The Jackson Laboratory. B10.Q/Ai mice were obtained from Taconic Farms through the National Institute of Allergy and Infectious Diseases. Homozygous Tyk2 null mice (17) were used after backcrossing to C57BL/6 background for six generations. All mice were housed under specific pathogen-free conditions at Brown University.
Linkage Analysis of B10.Q/J Susceptibility to T. gondii Infection. (B10.Q/J × BALB/c) × B10.Q/J F1 backcross mice were phenotyped individually with respect to IFN-γ production and resistance to T. gondii and genotyped by using simple sequence length polymorphism markers polymorphic between B10 and BALB/c from Research Genetics (Huntsville, AL) according to the manufacturer's protocol. Detailed procedures used for sequence determination of the Tyk2 coding region and AvaI restriction typing at the Tyk2 locus can be found in Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Cytokines, Antibodies, and Reagents. Murine rIL-12 (a gift of the Genetics Institute, Cambridge, MA) was used at a concentration of 50 ng/ml. Murine IFN-α was purchased from PBL Biomedical Laboratories (New Brunswick, NJ) and used at 1,000 units/ml. Human rIL-23 was purchased from R & D Systems and used at a concentration of 1 μg/ml. Antibodies to Stat1 (E-23); p-Stat1 (Tyr 701); Stat3 (C-20); p-Stat3 (B-7); Stat4 (C-20); Tyk2 (C-20, H-135, C-8); p-Tyk2 (Tyr 1054/1055); Janus kinase (Jak)2 (C-20); and appropriate horseradish peroxidase-conjugated secondary IgG antibodies were obtained from Santa Cruz Biotechnology. Additional anti-human Tyk2 Abs (polyclonal T10-2 and monoclonal R5-9) were obtained from BD Biosciences (San Diego) and were donated by S. Pellegrini (Pasteur Institute, Paris). Antibody to p-Stat4 (Y693) was purchased from Zymed. The antiphosphotyrosine antibody (p-Y) clone 4G10 was obtained from Upstate Biotechnology (Lake Placid, NY).
In Vitro Assays for Cytokine Signaling/Responsiveness. Cell extracts were prepared from splenocytes activated with Con A (1 μg/ml) for 3 days and serum starved for 4 h. In some cases, MG-132 (10 ng/ml) (Sigma) was added during the starvation period. Before lysis with RIPA buffer, cells were stimulated with or without cytokines at 37°C for the indicated times. Total cell lysate (1.5–2 mg) was used for immunoprecipitation and subsequent immunoblotting detection of p-Tyk2, Tyk2, p-Jak2, and Jak2 according to published methods (17). IFN-α responsiveness was assayed by using inhibition of vesicular stomatitis virus infection in primary cultures of lung fibroblasts (see Supporting Materials and Methods).
In Vitro Mutagenesis of Human Tyk2 and Complementation Assay. The human E783K mutation was introduced by PCR and by the overlap extension method, subsequently subcloned into pRcCMV-Tyk2B (18), and sequenced. Stable clones of E782K and R856G transfectants were generated and analyzed for IFN responsiveness (19), details of which are available as Supporting Materials and Methods.
Results
Linkage of B10.Q/J Phenotype to Chromosome 9 and Genetic Identity with Tyk2. To identify the B10.Q/J gene, we undertook a genetic approach to map the location of the susceptibility locus in the genome and to test potential candidate gene(s) present in the linked region. We have previously shown that B10.Q/J × BALB/c F1 progeny to be normal. In contrast, approximately half of the (B10.Q/J × BALB/c) × B10.Q/J backcross progeny regain the B10.Q-defective phenotype indicative of a single recessive locus control (3). To map this locus, we performed an initial genomewide scan involving 52 such F1 backcross mice and using 43 simple sequence length markers polymorphic for B10.Q and BALB/c. In this initial experiment, the B10.Q/J phenotype of increased pathogen burden displayed strongest association with a B10.Q allele of D9Mit191, a mouse chromosome 9 marker, with no additional significant linkages observed (unpublished data).
To improve the resolution of our mapping experiment, we analyzed a total of 189 F1 backcross progeny and 20 additional chromosome 9 markers. As shown in Table 1, significant logarithm of odds (LOD) scores were obtained over a proximal region of mouse chromosome 9. A peak LOD score of 22.1 was observed for D9Mit328 located 17 cM away from the centromere. However, the B10.Q phenotype appeared to map ≈15 cM proximal of D9Mit328 (31 recombination events in 189 back-cross mice). This observation and the lack of more polymorphic markers proximal to D9Mit328 led us to investigate the presence of candidate genes in this region using the UCSC genome database (http://genome.ucsc.edu). In a short segment of mouse chromosome 9 syntenic to human chromosome 19, we identified Tyk2, one of four Jaks involved in cellular signaling by cytokine receptors (20). Tyk2 maps 8 cM away from the centromere and 15 cM proximal to D9Mit328, the most centromeric marker analyzed in the cross (http://genome.ucsc.edu). Importantly, Tyk2-deficient mice exhibit impaired responsiveness to IL-12, similar to the B10.Q/J mice (17, 21). The location of Tyk2 on chromosome 9 and its critical role in IL-12 signaling prompted us to test Tyk2 as a candidate gene for the B10.Q/J locus.
Table 1. Linkage of B10.Q/J phenotype to chromosome 9.
| Locus | Map distance, cM* | LOD score† |
|---|---|---|
| Tyk2/Aval | 0.0 | 53.3 |
| D9Mit328 | 17.0 | 22.1 |
| D9Mit192 | 17.7 | 21.4 |
| D9Mit191 | 19.3 | 19.9 |
| D9Mit330 | 21.9 | 17.8 |
| D9Mit71 | 23.6 | 16.5 |
| D9Mit4 | 25.4 | 15.3 |
Map distance was calculated by using mapmaker 3.0 (Roswell Park Cancer Institute, Buffalo, NY) on the basis of Haldane function, which assumes no interference (multiple crossovers according to a Poisson distribution).
LOD score calculation was based on two-point linkage analysis by using mapmaker 3.0 with LOD threshold of 3.0 and distance threshold of 80 Haldane cM.
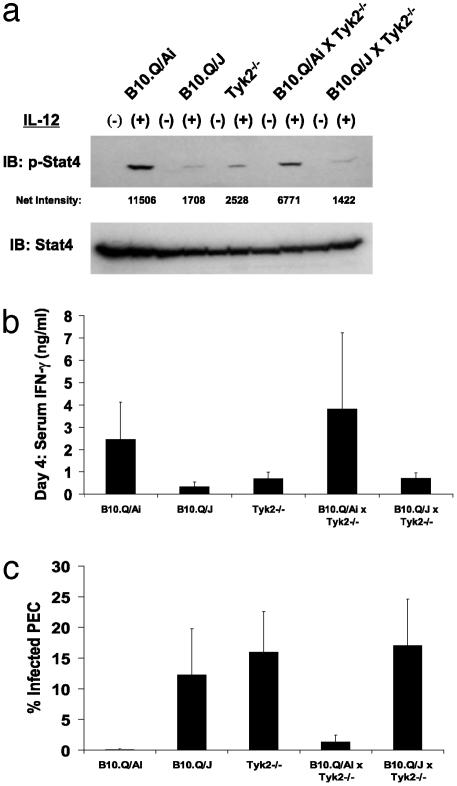
Thus, we tested for the ability of B10.Q/J mice to genetically complement a homozygous null mutation in the Tyk2 locus. As shown in Fig. 1a, B10.Q/J and Tyk2–/– lymphocytes displayed impaired IL-12-induced Stat4 activation compared with the wild-type B10.Q/Ai cells consistent with previous data (3). As expected, Stat4 activation was restored to near wild-type levels in B10.Q/Ai × Tyk–/– but not in B10.Q/J × Tyk2–/– cells (Fig. 1a). This analysis was extended by assessing the ability of the F1 progeny to mount an IFN-γ-dependent protective response to T. gondii challenge. When challenged with T. gondii, both B10.Q/J and Tyk2–/– mice, in contrast to the wild-type mice, were highly susceptible, exhibiting low IFN-γ levels and high parasite numbers (Fig. 1 b and c). In contrast to the parental Tyk2–/– mice, B10.Q/Ai × Tyk2–/– F1 progeny were now able to mount an effective immune response to the T. gondii challenge (Fig. 1 b and c). Importantly, B10.Q/J × Tyk2–/– F1 progeny remained susceptible to infection, as shown by low serum IFN-γ and high parasite counts (Fig. 1 b and c). This negative complementation test provides definitive evidence for genetic identity between Tyk2 and the B10.Q/J locus.
Fig. 1.
B10.Q/J mice fail to genetically complement Tyk2-deficient mice. (a) T cell blasts from wild-type, B10.Q/J, Tyk2–/–, B10.Q/Ai × Tyk2–/– F1, and B10.Q/J × Tyk2–/– F1 mice were assayed for IL-12-induced phosphorylation of Stat4. (b) IFN-γ levels were determined in sera obtained 4 days after T. gondii infection in parental strains, B10.Q/Ai × Tyk2–/– F1, and B10.Q/J × Tyk2–/– F1 mice. (c) The percentage of infected peritoneal cells of the same mice described in b were determined on day 5 postinfection. Results shown are representative of one of two experiments. In each experiment, three to four age- and sex-matched mice were used.
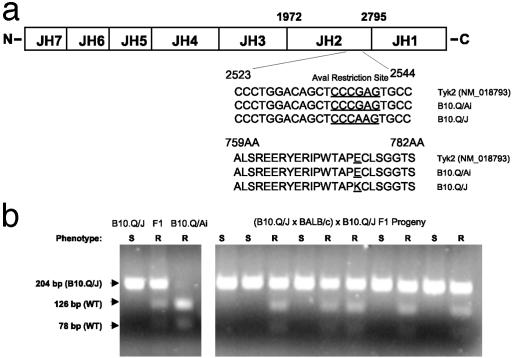
B10.Q/J Tyk2 Has a Single-Base Mutation Resulting in an E775K Substitution Within the Pseudokinase Domain of Tyk2. Jaks share a common structure, comprising of seven Jak homology (JH) domains (Fig. 2a). The N terminus of Jaks (JH7–JH3) is important for association with the intracellular tails of cytokine receptors and nuclear localization (22, 23), whereas the C-terminal JH1 domain contains the kinase domain (24–26). To investigate the precise nature of the mutation in the Tyk2 gene, Tyk2 cDNAs from the spleen of both B10.Q/Ai and B10.Q/J mice were amplified and sequenced. This analysis revealed a single missense mutation (G→A substitution) at position 2538 in the B10.Q/J Tyk2 coding region (Fig. 2a). The B10.Q/Ai strain Tyk2 sequence was completely identical to the published sequence derived from C57BL/6 strain (GenBank accession no. NM_018793) (Fig. 2a). In-frame translation of the B10.Q/J Tyk2 nucleotide sequence predicts a nonconservative substitution from glutamic acid (E) to lysine (K) at position 775 (Fig. 2a), occurring within a highly conserved APE motif of the JH2 (pseudokinase) domain of Tyk2 (18, 27, 28). The G→A substitution in the B10.Q/J strain results in the obliteration of an AvaI restriction site present in the wild-type Tyk2 genomic sequence (Fig. 2a), confirmed by data shown in Fig. 2b Left). Analysis of this restriction site polymorphism was able to predict the phenotype of all 179 F1 backcross mice genotyped (Fig. 2b shows a sampling of this analysis). Thus, in addition to confirming the presence of the E to K mutation in the genome, the complete segregation of the B10.Q/J phenotype with the Tyk2 polymorphism further substantiates the genetic identity of B10.Q/J locus to be Tyk2 (Table 1). However, our results do not exclude the unlikely existence of an additional mutation outside of the Tyk2 coding region.
Fig. 2.
B10.Q/J Tyk2 contains a single G→ A mutation in the JH2 domain. (a) Comparison of B10.Q/Ai and B10.Q/J Tyk2 cDNA sequences. Splenic cDNA from B10.Q/Ai and B10.Q/J mice were amplified by PCR by using primers (as described in Materials and Methods) spanning the entire coding region of Tyk2. B10.Q/J mice exhibit a G→ A base substitution at position 2538. An E775K amino acid substitution is predicted in the JH2 domain. Note: The G→ A polymorphism resulted in the loss of an AvaI restriction site present in the wild-type Tyk2 sequence. (b) A 204-bp Tyk2 fragment was amplified from genomic DNA of parental, F1, and F1 backcross progeny (as described in Materials and Methods) and subjected to AvaI restriction typing. The presence of the wild-type Tyk2 allele is indicated by the appearance of 126- and 78-bp AvaI digested fragments. The AvaI-resistant 204-bp PCR product corresponds to the mutant B10.Q/J Tyk2 allele. AvaI digestion patterns for individual parental and F1 mice (Left) and representative backcross mice (Right) are shown. The resistance or susceptibility phenotype of individual mice to T. gondii infection is indicated by R or S, respectively.
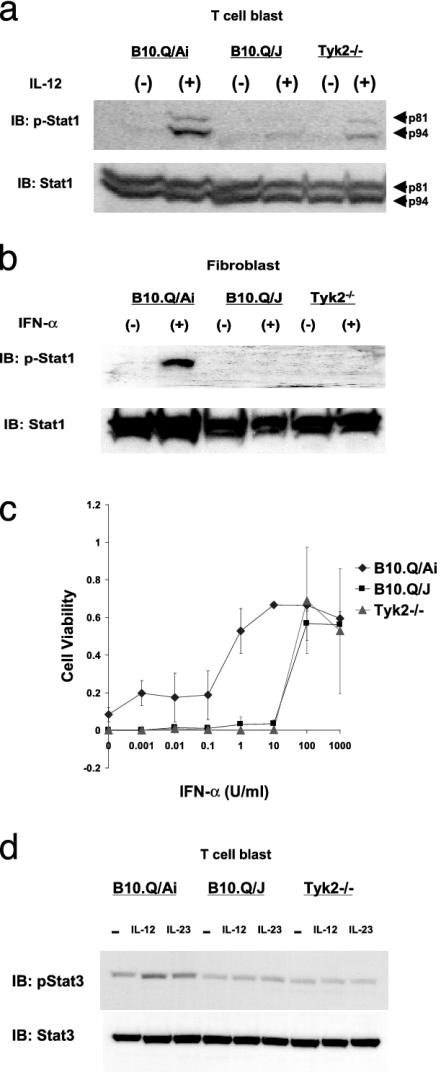
Impaired Activation of Stats by IL-12, Type I IFN, and IL-23 in B10.Q/J Cells. The activation of Tyk2 has been implicated in the signaling of various cytokines, including, IL-3, -6, -10, -12 and Type I IFNs (27). Nevertheless, mice deficient in Tyk2 function exhibited hyporesponsiveness mainly to IL-12 and Type I IFNs (17, 21). The nonconservative (E775K) mutation observed in the B10.Q/J Tyk2 gene might lead to a global or selective disruption of cellular responses to IL-12 and Type I IFNs. Fig. 3a demonstrates that the impaired IL-12 responsiveness of B10.Q/J lymphoblasts is not restricted to Stat4 phosphorylation (ref. 16 and Fig. 1a above) but also extends to Stat1 activation, indicative of a more global negative effect on signaling downstream of the IL-12 receptor.
Fig. 3.
B10.Q/J cells exhibit impaired cellular responses to IL-12, IFN-α, and IL-23. Impaired Stat1 phosphorylation in B10.Q/J cells after IL-12 (a) or IFN-α (b) exposure is shown. (a) T cell blasts derived from B10.Q/Ai, B10.Q/J, and Tyk2–/– mice were cultured with (+) or without (–) IL-12 (50 ng/ml) for 15 min. (b) Lung-derived fibroblasts from B10.Q/Ai, B10.Q/J, and Tyk2–/– mice were cultured with (+) or without (–) IFN-α (1,000 units/ml) for 15 min. Cells were analyzed for activation of Stat1 by direct Western blot with antiphosphotyrosine-Stat1 (p-Stat1) Ab. Protein loading was controlled by reprobing with normal Stat1 Ab. (c) Antiviral response of B10.Q/J fibroblasts. Fibroblasts from B10.Q/Ai, B10.Q/J, and Tyk2–/– mice were stimulated with IFN-α and cultured with vesicular stomatitis virus. After 48 h, surviving cells were quantified by crystal violet staining. (d) Lymphocytes from wild-type, B10.Q/J, and Tyk2–/– mice were analyzed for Stat3 activation after 30-min stimulation with IL-12 (50 ng/ml) or IL-23 (1 μg/ml) with p-Stat3 Ab. Protein loading was controlled by reprobing with normal Stat3 Ab. a and b are representative of one of three experiments, and c and d are representative of two experiments.
To evaluate the impact of the B10.Q/J mutation on IFN-α responsiveness, fibroblasts were derived from lungs of wild-type and B10.Q/J as well as Tyk2-deficient mice. As shown in Fig. 3b, in response to IFN-α treatment, Stat1 activation was impaired in B10.Q/J-derived fibroblasts, relative to similarly treated B10.Q/Ai cells. As an additional bioassay, we assessed the ability of IFN-α to elicit an antiviral response to vesicular stomatitis virus in the same fibroblasts. Fig. 3c demonstrates that B10.Q/J cells, similar to Tyk2–/– fibroblasts, required a 100-fold (relative to wild-type cells) higher concentration of IFN-α to achieve full protection, suggesting the existence of a Tyk2-independent but IFN-α/βR-dependent biologic response. Thus, the E775K mutation in B10.Q/J Tyk2 is associated with a hyporesponsive phenotype to both IL-12 and type I IFNs, indistinguishable from that induced by genetic ablation of Tyk2.
The defective response of the B10.Q/J cells may also extend to IL-23, an IL-12p40-containing heterodimeric cytokine reported to use the IL-12Rβ1 subunit and activate an common set of Jak-Stats, including Tyk2. To investigate the impact of the B10.Q/J mutation and the role of Tyk2 in IL-23 signaling, we used Stat3 phosphorylation as a readout, because IL-23 is a weak inducer of Stat4 activation (ref. 29 and unpublished data). As shown in Fig. 3d, in response to IL-23 treatment, wild-type T cell blasts displayed increased phospho-Stat3 levels, whereas in similarly treated B10.Q/J or Tyk2–/– lymphocytes the induction of Stat3 phosphorylation was not observed. The latter finding represents the first indication that Tyk2 plays an essential role in IL-23 signaling and further demonstrates that the B10.Q/J response defect extends to this cytokine. Overall, the above studies with all three cytokine responses demonstrate that B10.Q/J cells are functionally equivalent to Tyk2 null cells, leading us to investigate Tyk2 gene expression and function in the B10.Q/J strain.
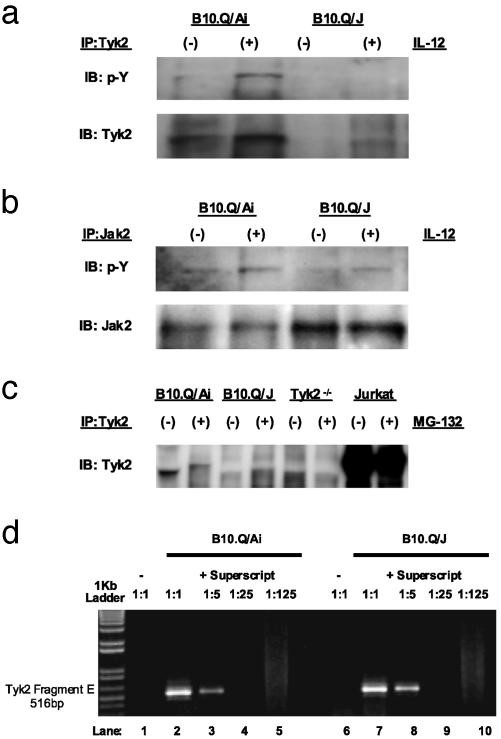
B10.Q/J Tyk2 Gene Expression and Protein Function. To address the issue of expression and function of the B10.Q/J Tyk2 gene product, IL-12-induced Tyk2 phosphorylation was assayed in T cell blasts derived from B10.Q/Ai and B10.Q/J mice. This assay was performed by immunoprecipitation of Tyk2 from cell extracts by using the C-20 Ab generated against residues 1170–1187 of human Tyk2. Precipitated material was then probed with either phosphotyrosine-specific or the same Tyk2-specific C-20 Ab. As shown in Fig. 4a, whereas the phosphorylated form of Tyk2 was observed in IL-12-treated B10.Q/Ai cells, we were consistently unable to detect the activated form of Tyk2 in similarly treated B10.Q/J cells. Identical negative results were obtained with an antibody specific to the phosphorylated form of human Tyk2 (unpublished data). Surprisingly, when the same immunoprecipitates were probed with the C-20 Ab, a 135-kDa band corresponding to mouse Tyk2 was visible only in B10.Q/Ai extracts (Fig. 4a). Attempts to visualize either wild-type or mutant forms of the mouse Tyk2 gene product by using five additional anti-human Tyk2 Abs (including T10-2) were not successful. Contrary to the complete absence of immunoreactive Tyk2 protein, B10.Q/J cells expressed normal levels of Jak2 protein and exhibited an IL-12-inducible Jak2 phosphorylation response, as observed in Tyk2–/– cells (Fig. 4b). Thus, the E775K mutation appears to result in either a loss in antigenicity for the C-20 anti-Tyk2 Ab or Tyk2 protein instability. To address the possibility that the lysine substitution enhances proteasomemediated degradation (30, 31) of mutant Tyk2, T cell blasts from wild-type and B10.Q/J were treated with proteasome inhibitor, MG-132, 8 h before cell lysis. As shown in Fig. 4c, MG-132 pretreatment failed to reveal Tyk2 immunoreactivity in B10.Q/J cells. In wild-type cells, treatment with proteasome inhibitor caused the appearance of a higher molecular weight Tyk2 band (Fig. 4c), presumably resulting from accumulation of the ubiquitinated form of the protein. Overall, the above results indicate that the mutant mouse Tyk2 protein, for unknown reasons, is poorly expressed in B10.Q/J cells, despite the fact Tyk2-specific transcripts are readily detectable in RNA preparations isolated from both wild-type and mutant B10.Q/J cells (Fig. 4d). To further examine the effect of the E775K mutation on Tyk2 function, the corresponding E782K mutation in the human Tyk2 cDNA was made and tested for functional complementation of the IFN-α unresponsive Tyk2 null 11,1 human cell line (19). Cells (11,1) were stably transfected with wild-type Tyk2, the E782K mutant, or a previously described kinase-inactive mutant R856G (18). The levels of mutant Tyk2 protein were monitored by Western blot by using the T10-2 mAb and were found to be comparable to wild-type Tyk2 (Fig. 5, which is published as supporting information on the PNAS web site). Pools of neoR cells were plated in hypoxanthine/aminopterin/thymidine medium supplemented with IFN-α and monitored for colony formation (19). Although >200 colonies arose from cells transfected with wild-type Tyk2, no colonies were obtained from the R856G or the E782K transfected pools, showing that the latter mutant, similar to the R856G mutant, failed to restore IFN-α responsiveness. The latter result indicates that lysine substitution of this conserved glutamic acid residue of the pseudokinase domain of human Tyk2 results in complete loss of function, consistent with our earlier observations that B10.Q/J mouse cells behave essentially as Tyk2 nulls in their responsiveness to IL-12, IL-23, and type I IFN.
Fig. 4.
Lack of immunoreactive Tyk2 expression in B10.Q/J cells despite the presence of Tyk2-specific transcript. (a and b) T cell blasts from B10.Q/Ai and B10.Q/J mice were cultured with (+) or without (–) IL-12 (50 ng/ml) and analyzed for Tyk2 activation (a) or Jak2 activation (b) by immunoprecipitation with Tyk2- or Jak2-specific Abs and subsequent immunoblotting of precipitates with phosphotyrosine-specific (p-Y) Ab. The same blots were reprobed with Tyk2- and Jak2-specific Abs to control for protein loading. (c) T cell blasts from B10.Q/Ai, B10.Q/J, and Tyk2–/– mice were cultured for 8 h in the presence (+) or absence (–) of proteasome inhibitor MG-132 (10 ng/ml). Total protein extracts were then immunoprecipitated with Tyk2-specific Ab followed by immunoblotting with the same Tyk2 Ab. Jurkat T cells were included to serve as positive control for immunoprecipitation and immunoblotting. (d) B10.Q/J cells have equivalent amounts of Tyk2-specific transcripts compared with wild-type cells. cDNA was prepared from the spleens of B10.Q/Ai or B10.Q/J mice followed by PCR with primers specific for the 3′ terminus of Tyk2 (Fragment E: 2143–2580). cDNA primed with random hexamers in the presence of reverse transcriptase (RT) from both strain of mice was diluted serially (1:5) (lanes 2–5 and 7–10). cDNA primed in the absence of RT lack Tyk2-specific transcripts (lanes 1 and 6).
Discussion
In the present study, we have used linkage analysis to identify the underlying genetic defect responsible for the altered immune phenotype of the B10.Q/J strain of mice. This B10.Q substrain, from The Jackson Laboratory, was serendipitously discovered to be naturally refractory to collagen-induced arthritis induction and exquisitely sensitive to T. gondii infection (3, 16), apparently resulting from impaired IL-12 responsiveness of B10.Q/J lymphocytes. We now provide definitive genetic and functional evidence that the gene mutated in the defective B10.Q/J mice is indeed Tyk2, a Jak critically involved in IL-12 cell signaling. To our knowledge, this is the first report, in mice or humans, of a spontaneously occurring mutation in Tyk2, functionally linked to altered disease susceptibility. Our earlier in vivo characterization of T. gondii infection and collagen-induced arthritis development in the mutant B10.Q/J strain provides a clearer picture of the consequences of Tyk2 deficiency on infectious and inflammatory disease processes, not previously reported in the initial studies of Tyk2 knockout animals (21, 25). Our studies further suggest that polymorphisms at the Tyk2 locus could act as a modifier of autoimmune and infectious disease susceptibilities. Of specific interest is a cohort of children with increased susceptibility to avirulent bacterial infections. In addition to the previously reported alterations in the genes encoding for IL-12 subunits (32), IL-12 receptor chains (8, 9, 33, 34), the IFN-γ receptor (35, 36), or Stat1 (37) mutation(s) in the Tyk2 gene may underlie the phenotype of a subset of patients with still unexplained immunodeficiency (38, 39).
Although the Tyk2 mutation in B10.Q/J mice evidently impairs cellular responses to IL-12, IL-23, and IFN-α, precisely how the G→A transition affects the fate and function of Tyk2 in murine cells remains unclear. This point mutation is predicted to introduce a nonconservative amino acid (E775K) substitution in the Tyk2 JH2 domain. Unexpectedly, the B10.Q/J Tyk2 protein could not be detected by using either Tyk2- or phospho-Tyk2-specific Ab reagents (Fig. 4a and unpublished data), even in the presence of a proteasome inhibitor and despite detectable expression of Tyk2 mRNA. However, when the analogous E782K mutation was introduced in the human Tyk2 cDNA, protein expression in transfected cells was readily detectable, thus permitting functional evaluation of this pseudokinase domain mutant. Although devoid of any intrinsic kinase activity, the JH2 domain has recently been demonstrated to play a critical role in modulating Jak activation and function (18, 40, 41). Our preliminary experiments indicate that the E782K mutant, just like the R856G variant (18), is basally hyperphosphorylated (Fig. 5) and fails to complement IFN-α responsiveness in Tyk2-deficient 11,1 cells. Like E782, R856 is localized in the pseudokinase domain. These two residues are conserved throughout the tyrosine kinase superfamily and are thought to form an internal salt bridge, thus providing structural stability (18, 28). Given this structural relationship and the functional similarities, it is likely that the E782K mutant will also have poor kinase activity on receptor substrates (18). Further comparative studies of the human and mouse forms of the E and R Tyk2 mutants could resolve whether the apparent disparity in mutant Tyk2 protein expression and stability reflects a real species difference or stems from technical issues related to the use of anti-human Tyk2 Ab reagents.
The original descriptions of the Tyk2–/– mice (17, 21) and our characterization of the B10.Q/J spontaneous mutant indicate that a certain level of cytokine signaling and biologic effect can occur in the absence of Tyk2. Activation of Stat1, Stat2, and IRF-1 still occurs in Tyk2–/– cells on IFN treatment, and Tyk2–/– mice show only partial susceptibility to vaccinia challenge (17, 21). Similarly, a residual Stat4 phosphorylation response to IL-12 has been observed in both Tyk2–/– and B10.Q/J cells. We have previously demonstrated that this Tyk2-independent IL-12 response is biologically significant, because the in vivo rescue of B10.Q/J mice from T. gondii susceptibility by IL-18 treatment required endogenously produced IL-12 (3). Although the precise mechanism(s) of the IL-18 effect remains to be elucidated, we speculate that signals downstream of the IL-18R can effectively cooperate with partial IL-12 signaling presumably mediated by Jak2. One possible mechanism might involve IL-18-induced and p38-mediated phosphorylation of Stat4 at S721, an event recently shown to be critical for IL-12-induced IFN-γ production (42).
Our present findings and our earlier report that the B10.Q/J mouse is refractory to collagen-induced arthritis (16) clearly indicate a critical role for Tyk2 in autoimmune and inflammatory disease pathogenesis. The greater resistance of the B10.Q/J strain, relative to IFN-γR (43) or IL-12 knockout mice (12), could be explained by the pleiotropic role of Tyk2 in signaling by multiple cytokines, including IL-12, IL-23, and type I IFNs. Even though IL-12 has been shown to be the most potent differentiation factor for TH1 self-reactive lymphocytes, IL-12p35-deficient animals still develop significant autoimmune disease. In contrast, IL-23p19 knockout animals exhibited complete resistance to experimental autoimmune encephalomyelitis, an autoimmune demyelinating disease of the CNS, despite the presence of TH1 cells (44). Thus, disease expression requires both IL-12 and IL-23 operating in the inductive and effector phases of the autoimmune response, respectively. Our results predict that the B10.Q/J autoimmune response is blunted at both stages. It remains unclear whether hyporesponsiveness to type I IFNs further contributes to the resistance of B10.Q/J mice to collagen-induced arthritis. Nonetheless, central roles for type I IFNs and Tyk2 in the pathogenesis of both systemic lupus erythematosus (45) and lipopolysaccharide-induced septic shock (46) have recently been described. Thus, inhibitory targeting of Tyk2 activity represents an attractive treatment strategy for autoimmune and inflammatory diseases.
Finally, our successful identification of the underlying genetic lesion of the B10.Q/J strain illustrates the power and sensitivity of using T. gondii susceptibility as a biologic endpoint in revealing the most essential components of cell-mediated immunity. This we attribute to the inherently high virulence potential of T. gondii parasites, which maximizes disease penetrance and reduces the level of genetic redundancy to a minimum.
Supplementary Material
Acknowledgments
We thank E. Chin (Brown University), C. Hunter (University of Pennsylvania, Philadelphia), and R. Kastelein (DNAX) for providing reagents; and C. Biron, J. Louten, and M. Dalod (Brown University) for assistance with vesicular stomatitis virus infections. We are also grateful to A. VanSlyke for technical assistance. G.S.Y. thanks R. Ortmann, E. Shevach, A. Sher, and W. Pavan for previous collaboration and providing encouragement. This work was supported by National Institutes of Health Grant AI50618 (to G.S.Y.) and, in part, National Institutes of Health Grant RR 015578 (to the Center for Genetics and Genomics, Brown University) and the Rhode Island Foundation. M.H.S. was supported by Predoctoral (Graduate Assistance in Areas of National Need) Training Grant P200A000117 from the U.S. Department of Education. M.K. was supported by the Austrian Science Fund, and M.M. was supported by the Austrian Ministry of Education, Science, and Culture. S.P. was supported by grants from the Association pour la Recherche sur le Cancer.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Stat, signal transducer and activator of transcription; Jak, Janus kinase; JH, Jaks homology; LOD, logarithm of odds.
References
- 1.Scharton-Kersten, T. M., Yap, G., Magram, J. & Sher, A. (1997) J. Exp. Med. 185, 1261–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yap, G., Pesin, M. & Sher, A. (2000) J. Immunol. 165, 628–631. [DOI] [PubMed] [Google Scholar]
- 3.Yap, G. S., Ortmann, R., Shevach, E. & Sher, A. (2001) J. Immunol. 166, 5720–5725. [DOI] [PubMed] [Google Scholar]
- 4.Cooper, A. M., Magram, J., Ferrante, J. & Orme, I. M. (1997) J. Exp. Med. 186, 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mattner, F., Magram, J., Ferrante, J., Launois, P., Di Padova, K., Behin, R., Gately, M. K., Louis, J. A. & Alber, G. (1996) Eur. J. Immunol. 26, 1553–1559. [DOI] [PubMed] [Google Scholar]
- 6.Thierfelder, W. E., van Deursen, J. M., Yamamoto, K., Tripp, R. A., Sarawar, S. R., Carson, R. T., Sangster, M. Y., Vignali, D. A., Doherty, P. C., Grosveld, G. C., et al. (1996) Nature 382, 171–174. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan, M. H., Sun, Y. L., Hoey, T. & Grusby, M. J. (1996) Nature 382, 174–177. [DOI] [PubMed] [Google Scholar]
- 8.de Jong, R., Altare, F., Haagen, I. A., Elferink, D. G., Boer, T., van Breda Vriesman, P. J., Kabel, P. J., Draaisma, J. M., van Dissel, J. T., et al. (1998) Science 280, 1435–1438. [DOI] [PubMed] [Google Scholar]
- 9.Altare, F., Durandy, A., Lammas, D., Emile, J. F., Lamhamedi, S., Le Deist, F., Drysdale, P., Jouanguy, E., Doffinger, R., Bernaudin, F., et al. (1998) Science 280, 1432–1435. [DOI] [PubMed] [Google Scholar]
- 10.Gollob, J. A., Veenstra, K. G., Jyonouchi, H., Kelly, A. M., Ferrieri, P., Panka, D. J., Altare, F., Fieschi, C., Casanova, J. L., Frank, D. A., et al. (2000) J. Immunol. 165, 4120–4126. [DOI] [PubMed] [Google Scholar]
- 11.Gately, M. K., Renzetti, L. M., Magram, J., Stern, A. S., Adorini, L., Gubler, U. & Presky, D. H. (1998) Annu. Rev. Immunol. 16, 495–521. [DOI] [PubMed] [Google Scholar]
- 12.McIntyre, K. W., Shuster, D. J., Gillooly, K. M., Warrier, R. R., Connaughton, S. E., Hall, L. B., Arp, L. H., Gately, M. K. & Magram, J. (1996) Eur. J. Immunol. 26, 2933–2938. [DOI] [PubMed] [Google Scholar]
- 13.Trembleau, S., Penna, G., Bosi, E., Mortara, A., Gately, M. K. & Adorini, L. (1995) J. Exp. Med. 181, 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neurath, M. F., Fuss, I., Kelsall, B. L., Stuber, E. & Strober, W. (1995) J. Exp. Med. 182, 1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leonard, J. P., Waldburger, K. E. & Goldman, S. J. (1995) J. Exp. Med. 181, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ortmann, R., Smeltz, R., Yap, G., Sher, A. & Shevach, E. M. (2001) J. Immunol. 166, 5712–5719. [DOI] [PubMed] [Google Scholar]
- 17.Karaghiosoff, M., Neubauer, H., Lassnig, C., Kovarik, P., Schindler, H., Pircher, H., McCoy, B., Bogdan, C., Decker, T., Brem, G., et al. (2000) Immunity 13, 549–560. [DOI] [PubMed] [Google Scholar]
- 18.Yeh, T. C., Dondi, E., Uze, G. & Pellegrini, S. (2000) Proc. Natl. Acad. Sci. USA 97, 8991–8996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pellegrini, S., John, J., Shearer, M., Kerr, I. M. & Stark, G. R. (1989) Mol. Cell. Biol. 9, 4605–4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velazquez, L., Fellous, M., Stark, G. R. & Pellegrini, S. (1992) Cell 70, 313–322. [DOI] [PubMed] [Google Scholar]
- 21.Shimoda, K., Kato, K., Aoki, K., Matsuda, T., Miyamoto, A., Shibamori, M., Yamashita, M., Numata, A., Takase, K., Kobayashi, S., et al. (2000) Immunity 13, 561–571. [DOI] [PubMed] [Google Scholar]
- 22.Gauzzi, M. C., Barbieri, G., Richter, M. F., Uze, G., Ling, L., Fellous, M. & Pellegrini, S. (1997) Proc. Natl. Acad. Sci. USA 94, 11839–11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen, M., Cheng, A., Chen, Y. Q., Hymel, A., Hanson, E. P., Kimmel, L., Minami, Y., Taniguchi, T., Changelian, P. S. & O'Shea, J. J. (1997) Proc. Natl. Acad. Sci. USA 94, 6910–6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gauzzi, M. C., Velazquez, L., McKendry, R., Mogensen, K. E., Fellous, M. & Pellegrini, S. (1996) J. Biol. Chem. 271, 20494–20500. [DOI] [PubMed] [Google Scholar]
- 25.Zhou, Y. J., Hanson, E. P., Chen, Y. Q., Magnuson, K., Chen, M., Swann, P. G., Wange, R. L., Changelian, P. S. & O'Shea, J. J. (1997) Proc. Natl. Acad. Sci. USA 94, 13850–13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng, J., Witthuhn, B. A., Matsuda, T., Kohlhuber, F., Kerr, I. M. & Ihle, J. N. (1997) Mol. Cell. Biol. 17, 2497–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leonard, W. J. & O'Shea, J. J. (1998) Annu. Rev. Immunol. 16, 293–322. [DOI] [PubMed] [Google Scholar]
- 28.Hanks, S. & Hunter, T. (1995) in FactsBook Series, ed. Hardie, G. & Hanks, S. (Academic, London), pp. 7–47.
- 29.Parham, C., Chirica, M., Timans, J., Vaisberg, E., Travis, M., Cheung, J., Pflanz, S., Zhang, R., Singh, K. P., Vega, F., et al. (2002) J. Immunol. 168, 5699–5708. [DOI] [PubMed] [Google Scholar]
- 30.Baldi, L., Brown, K., Franzoso, G. & Siebenlist, U. (1996) J. Biol. Chem. 271, 376–379. [DOI] [PubMed] [Google Scholar]
- 31.Hou, D., Cenciarelli, C., Jensen, J. P., Nguygen, H. B. & Weissman, A. M. (1994) J. Biol. Chem. 269, 14244–14247. [PubMed] [Google Scholar]
- 32.Altare, F., Lammas, D., Revy, P., Jouanguy, E., Doffinger, R., Lamhamedi, S., Drysdale, P., Scheel-Toellner, D., Girdlestone, J., Darbyshire, P., et al. (1998) J. Clin. Invest. 102, 2035–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dorman, S. E. & Holland, S. M. (1998) J. Clin. Invest. 101, 2364–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altare, F., Ensser, A., Breiman, A., Reichenbach, J., Baghdadi, J. E., Fischer, A., Emile, J. F., Gaillard, J. L., Meinl, E. & Casanova, J. L. (2001) J. Infect. Dis. 184, 231–236. [DOI] [PubMed] [Google Scholar]
- 35.Jouanguy, E., Altare, F., Lamhamedi, S., Revy, P., Emile, J. F., Newport, M., Levin, M., Blanche, S., Seboun, E., Fischer, A., et al. (1996) N. Engl. J. Med. 335, 1956–1961. [DOI] [PubMed] [Google Scholar]
- 36.Newport, M. J., Huxley, C. M., Huston, S., Hawrylowicz, C. M., Oostra, B. A., Williamson, R. & Levin, M. (1996) N. Engl. J. Med. 335, 1941–1949. [DOI] [PubMed] [Google Scholar]
- 37.Dupuis, S., Dargemont, C., Fieschi, C., Thomassin, N., Rosenzweig, S., Harris, J., Holland, S. M., Schreiber, R. D. & Casanova, J. L. (2001) Science 293, 300–303. [DOI] [PubMed] [Google Scholar]
- 38.Ottenhoff, T. H., Verreck, F. A., Lichtenauer-Kaligis, E. G., Hoeve, M. A., Sanal, O. & van Dissel, J. T. (2002) Nat. Genet. 32, 97–105. [DOI] [PubMed] [Google Scholar]
- 39.Casanova, J. L. & Abel, L. (2002) Annu. Rev. Immunol. 20, 581–620. [DOI] [PubMed] [Google Scholar]
- 40.Saharinen, P., Takaluoma, K. & Silvennoinen, O. (2000) Mol. Cell. Biol. 20, 3387–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen, M., Cheng, A., Candotti, F., Zhou, Y. J., Hymel, A., Fasth, A., Notarangelo, L. D. & O'Shea, J. J. (2000) Mol. Cell. Biol. 20, 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morinobu, A., Gadina, M., Strober, W., Visconti, R., Fornace, A., Montagna, C., Feldman, G. M., Nishikomori, R. & O'Shea, J. J. (2002) Proc. Natl. Acad. Sci. USA 99, 12281–12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manoury-Schwartz, B., Chiocchia, G., Bessis, N., Abehsira-Amar, O., Batteux, F., Muller, S., Huang, S., Boissier, M. C. & Fournier, C. (1997) J. Immunol. 158, 5501–5506. [PubMed] [Google Scholar]
- 44.Cua, D. J., Sherlock, J., Chen, Y., Murphy, C. A., Joyce, B., Seymour, B., Lucian, L., To, W., Kwan, S., Churakova, T., et al. (2003) Nature 421, 744–748. [DOI] [PubMed] [Google Scholar]
- 45.Santiago-Raber, M. L., Baccala, R., Haraldsson, K. M., Choubey, D., Stewart, T. A., Kono, D. H. & Theofilopoulos, A. N. (2003) J. Exp. Med. 197, 777–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karaghiosoff, M., Steinborn, R., Kovarik, P., Kriegshauser, G., Baccarini, M., Donabauer, B., Reichart, U., Kolbe, T., Bogdan, C., Leanderson, T., et al. (2003) Nat. Immunol. 4, 471–477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.