Abstract
Cbl-associated protein (CAP) is an adaptor protein that interacts with both signaling and cytoskeletal proteins. Here, we characterize the expression, localization and potential function of CAP in striated muscle. CAP is markedly induced during myoblast differentiation, and colocalizes with vinculin during costamerogenesis. In adult mice, CAP is enriched in oxidative muscle fibers, and it is found in membrane anchorage complexes, including intercalated discs, costameres, and myotendinous junctions. Using both yeast two-hybrid and proteomic approaches, we identified the sarcomeric protein filamin C (FLNc) as a binding partner for CAP. When overexpressed, CAP recruits FLNc to cell–extracellular matrix adhesions, where the two proteins cooperatively regulate actin reorganization. Moreover, overexpression of CAP inhibits FLNc-induced cell spreading on fibronectin. In dystrophin-deficient mdx mice, the expression and membrane localization of CAP is increased, concomitant with the elevated plasma membrane content of FLNc, suggesting that CAP may compensate for the reduced membrane linkage of the myofibrils due to the loss of the dystroglycan–sarcoglycan complex in these mice. Thus, through its interaction with FLNc, CAP provides another link between the myofibril cytoskeleton and the plasma membrane of muscle cells, and it may play a dynamic role in the regulation and maintenance of muscle structural integrity.
INTRODUCTION
Striated muscle contains highly organized cytoskeletal networks that are critical for contractile activity (Au, 2004). The basic contractile units of the myofibril are sarcomeres. Z-discs comprise the border of individual sarcomeres, where antiparallel actin filaments that span these units are cross-linked. The contractile cytoskeleton of the myofibril is tethered to the muscle plasma membrane, or sarcolemma, at specialized membrane anchorage sites (Clark et al., 2002). Z-discs are attached to the sarcolemma at costameres, and protein complexes in these structures link the myofibril cytoskeleton to the extracellular matrix (ECM). Skeletal muscle myofibrils terminate at the myotendinous junction, whereas myofibrils of cardiomyocytes terminate at intercalated discs, where adjacent myocytes are connected to each other. All three types of membrane linkage share structural and molecular similarities, and they are important for transmitting chemical and mechanical signals across the sarcolemma, and in maintaining the structural integrity of muscle cells. Moreover, mutations in the costameric transmembrane adhesion complexes, including the dystrophin–glycoprotein complex (DGC) and the integrin–focal adhesion complex, account for a large percentage of human myopathies (Durbeej and Campbell, 2002; Spence et al., 2002; Dalkilic and Kunkel, 2003; Davies and Nowak, 2006).
Filamin C (FLNc) is an actin-binding protein that interacts with δ- and γ-sarcoglycan in the DGC (Thompson et al., 2000). A mutation in FLNc has been associated with a novel form of myofibrillar myopathy with clinical features of the late-onset limb girdle muscular dystrophy (Vorgerd et al., 2005). A recent study using knockout mice verified a crucial role for FLNc in muscle development and maintenance of muscle structural integrity (Dalkilic et al., 2006). FLNc also binds directly to the Z-disk proteins FATZ and myotilin (Gontier et al., 2005), resulting in its two-pool-distribution in muscle cells. The majority of FLNc is concentrated at Z-discs, whereas the remainder is found under the sarcolemmal membrane within the costamere region (Thompson et al., 2000). FLNc was also identified as a component of intercalated discs and myotendinous junctions (van der Ven et al., 2000). Interestingly, FLNc localization is abnormal in a number of muscle diseases (Sewry et al., 2002; Bonnemann et al., 2003; Beatham et al., 2004). For example, in Duchenne muscular dystrophy patients, the amount of FLNc at the sarcolemma is markedly increased, suggesting that FLNc may participate in a compensatory response to sarcolemmal damage, with the loss of dystrophin (Hoffman et al., 1987; Thompson et al., 2000).
The filamin family includes three isoforms, filamin A, B, and C (van der Flier and Sonnenberg, 2001). Although FLNa and FLNb are ubiquitously expressed in cells and tissues, FLNc is specifically found in striated muscles (Thompson et al., 2000). All filamins cross-link and stabilize three-dimensional networks of actin filaments, and they link them to cell membranes. These proteins contain an N-terminal actin-binding domain (ABD), followed by a long rod-shaped domain with 24 immunoglobulin (Ig)-like repeats. Filamins have been proposed to regulate actin polymerization, mechanoprotection, signal transduction, and cellular migration (Stossel et al., 2001; Feng and Walsh, 2004; Popowicz et al., 2006). Numerous studies have demonstrated an essential role for FLNa in cell motility and membrane stability (Robertson, 2005). FLNc has also been suggested to be involved in the regulation of cell morphology and the migration of myogenic progenitor cells (Tu et al., 2003; Goetsch et al., 2005).
CAP is an adaptor protein containing a SoHo domain in its N terminus and three tandem SH3 domains at the C terminus (Ribon et al., 1998b). CAP exists as multiple isoforms that arise through alternative splicing of the SORBS1 gene in different tissues and cell lines (Ribon et al., 1998b). These isoforms were previously given different names by various groups (Mandai et al., 1999; Zhang et al., 2003). Here, we use the name CAP to generically refer to these isoforms. Our previous studies have shown that the SoHo domain mediates the interaction of CAP with flotillin in lipid rafts, which may play a role in insulin-stimulated glucose uptake in 3T3-L1 adipocytes (Kimura et al., 2001; Liu et al., 2005). The C-terminal Src homology (SH)3 domains in CAP mediate its interaction with a variety of cytoskeletal or signaling molecules including Cbl, vinculin, and paxillin (Ribon et al., 1998b; Mandai et al., 1999). We recently demonstrated that CAP localizes to cell–ECM adhesion sites in fibroblasts through its association with the focal adhesion protein vinculin and that it negatively regulates cell spreading and migration by stabilizing focal adhesion complexes (Zhang et al., 2006). The enrichment of CAP in heart and skeletal muscle (Ribon et al., 1998b) prompted us to further characterize and explore its potential functions in these tissues. In this report, we identify FLNc as a novel binding partner for CAP, and we demonstrate that the CAP–FLNc interaction may provide another link between the myofibril cytoskeleton and the plasma membrane of the muscle cells, playing a dynamic role in the regulation and maintenance of muscle structural integrity.
MATERIALS AND METHODS
Antibodies and Reagents
CAP, insulin receptor (IR), green fluorescent protein (GFP), hemagglutinin (HA), and Myc antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). α-actinin antibodies were obtained from Sigma-Aldrich (St. Louis, MO). Filamin C antibody was a gift from Dr. Louis M. Kunkel. Myosin antibodies were from Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA). Vinculin and CAP antibodies were from Upstate Biotechnology (Lake Placid, NY), and Paxillin, caveolin 3, and integrin β1 antibodies were purchased from BD Biosciences (San Jose, CA). γ-Sarcoglycan antibodies were from Vision BioSystems (Newcastle, United Kingdom). The Akt antibodies were from Cell Signaling Technology (Beverly, MA). The Alexa Fluor secondary antibodies and phalloidin were from Molecular Probes (Eugene, OR). NADH and nitro blue tetrazolium and human plasma fibronectin were purchased from Sigma-Aldrich. Protein A/G-agarose beads were from Santa Cruz Biotechnology.
Animals
C57BL/6, mdx (C57BL/10ScSn-Dmdmdx/J), and control (C57BL/10ScSnJ) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All mice used in this study were housed in the Unit for Laboratory Animal Medicine at the University of Michigan.
Plasmids and Mutagenesis
The CAP cDNA used in our experiments represents the CAP2 isoform that was described previously (Zhang et al., 2003). Myc, GFP, glutathione transferase (GST)-tagged wild-type and mutant CAP constructs were described previously (Zhang et al., 2006). Full-length filamin C cDNA was cloned by reverse transcription-polymerase chain reaction (PCR) from mouse muscle mRNA, and it was subsequently subcloned into the pkH3 vector at EcoRI site. Fragments of filamin C were generated by PCR by using the full-length cDNA as template, and subcloned into the pGEX vector. The filamin A fragment was cloned form muscle mRNA and subcloned into pGEX vector. The HA-FLNc ΔR2 mutant was generated by PCR-mediated internal deletion. All mutations and cloning products were confirmed by sequencing.
Cell Culture and Transient Transfection
All cells were cultured in DMEM containing 10% fetal bovine serum (FBS), 100 U/ml penicillin G sodium, and 100 μg/ml streptomycin sulfate. L6 myoblasts were induced to differentiate by switching to DMEM containing 1% FBS, and C2C12 differentiation was induced by switching to DMEM with 4% horse serum. COS-1 cells were transfected using FuGENE 6 reagent (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions. L6 cells were transfected using Lipofectamine 2000 (Invitrogen).
Isolation of Neonatal Cardiomyocytes
Hearts were isolated from 3-d-old mouse pups and digested by collagenase. Dissociated cells were subjected to differential preplating to enrich for cardiomyocytes, and then they were plated in DMEM/F-12 with 10% FBS onto fibronectin-coated coverslips, at a density of 0.5–1 × 106 cells/cm2. Media were changed after 24 h of plating.
Immunofluorescence and Confocal Microscopy
Heart or various skeletal muscles was isolated and embedded in O.C.T. (Tissue-Tek; QIAGEN, Valencia, CA), snap-frozen in isopentane cooled in liquid nitrogen, and stored at −80°C. The frozen samples were cut into 10-μm sections, fixed in methanol for 5 min, followed by immunostaining. Cells grown on glass coverslips were fixed with 10% Formalin for 20 min, followed by permeabilization with 0.5% Triton X-100 for 5 min. Tissue sections or cells on coverslips were blocked with 1% bovine serum albumin, 1% ovalbumin, and 2% goat serum for 1 h, and then incubated with primary antibodies for 1 h followed by Alexa Fluor secondary antibodies for 0.5 h. Sections were mounted with Vectashield (Vector Laboratories, Burlingame, CA). Cells were imaged using a confocal fluorescence microscope (Olympus IX SLA; Olympus, Melville, NY). Images were then imported into Photoshop (Adobe Systems, Mountain View, CA) for processing.
For quantification of the membrane staining, all images were captured under the same laser intensity and Photomultiplier tube (PMT) setting. The staining intensity on the membrane was scored for 20 myofibers from each group, by using Olympus FluoView software.
Muscle Fiber Typing
Serial sections of skeletal muscle were subjected to either immunostaining with anti-CAP and anti-myosin (slow or fast) antibodies, or NADH-tetrazolium reductase (TR) staining. For NADH-TR staining, sections were incubated with 1 mg/ml NADH and 1 mg/ml nitro blue tetrazolium at 37°C for 30 min. Then they were rinsed with water, destained with acetone, mounted with Vectashield, and photographed.
Immunoprecipitation and Immunoblotting
For immunoprecipitation studies, cells were washed twice with ice-cold phosphate-buffered saline, and then they were lysed for 30 min at 4°C with buffer containing 50 mM Tris-HCl, pH 8.0, 135 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM sodium pyrophosphate, 1 mM Na3VO4, 10 mM NaF, and protease inhibitors (Roche Diagnostics). Lysates were clarified by centrifugation at 14,000 × g for 10 min. The supernatants were incubated with the indicated antibodies for 2 h at 4°C. Immune complexes were precipitated with protein A/G-agarose for 1 h at 4°C, and then they were washed extensively with lysis buffer before solubilization in SDS sample buffer. Bound proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membranes. Individual proteins were detected with the specific antibodies and visualized by blotting with horseradish peroxidase-conjugated secondary antibodies. For other studies, tissue or cells were lysed in radioimmunoprecipitation assay buffer (above-mentioned lysis buffer, including 0.5% sodium deoxycholate and 0.1% SDS) and followed by SDS-PAGE and immunoblotting.
GST Pull-Down Assay
GST fusion proteins were expressed in the Escherichia coli strain BL21 and purified as described previously (Liu et al., 2000). Cells were lysed as described above for immunoprecipitation. Lysates were incubated with GST fusion proteins immobilized on glutathione-Sepharose beads (GE Healthcare, Chalfont St. Giles, United Kingdom) for 1 h at 4°C. The beads were washed extensively with lysis buffer, and the bound proteins were solubilized in SDS sample buffer and analyzed by immunoblotting.
Triton X-100–soluble and – insoluble Fractionation
Cells were washed with cell solubilization buffer (CSB) containing 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid), 50 mM KCl, 10 mM EGTA, 3 mM MgCl2, 2 M glycerol, 2 mM NaF, 1 mM Na3VO4, and protease inhibitors, and then they were incubated for exactly 5 min at 4°C in CSB containing 1% Triton X-100. This soluble fraction was collected, and the plates were washed once with CSB. The remaining cytoskeletal fraction was lysed in extraction buffer containing 20 mM Tris-HCl, 300 mM NaCl, 30 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, and protease inhibitors. The triton-insoluble fraction was passed through a 28-gauge syringe 10 times before protein quantification and Western blot analysis.
Cell Spreading Assay
Serum-starved cells were collected by trypsinization, washed, counted, and resuspended in DMEM. Cells were kept in suspension for 1 h, and then 5 × 105 cells were added to 35-mm tissue culture dishes that were precoated with fibronectin (BD Biosciences). Cells were allowed to spread for the indicated times at 37°C, chilled on ice for 10 min, and then photographed. Spread cells were defined as cells with extended processes, lacking a rounded morphology and not phase-bright, whereas nonspread cells were rounded and phase-bright under microscope. Five random microscopic fields were counted per plate.
RESULTS
CAP Expression Is Induced during Myogenesis
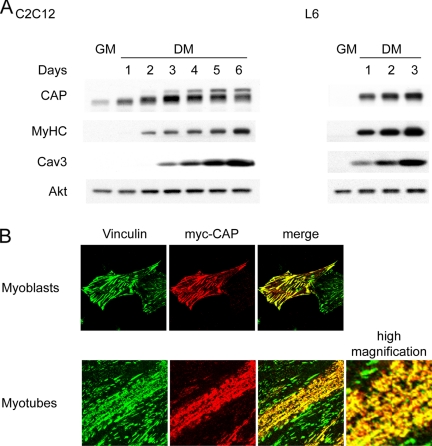
We first examined CAP expression during myoblast differentiation in well characterized muscle cell lines. Immunoblot analysis showed that in both C2C12 and L6 cells, CAP is expressed at low levels in undifferentiated myoblasts and up-regulated upon differentiation (Figure 1A). The increase in CAP protein levels is accompanied by increases in the myogenic markers myosin heavy chain and caveolin 3. Multiple spliced variants of CAP have been identified in different tissues and cell lines (Ribon et al., 1998b; Mandai et al., 1999; Zhang et al., 2003). Interestingly, several higher-molecular-weight isoforms appear during C2C12 myogenesis, whereas only one major isoform is expressed in L6 cells. The anti-CAP antibody used in immunoblotting and immunostaining experiments recognizes an N-terminal 16 amino acid region of CAP, which exists in all splice variants. The specificity of this antibody was confirmed by Western blot of muscle lysates from wild-type and CAP null mice (Lesniewski et al., 2007; Supplemental Figure S1).
Figure 1.
Expression and subcellular localization of CAP during myoblast differentiation. (A) CAP was dramatically induced during myoblast differentiation. Western blot of CAP protein level in C2C12 or L6 myoblast growing in growth media (GM) or after switching to differentiation media (DM). (B) L6 myoblasts were transfected with myc-CAP and stained with anti-vinculin and anti-myc antibodies. Myotubes were costained with vinculin (green) and endogenous CAP (red).
We next examined the localization of endogenous CAP in myoblasts versus myotubes. Our previous studies have shown that CAP localizes to focal adhesions in fibroblasts through its interaction with vinculin (Zhang et al., 2006). As expected, CAP is costained with vinculin at adhesion sites when overexpressed in L6 myoblasts (Figure 1B). Upon serum withdrawal, myoblasts fuse to form myotubes. During this process, vinculin redistributes to longitudinal aggregates that represent nascent costameres. As shown in Figure 1B, endogenous CAP colocalizes with vinculin in these structures.
CAP Localizes to Intercalated Discs, Costameres, and Myotendinous Junctions in Striated Muscles
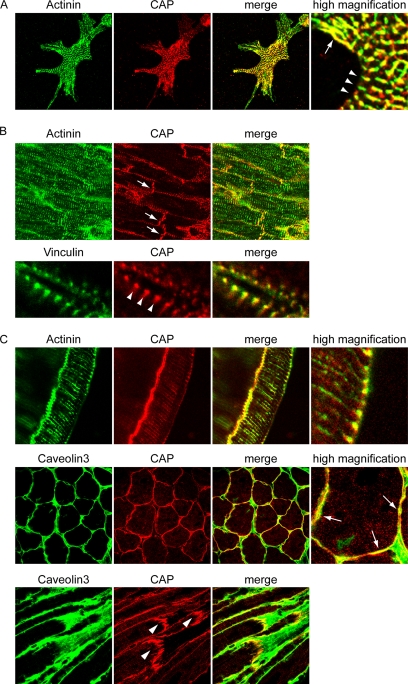
To confirm the localization of CAP in membrane anchorage complexes in vivo, we examined the subcellular localization of CAP in cardiomyocytes and mouse muscle sections (Figure 2). Neonatal cardiomyocytes were isolated from 3-d-old mice and plated on fibronectin-coated coverslips. Cells were fixed and costained with anti-α-actinin and anti-CAP antibodies. Confocal images were captured at the cell–substratum interface. CAP displayed clear focal adhesion (arrows) and costameric (arrowheads) localization that registered with overlying Z-disk staining of α-actinin. In heart sections, the majority of CAP immunostaining was observed at intercalated discs as well as along the lateral membranes of individual cardiomyocytes. The high magnification pictures clearly demonstrate that CAP colocalizes with vinculin at costameres along the membrane.
Figure 2.
Localization of CAP in striated muscle. (A) Mouse neonatal cardiomyocytes were costained with α-actinin and CAP. CAP localizes to focal adhesions (arrows) and costameres (arrowheads). (B) Mouse heart sections were stained with CAP and α-actinin or vinculin. CAP localizes to intercalated discs (arrows) and costameres (arrowheads). (C) Cross or longitudinal sections of mouse soleus muscle were stained with CAP and α-actinin or caveolin 3. CAP localizes to the sarcolemma membranes, with concentrated regions on the cross sections (arrows). Staining of the longitudinal sections showed CAP localization at costameres and MTJs (arrowheads).
In soleus muscle, CAP displayed a punctate pattern along the perimeter of the myofibers, colocalizing with caveolin 3. Consistent with this observation, CAP predominantly localizes to the sarcolemma membrane in the longitudinal sections, with concentrated localization at the costamere region. In addition, there is weaker staining of CAP at Z-discs where it costained with α-actinin. CAP is also localized and enriched at myotendinous junctions at the end of myofibers. Together, these results revealed a complex pattern of CAP localization that coincides with all the myofibril anchorage structures in striated muscles.
CAP Is Highly Expressed in Oxidative Muscle Fibers
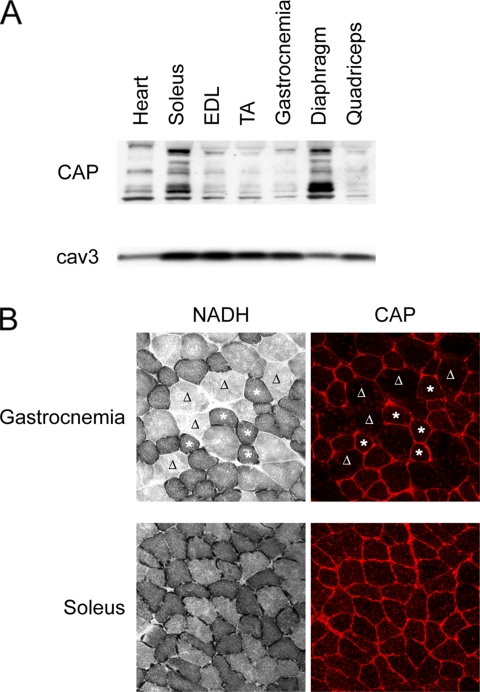
During the immunostaining studies of CAP localization in muscle, we noticed that CAP expression seemed to be fiber-type specific. We first examined the protein levels of CAP in different striated muscles with various fiber compositions. As shown in Figure 3A, CAP is expressed at high levels in soleus and diaphragm, whereas the TA, EDL, and quadriceps muscles have very low levels of CAP. As stated, the numerous bands seen represent multiple splicing variants of CAP that exist in muscle lysates (also see Supplemental Figure S1). Skeletal muscle fibers are classified into type I and type II (a, b, and x) according to the expression of the myosin heavy chain isoforms (Schiaffino and Reggiani, 1994). Based on their metabolic enzyme profiles, muscle fibers can also be classified as oxidative versus glycolytic fibers. Soleus and diaphragm muscles are rich in type I and type IIa fibers that contain more mitochondria and exhibit relatively higher rates of oxidative metabolism. To examine whether the expression of CAP is associated with the contractile and/or metabolic property of muscle fibers, consecutive sections of gastrocnemius muscles were stained for NADH-TR and CAP. As shown in Figure 3B, CAP is highly expressed in oxidative fibers that stained strongly for NADH-TR. Analysis of soleus and diaphragm muscle sections revealed that although these muscles contain different portions of type I versus type II fibers, the majority of the fibers stained positively for both CAP and NADH-TR (Figure 3B; data not shown). This is consistent with the high level of CAP protein detected by Western blot analysis. Therefore, CAP expression correlates with the oxidative activity of the muscle fibers.
Figure 3.
CAP is highly expressed in oxidative muscle fibers. (A) Western blot of CAP protein level in various striated muscles. (B) Serial cryosections of gastrocnemium (top) and soleus (bottom) muscles were stained for NADH-TR or CAP. Asterisks indicate examples of muscle fibers with high intensity of CAP staining and positive for NADH-TR staining on two consecutive sections of gastrocnemia muscle. Δ indicates fibers that are negative for either staining.
CAP Interacts with Muscle-specific Filamin
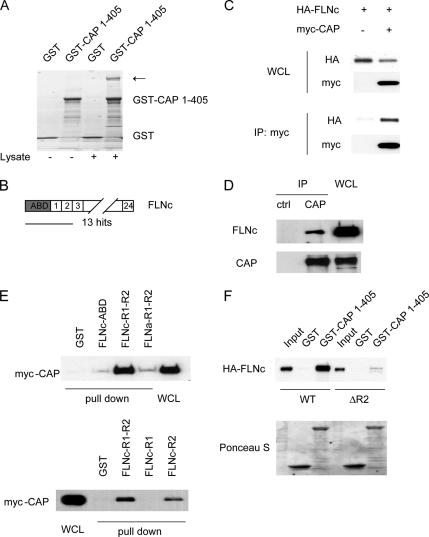
To understand the potential function of CAP in muscle, we decided to identify novel binding proteins using a proteomics approach. We prepared a GST-CAP fusion protein in which the three SH3 domains of the protein were deleted, to eliminate the many previously known partners that bind via the SH3 domains (Ribon et al., 1998b; Zhang et al., 2006). Cell lysates were precipitated with either GST alone or GST-CAP1-405, and the precipitates were subjected to SDS-PAGE, followed by staining with Coomassie Blue (Figure 4A). Compared with the no-lysate or GST alone controls, a few specific bands appeared in the GST-CAP1-405 precipitates (lane 4). Mass spectrometry analysis of isolated proteins revealed that one of the major bands was FLNc (indicated by an arrow). This finding agreed with the results obtained from a yeast two-hybrid screen of a 3T3-L1 cDNA library by using CAP1-405 as bait. From this screen, 13 of the 30 interacting proteins identified encoded an N-terminal fragment of FLNc (Figure 4B).
Figure 4.
CAP interacts with FLNc. (A) GST-CAP1-405 was used to affinity purify associating proteins. Cell lysates were subjected to pull down by GST or GST-CAP1-405, followed by SDS-PAGE, and stained with Coomassie Blue. Bands were cut out and analyzed by mass spectrometry. The arrow indicates the presence of FLNc specifically in GST-CAP1-405 pull down (lane 4). (B) FLNc is a yeast two-hybrid hit of CAP1-405. (C) HA-FLNc coimmunoprecipitated with myc-CAP when overexpressed in COS-1 cells. WCL, whole cell lysates. (D) Coimmunoprecipitation of endogenous FLNc with CAP in C2C12 cells. (E) CAP interacts with FLNc through the second Ig repeat (FLNc-R2). COS-1 cells were transfected with myc-CAP. Cell lysates were subjected to pull down experiments by GST-fusion fragments of filamins. (F) GST alone or GST-CAP1-405 were used to pull down in vitro translated HA-FLNc or the ΔR2 mutant. The precipitates were subjected to SDS-PAGE and Western blot analysis. Ponceau S staining shows the amount of GST fusion proteins used.
To confirm the interaction between CAP and FLNc, coimmunoprecipitation experiments were performed from cell lysates. HA-tagged FLNc was cotransfected into COS-1 cells with empty vector or myc-tagged CAP. Cell lysates were immunoprecipitated with anti-myc antibodies. As shown in Figure 4C, HA-FLNc coprecipitated with myc-CAP. To determine whether the endogenous proteins also interact with each other, C2C12 cell lysates were subjected to immunoprecipitation with anti-CAP antibodies or control rabbit IgG, followed by Western blot analysis. Endogenous FLNc was specifically coimmunoprecipitated with CAP (Figure 4D).
We next characterized the domains crucial for the interaction between CAP and FLNc. The cDNA recovered from the yeast two-hybrid screen encodes N-terminal sequences in FLNc that contain the ABD and the first two Ig repeats (R1 and R2; Figure 4B). To delineate the CAP-binding site in FLNc, we generated GST-fusion proteins of either the actin-binding domain or the first two repeats. COS-1 cells were transfected with myc-CAP, and cell lysates were incubated with GST fusion proteins. The precipitates were analyzed by Western blotting. As shown in Figure 4E, Myc-CAP was pulled down by GST-FLNc-R1-R2, but not GST-FLNc-ABD or GST alone. In contrast, a GST fusion protein containing the first two repeats of filamin A (GST-FLNa-R1-R2) did not pull down myc-CAP in the same experiments. To further determine which of the two repeats interacts with CAP, GST fusion proteins containing the single repeat were generated and used in a similar experiment. GST-FLNc-R2, but not R1, was able to precipitate myc-CAP. Together, these data demonstrated that CAP binds to the second Ig repeat and further that this interaction is specific for FLNc.
To establish that the interaction between CAP and FlnC was direct, HA-tagged wild-type (WT) and a mutant form of FLNc with the second repeat region deleted (ΔR2) were in vitro translated, and then they were incubated with the same GST-CAP1-405 fusion proteins used in the proteomics study. WT FLNc, but not the ΔR2 mutant, was precipitated by GST-CAP1-405, indicating the direct binding of FLNc to CAP through its second Ig repeat (Figure 4F).
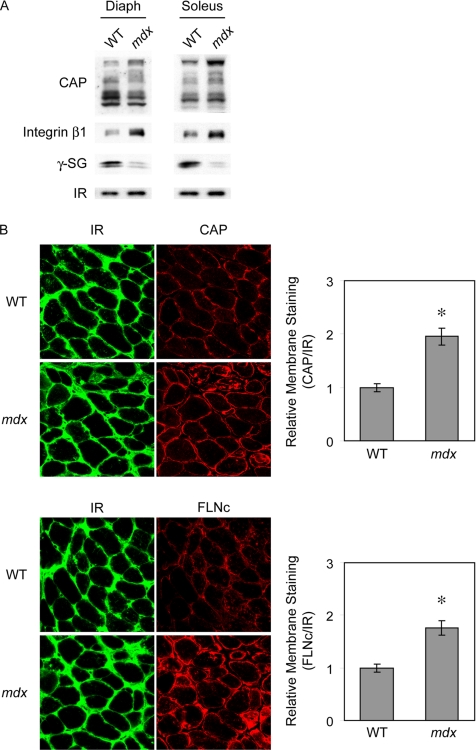
Analysis of CAP and FLNc in Mdx Mice
Many sarcolemmal proteins are involved in the pathogenesis of muscular dystrophies, and many of these are known to bind FLNc (Dalkilic and Kunkel, 2003). Indeed, in Duchenne muscular dystrophy (DMD), FLNc is significantly increased at the sarcolemma potentially to compensate for the loss of the dystrophin-associated protein complex (Thompson et al., 2000). To test the hypothesis that CAP might also be altered in dystrophic muscle, we examined the expression and localization of CAP in the mouse model for DMD, the dystrophin-deficient mdx mice (Sicinski et al., 1989). As shown in Figure 5A, the protein levels of the longer isoforms of CAP are increased in mdx mice compared with control mice. Consistent with previous reports, we also observed increased expression of integrin β1 in mdx mice, whereas γ-sarcoglycan levels were reduced (Hodges et al., 1997; Rafael and Brown, 2000; Cote et al., 2002). Immunofluorescence microscopy revealed a significant increase of CAP on the membrane of myofibers in the mdx mice compared with controls (Figure 5B and Supplemental Figure S2). As expected, the membrane staining of FLNc is also increased in mdx mice. Quantification of the staining intensity revealed an approximate twofold increase for both CAP and FLNc on the membrane. These results suggest that CAP plays a dynamic role in muscle, and it is potentially involved in muscular dystrophy. CAP might be responsible for the redistribution of FLNc at the mdx muscle membrane.
Figure 5.
Expression and localization of CAP in mdx mice. (A) Western blot analysis of CAP protein levels in diaphragm (Diaph) and Soleus from WT or mdx mice. (B) Immunostaining of the cross sections of diaphragm from WT or mdx mice for IR (green) and CAP or FLNc (red). The graphs on the right show the quantitation of membrane staining of CAP and FLNc relative to IR. The data represent mean ± SE. *p < 0.001.
CAP Recruits FLNc to Cell–ECM Adhesion Sites
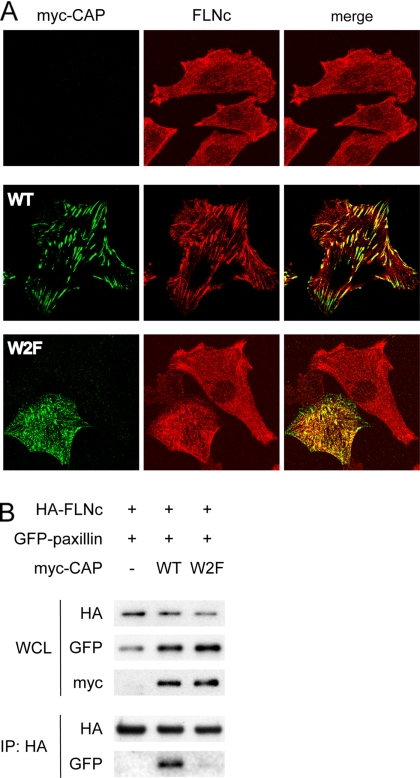
To test the hypothesis that CAP may regulate the cellular distribution of FLNc, we evaluated the effects of ectopically expressed CAP on the localization of FLNc by immunofluorescence studies. L6 myoblasts were transfected with myc-tagged wild-type or the W2F mutant of CAP. In the W2F mutant, two tryptophan residues in the first two SH3 domains were substituted with phenylalanine, rendering it unable to bind to vinculin and paxillin and thus losing its focal adhesion localization (Zhang et al., 2006). Overexpression of wild-type CAP induced a strong accumulation of endogenous FLNc at focal adhesion sites, whereas the W2F mutant failed to do so (Figure 6A). Thus, CAP functions as an adaptor protein to recruit FLNc to cell–ECM anchorage sites.
Figure 6.
CAP recruits FLNc to cell-ECM adhesions. (A) L6 myoblasts were transiently transfected with myc-tagged wild-type CAP or the W2F mutant and costained with anti-myc (green) and anti-FLNc (red) antibodies. (B) FLNc coprecipitated with paxillin in the presence of CAP. COS-1 cells were transfected with HA-FLNc, GFP-paxillin, and either empty vector, myc-tagged WT CAP, or the W2F mutant. Cells were lysed and immunoprecipitated with anti-HA antibody, and the coprecipitated GFP-paxillin was detected by Western blot analysis.
To further confirm that CAP may connect FLNc to focal adhesion components, immunoprecipitation experiments were perform to test the potential association between FLNc and paxillin when CAP is present. COS-1 cells were transfected with HA-FLNc, GFP-paxillin and an empty vector, myc-tagged wild-type, or the W2F mutant of CAP. As shown in Figure 6B, HA-FLNc was coprecipitated with GFP-paxillin only in the presence of wild-type CAP, but not the W2F mutant that cannot bind to paxillin, indicating that CAP bridges the interaction between these two proteins.
CAP and FLNc Induce Actin Reorganization and Affect Cell Spreading on Fibronectin
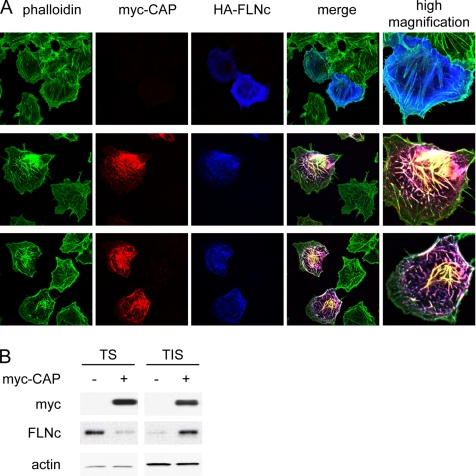
Extensive evidence demonstrates that filamins cross-link actin filaments and regulate cell shape and motility in response to various stimuli. Our recent studies have revealed a role for CAP in the regulation of the actin cytoskeleton and cell motility (Zhang et al., 2006). To investigate the functional significance of the CAP–FLNc interaction, we examined the effects of ectopic expression of these proteins in COS-1 cells. Cells were transfected with HA-FLNc and myc-CAP followed by immunofluorescence staining with anti-HA, anti-myc antibodies and phalloidin to visualize actin. Overexpressed FLNc exhibited lamellipodial meshwork and diffused cytoplasmic localization at the cell periphery, and it was also found along actin stress fibers. In cells doubly transfected with both HA-FLNc and myc-CAP, F-actin forms an intensive network, with FLNc and CAP decorating these enlarged actin bundles (Figure 7A). To determine whether CAP induced a redistribution of FLNc to actin cytoskeletal structures, cell lysates were separated into Triton X-100–soluble and insoluble fractions. Western blot analysis showed that overexpression of CAP induced a redistribution of FLNc to Triton X-100–insoluble fractions (Figure 7B). Thus, overexpression of CAP and FLNc together significantly induces the formation of extra-large actin bundles, suggesting that CAP enhances the actin cross-linking function of FLNc.
Figure 7.
Effects of CAP and FLNc on actin reorganization. (A) Overexpression of CAP and FLNc induces actin network formation. COS-1 cells were transfected with myc-CAP and HA-FLNc and costained with phalloidin (green), anti-myc (red), and anti-HA (blue) antibodies. (B) CAP induced redistribution of FLNc to detergent-insoluble structures. COS-1 cells were transfected with empty vector or myc-CAP. Triton-soluble (TS) and -insoluble (TIS) fractions were separated as described in Materials and Methods. Equal amount of proteins were loaded on a SDS–PAGE gel, and the levels of FLNc in each fraction were detected by Western blot. Actin was used as a loading control.
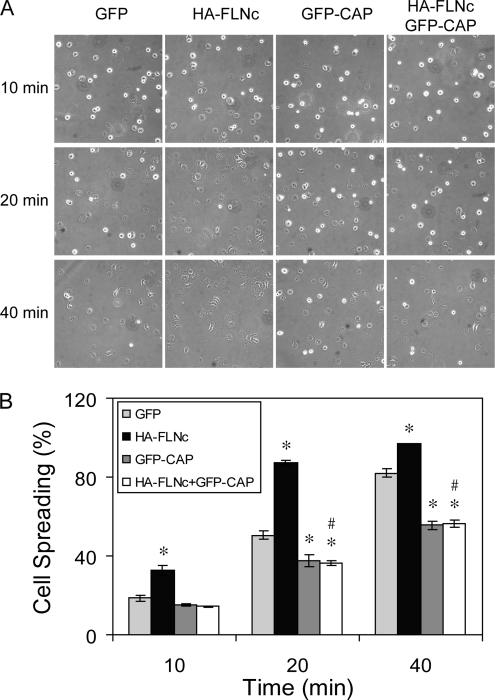
The importance of the CAP–FLNc interaction in regulating cell morphology was examined by evaluating cell spreading in response to the ECM. COS-1 cells were transiently transfected with GFP or GFP-CAP, with or without HA-FLNc, and then they were subsequently replated on fibronectin for 10, 20, or 40 min. More than 90% of cells were transfected when examined by fluorescence microscopy (data not shown). As shown in Figure 8, CAP overexpression impaired cell spreading, as reported previously (Zhang et al., 2006). In contrast, FLNc overexpression enhanced cell spreading at each time point, as shown by fewer phase-bright cells in the fields. Coexpression of CAP reversed the effect of FLNc, exhibiting a spreading defect similar to cells transfected with CAP alone. All the cells were able to spread after a prolonged incubation (data not shown). Together, these data suggest that CAP negatively regulates FLNc-mediated actin reorganization that results in cell spreading.
Figure 8.
Overexpression of CAP inhibits FLNc induced cell spreading on fibronectin. COS-1 cells were transfected with GFP control, HA-FLNc, GFP-CAP, or both. Cells were then plated on fibronectin-coated plates for indicated time and photographed. A representative field of each time points was shown (A), and the percentage of spread cells was calculated from five fields (B). More than 90% of cells were transfected when examined by GFP fluorescence and HA staining of an aliquot of the cells (data not shown). Results represent one of three experiments. The data represent mean ± SE. *p <0.05, compared with GFP; #p <0.02, compared with HA-FLNc.
DISCUSSION
In striated muscle, the specialized linkages between the subsarcolemmal cytoskeleton and the extracellular matrix are crucial to the transduction of contractile force, the organization of myofibrils, and the integrity of the membrane during contraction. Despite the importance of these connections, the molecular mechanisms that regulate membrane anchorage complexes remain poorly defined. In this study, we have identified CAP as an FLNc-associated protein that participates in linking the cytoskeleton to the sarcolemma in muscle cells.
Costameres were originally described as electron-dense plaques rich in the focal adhesion protein vinculin (Koteliansky and Gneushev, 1983; Pardo et al., 1983a,b; Shear and Bloch, 1985; Bloch et al., 2002). Indeed, these structures share many of the features of focal adhesions, and they are considered a striated muscle-specific elaboration of focal adhesions (Ervasti, 2003; Samarel, 2005). Our previous studies identified CAP as a component of the cell–ECM adhesion complex in fibroblasts. CAP interacts with both paxillin and vinculin, and the latter is crucial in anchoring CAP at adhesion sites (Zhang et al., 2006). CAP was also reported to localize to cell–cell junctions in epithelial cells (Mandai et al., 1999). These findings prompted us to explore whether CAP is also localized at membrane anchorage structures in muscle cells, including costameres, myotendinous junctions (MTJs), and intercalated discs.
During myoblast differentiation, CAP localization changes from focal adhesions to pre- and nascent costameres. Similar observations have been reported from studies of ponsin (another name of one CAP isoform) in human skeletal muscle cells (Gehmlich et al., 2007). In a recent study using an in vitro myoblast differentiation model, several focal adhesion components, including paxillin, vinculin, focal adhesion kinase (FAK), and integrin, were found to redistribute to costameres upon muscle differentiation (Quach and Rando, 2006). In addition, focal adhesion signaling through FAK is essential for costamerogenesis and myofibrillogenesis. The similar redistribution of CAP, together with the significant up-regulation of the CAP protein, suggests a potential role for CAP in the formation of costameres during myogenesis. Moreover, the increased expression of certain isoforms of CAP suggests different isoforms may have specific functions in mature muscle cells that have yet to be defined.
Using two independent approaches, we identified a novel interaction between CAP and the muscle-specific filamin FLNc. This interaction is mediated through the direct binding of the N-terminal part of CAP to the second repeat of FLNc. The N-terminal domain of FLNc binds to actin and the last repeat at the C terminus mediates its homodimerization. FLNc dimers cross-link F-actin to form parallel bundles or orthogonal networks. Our previous studies revealed that CAP binds to F-actin in an in vitro cosedimentation assay (Zhang et al., 2006). Therefore, we propose the existence of a CAP–FLNc–actin tertiary structure, where CAP helps to cross-link and stabilize the F-actin network. Indeed, cooverexpression of CAP and FLNc induced the formation of an intensive network of actin fibers, as shown in Figure 7A.
Filamins play an essential role in the modulation of cell shape and motility, and loss of function mutations in FLNa produce defects in neuronal migration (Fox et al., 1998). Although overlapping functions are suggested among filamin family members, few studies have examined the role of FLNc in the regulation of cell morphology and motility. FLNc has been suggested to be involved in migfillin-mediated cell shape regulation (Tu et al., 2003). Here, we show that overexpression of FLNc enhances cell spreading on fibronectin and that this function of FLNc is inhibited by CAP. Our recent study showed that CAP negatively regulates cell spreading via stabilizing cell–ECM adhesion structures (Zhang et al., 2006). We demonstrate here that CAP causes an accumulation of FLNc at adhesion sites, which may restrict the availability of FLNc at the periphery to dynamically regulate actin structures required for cell motility. The exact signaling pathway involved in the coordination between CAP and FLNc in modulating adhesion-mediated cytoskeleton rearrangement requires further study.
We propose a model in which CAP operates as a bifunctional adapter protein that binds to both filamin via N-terminal sequences, and cell adhesion proteins such as vinculin and paxillin via its C-terminal SH3 domains. Therefore, CAP may be able to reorganize the actin cytoskeleton by bringing filamin cross-linked actin to cell–ECM adhesion sites. In muscle cells, costameres contain two separate but synergistic cell—ECM-interacting complexes: the DGC and the vinculin–talin–integrin anchorage system (Anastasi et al., 2003, 2004). FLNc was initially identified as a binding partner of δ- and γ-sarcoglycans in the DGC complex (Thompson et al., 2000). In mdx mice, where the dystroglycan-associated protein dystrophin is deleted, the whole DGC complex is destabilized and degraded (Ervasti and Campbell, 1991; Ohlendieck and Campbell, 1991). Interestingly, membrane associated FLNc is greatly increased in mdx muscles despite an 80% decrease of sarcoglycans, suggesting another interaction/signal that regulates the localization of FLNc on the membrane.
Identification of the interaction between CAP and FLNc could potentially function as this second link of FLNc to the plasma membrane. CAP is a component of the integrin–focal adhesion complex through its binding to vinculin. Our data demonstrate that FLNc is recruited to cell–ECM adhesions by overexpression of CAP. Moreover, membrane staining of CAP is significantly increased in mdx muscles, suggesting that CAP may be responsible for the elevated FLNc on the membrane. Previous studies have shown an increase of α7β1 integrin in DMD patients and mdx mice, and overexpression of α7β1 integrin may compensate for the absence of the DGC complex and reduce the development of severe muscle disease in transgenic mice (Hodges et al., 1997; Burkin et al., 2001). By recruiting FLNc to the integrin–vinculin complex, CAP may function as an additional link that brings the myofibril cytoskeleton to the sarcolemma.
Another interesting finding from this study is that CAP is highly expressed in oxidative muscle fibers. Skeletal fibers are generally classified into type I and type II species that display marked differences in contraction, endurance, and metabolism. Type I fibers are rich in mitochondria that provide a slow but stable and long-lasting supply of ATP via oxidative metabolism. While type IIb fibers have the lowest content of mitochondria, these muscles have high levels of glycolytic enzymes, providing a rapid source of ATP independent of oxygen; therefore, they are more susceptible to fatigue. The metabolic and contractile properties of type IIa and IIx fibers lie in between. The expression of the CAP gene is transcriptionally induced by peroxisome proliferator-activated receptor (PPARγ) activators (Ribon et al., 1998a; Baumann et al., 2000a). Interestingly, both PPARγ and its coactivator PGC-1α induce a type I gene expression profile and a transition of muscle fibers from type IIb to type IIa and type I in transgenic mice (Lin et al., 2002; Wang et al., 2004). Moreover, PGC-1β drives the formation of oxidative IIx fibers (Arany et al., 2007). Therefore, the transcriptional regulation by PPARγ may account for the enriched expression of CAP in type I and IIa oxidative fibers, consistent with a role for CAP in the regulation of insulin-mediated glucose metabolism (Baumann et al., 2000b; Lesniewski et al., 2007). Interestingly, fiber type analyses in mdx mice have shown that the diaphragm muscle in these mice responds to progressive degeneration with a transition to a slower twitch phenotype. By 24 mo, there was a sevenfold increase in the slow twitch type I fiber and the type IIb/x fibers were almost gone in the mdx diaphragm. These changes were associated with reduced power output and marked increase in muscle endurance, both of which help to preserve the contractility and survival of the muscles (Petrof et al., 1993). Therefore, the enrichment of CAP in oxidative muscles and the increased expression and membrane localization in mdx muscles suggest that CAP may provide a link between the regulation of muscle structural integrity and metabolism that together contribute to the endurance and strength of skeletal muscle.
In summary, the identification of the interaction between CAP and filamin C furthers our understanding of the composition and regulation of membrane linkage complexes in skeletal muscle. Increased expression of CAP during myotube formation and its function in the regulation of FLNc localization and actin rearrangement suggest a dynamic role of CAP in the formation and maintenance of muscle structural integrity under normal and disease conditions. Further studies on the potential involvement of CAP in various myopathies may help us to better understand the underlying molecular mechanisms and contribute to novel approaches for treatment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Louis M. Kunkel for the FLNc antibody and Dr. Carey Lumeng for valuable discussions. This work was supported by a National Institutes of Health grants DK-61618 and DK-60591. M.Z. was supported by National Institute of Diabetes and Digestive and Kidney Diseases postdoctoral National Research Service Award fellowship F32 DK064551.
Abbreviations used:
- CAP
cbl-associated protein
- DGC
dystrophin-glycoprotein complex
- ECM
extracellular matrix
- FLNc
filamin C.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-06-0628) on September 26, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Anastasi G., et al. Distribution and localization of vinculin-talin-integrin system and dystrophin-glycoprotein complex in human skeletal muscle. Immunohistochemical study using confocal laser scanning microscopy. Cells Tissues Organs. 2003;175:151–164. doi: 10.1159/000074631. [DOI] [PubMed] [Google Scholar]
- Anastasi G., Cutroneo G., Rizzo G., Arco A., Santoro G., Bramanti P., Vitetta A. G., Pisani A., Trimarchi F., Favaloro A. Sarcoglycan and integrin localization in normal human skeletal muscle: a confocal laser scanning microscope study. Eur. J. Histochem. 2004;48:245–252. [PubMed] [Google Scholar]
- Arany Z., Lebrasseur N., Morris C., Smith E., Yang W., Ma Y., Chin S., Spiegelman B. M. The transcriptional coactivator PGC-1beta drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 2007;5:35–46. doi: 10.1016/j.cmet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Au Y. The muscle ultrastructure: a structural perspective of the sarcomere. Cell Mol. Life Sci. 2004;61:3016–3033. doi: 10.1007/s00018-004-4282-x. [DOI] [PubMed] [Google Scholar]
- Baumann C. A., Chokshi N., Saltiel A. R., Ribon V. Cloning and characterization of a functional peroxisome proliferator activator receptor-gamma-responsive element in the promoter of the CAP gene. J. Biol. Chem. 2000a;275:9131–9135. doi: 10.1074/jbc.275.13.9131. [DOI] [PubMed] [Google Scholar]
- Baumann C. A., Ribon V., Kanzaki M., Thurmond D. C., Mora S., Shigematsu S., Bickel P. E., Pessin J. E., Saltiel A. R. CAP defines a second signalling pathway required for insulin-stimulated glucose transport. Nature. 2000b;407:202–207. doi: 10.1038/35025089. [DOI] [PubMed] [Google Scholar]
- Beatham J., Romero R., Townsend S. K., Hacker T., van der Ven P. F., Blanco G. Filamin C interacts with the muscular dystrophy KY protein and is abnormally distributed in mouse KY deficient muscle fibres. Hum. Mol. Genet. 2004;13:2863–2874. doi: 10.1093/hmg/ddh308. [DOI] [PubMed] [Google Scholar]
- Bloch R. J., Capetanaki Y., O'Neill A., Reed P., Williams M. W., Resneck W. G., Porter N. C., Ursitti J. A. Costameres: repeating structures at the sarcolemma of skeletal muscle. Clin. Orthop. Relat. Res. 2002:S203–210. doi: 10.1097/00003086-200210001-00024. [DOI] [PubMed] [Google Scholar]
- Bonnemann C. G., et al. Filamin C accumulation is a strong but nonspecific immunohistochemical marker of core formation in muscle. J. Neurol. Sci. 2003;206:71–78. doi: 10.1016/s0022-510x(02)00341-6. [DOI] [PubMed] [Google Scholar]
- Burkin D. J., Wallace G. Q., Nicol K. J., Kaufman D. J., Kaufman S. J. Enhanced expression of the alpha 7 beta 1 integrin reduces muscular dystrophy and restores viability in dystrophic mice. J. Cell Biol. 2001;152:1207–1218. doi: 10.1083/jcb.152.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K. A., McElhinny A. S., Beckerle M. C., Gregorio C. C. Striated muscle cytoarchitecture: an intricate web of form and function. Annu. Rev. Cell Dev. Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- Cote P. D., Moukhles H., Carbonetto S. Dystroglycan is not required for localization of dystrophin, syntrophin, and neuronal nitric-oxide synthase at the sarcolemma but regulates integrin alpha 7B expression and caveolin-3 distribution. J. Biol. Chem. 2002;277:4672–4679. doi: 10.1074/jbc.M106879200. [DOI] [PubMed] [Google Scholar]
- Dalkilic I., Kunkel L. M. Muscular dystrophies: genes to pathogenesis. Curr. Opin. Genet. Dev. 2003;13:231–238. doi: 10.1016/s0959-437x(03)00048-0. [DOI] [PubMed] [Google Scholar]
- Dalkilic I., Schienda J., Thompson T. G., Kunkel L. M. Loss of filamin C (FLNc) results in severe defects in myogenesis and myotube structure. Mol. Cell Biol. 2006;26:6522–6534. doi: 10.1128/MCB.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies K. E., Nowak K. J. Molecular mechanisms of muscular dystrophies: old and new players. Nat. Rev. Mol. Cell Biol. 2006;7:762–773. doi: 10.1038/nrm2024. [DOI] [PubMed] [Google Scholar]
- Durbeej M., Campbell K. P. Muscular dystrophies involving the dystrophin-glycoprotein complex: an overview of current mouse models. Curr. Opin. Genet. Dev. 2002;12:349–361. doi: 10.1016/s0959-437x(02)00309-x. [DOI] [PubMed] [Google Scholar]
- Ervasti J. M. Costameres: the Achilles' heel of Herculean muscle. J. Biol. Chem. 2003;278:13591–13594. doi: 10.1074/jbc.R200021200. [DOI] [PubMed] [Google Scholar]
- Ervasti J. M., Campbell K. P. Membrane organization of the dystrophin-glycoprotein complex. Cell. 1991;66:1121–1131. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- Feng Y., Walsh C. A., et al. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat. Cell Biol. 2004;6:1034–1038. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- Fox J. W., et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- Gehmlich K., Pinotsis N., Hayess K., van der Ven P. F., Milting H., El Banayosy A., Korfer R., Wilmanns M., Ehler E., Furst D. O. Paxillin and ponsin interact in nascent costameres of muscle cells. J. Mol. Biol. 2007;369:665–682. doi: 10.1016/j.jmb.2007.03.050. [DOI] [PubMed] [Google Scholar]
- Goetsch S. C., Martin C. M., Embree L. J., Garry D. J. Myogenic progenitor cells express filamin C in developing and regenerating skeletal muscle. Stem Cells Dev. 2005;14:181–187. doi: 10.1089/scd.2005.14.181. [DOI] [PubMed] [Google Scholar]
- Gontier Y., Taivainen A., Fontao L., Sonnenberg A., van der Flier A., Carpen O., Faulkner G., Borradori L. The Z-disc proteins myotilin and FATZ-1 interact with each other and are connected to the sarcolemma via muscle-specific filamins. J. Cell Sci. 2005;118:3739–3749. doi: 10.1242/jcs.02484. [DOI] [PubMed] [Google Scholar]
- Hodges B. L., Hayashi Y. K., Nonaka I., Wang W., Arahata K., Kaufman S. J. Altered expression of the alpha7beta1 integrin in human and murine muscular dystrophies. J. Cell Sci. 1997;110:2873–2881. doi: 10.1242/jcs.110.22.2873. [DOI] [PubMed] [Google Scholar]
- Hoffman E. P., Brown R. H., Jr, Kunkel L. M. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Kimura A., Baumann C. A., Chiang S. H., Saltiel A. R. The sorbin homology domain: a motif for the targeting of proteins to lipid rafts. Proc. Natl. Acad. Sci. USA. 2001;98:9098–9103. doi: 10.1073/pnas.151252898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koteliansky V. E., Gneushev G. N. Vinculin localization in cardiac muscle. FEBS Lett. 1983;159:158–160. doi: 10.1016/0014-5793(83)80437-2. [DOI] [PubMed] [Google Scholar]
- Lesniewski L. A., Hosch S. E., Neels J. G., de Luca C., Pashmforoush M., Lumeng C. N., Chiang S. H., Scadeng M., Saltiel A. R., Olefsky J. M. Bone marrow-specific Cap gene deletion protects against high-fat diet-induced insulin resistance. Nat. Med. 2007;13:455–462. doi: 10.1038/nm1550. [DOI] [PubMed] [Google Scholar]
- Lin J., et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Liu J., Deyoung S. M., Zhang M., Dold L. H., Saltiel A. R. The stomatin/prohibitin/flotillin/HflK/C domain of flotillin-1 contains distinct sequences that direct plasma membrane localization and protein interactions in 3T3–L1 adipocytes. J. Biol. Chem. 2005;280:16125–16134. doi: 10.1074/jbc.M500940200. [DOI] [PubMed] [Google Scholar]
- Liu J., Wu J., Oliver C., Shenolikar S., Brautigan D. L. Mutations of the serine phosphorylated in the protein phosphatase-1-binding motif in the skeletal muscle glycogen-targeting subunit. Biochem. J. 2000;346:77–82. [PMC free article] [PubMed] [Google Scholar]
- Mandai K., Nakanishi H., Satoh A., Takahashi K., Satoh K., Nishioka H., Mizoguchi A., Takai Y. Ponsin/SH3P 12, an l-afadin- and vinculin-binding protein localized at cell-cell and cell-matrix adherens junctions. J. Cell Biol. 1999;144:1001–1017. doi: 10.1083/jcb.144.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlendieck K., Campbell K. P. Dystrophin-associated proteins are greatly reduced in skeletal muscle from mdx mice. J. Cell Biol. 1991;115:1685–1694. doi: 10.1083/jcb.115.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo J. V., Siliciano J. D., Craig S. W. Vinculin is a component of an extensive network of myofibril-sarcolemma attachment regions in cardiac muscle fibers. J. Cell Biol. 1983a;97:1081–1088. doi: 10.1083/jcb.97.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo J. V., Siliciano J. D., Craig S. W. A vinculin-containing cortical lattice in skeletal muscle: transverse lattice elements (“costameres”) mark sites of attachment between myofibrils and sarcolemma. Proc. Natl. Acad. Sci. USA. 1983b;80:1008–1012. doi: 10.1073/pnas.80.4.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrof B. J., Stedman H. H., Shrager J. B., Eby J., Sweeney H. L., Kelly A. M. Adaptations in myosin heavy chain expression and contractile function in dystrophic mouse diaphragm. Am. J. Physiol. 1993;265:C834–C841. doi: 10.1152/ajpcell.1993.265.3.C834. [DOI] [PubMed] [Google Scholar]
- Popowicz G. M., Schleicher M., Noegel A. A., Holak T. A. Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem. Sci. 2006;31:411–419. doi: 10.1016/j.tibs.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Quach N. L., Rando T. A. Focal adhesion kinase is essential for costamerogenesis in cultured skeletal muscle cells. Dev. Biol. 2006;293:38–52. doi: 10.1016/j.ydbio.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Rafael J. A., Brown S. C. Dystrophin and utrophin: genetic analyses of their role in skeletal muscle. Microsc. Res. Tech. 2000;48:155–166. doi: 10.1002/(SICI)1097-0029(20000201/15)48:3/4<155::AID-JEMT4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Ribon V., Johnson J. H., Camp H. S., Saltiel A. R. Thiazolidinediones and insulin resistance: peroxisome proliferatoractivated receptor gamma activation stimulates expression of the CAP gene. Proc. Natl. Acad. Sci. USA. 1998a;95:14751–14756. doi: 10.1073/pnas.95.25.14751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribon V., Printen J. A., Hoffman N. G., Kay B. K., Saltiel A. R. A novel, multifuntional c-Cbl binding protein in insulin receptor signaling in 3T3–L1 adipocytes. Mol. Cell Biol. 1998b;18:872–879. doi: 10.1128/mcb.18.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson S. P. Filamin A: phenotypic diversity. Curr. Opin. Genet. Dev. 2005;15:301–307. doi: 10.1016/j.gde.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Samarel A. M. Costameres, focal adhesions, and cardiomyocyte mechanotransduction. Am. J. Physiol. 2005;289:H2291–H2301. doi: 10.1152/ajpheart.00749.2005. [DOI] [PubMed] [Google Scholar]
- Schiaffino S., Reggiani C. Myosin isoforms in mammalian skeletal muscle. J. Appl. Physiol. 1994;77:493–501. doi: 10.1152/jappl.1994.77.2.493. [DOI] [PubMed] [Google Scholar]
- Sewry C. A., et al. The spectrum of pathology in central core disease. Neuromuscul. Disord. 2002;12:930–938. doi: 10.1016/s0960-8966(02)00135-9. [DOI] [PubMed] [Google Scholar]
- Shear C. R., Bloch R. J. Vinculin in subsarcolemmal densities in chicken skeletal muscle: localization and relationship to intracellular and extracellular structures. J. Cell Biol. 1985;101:240–256. doi: 10.1083/jcb.101.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicinski P., Geng Y., Ryder-Cook A. S., Barnard E. A., Darlison M. G., Barnard P. J. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- Spence H. J., Chen Y. J., Winder S. J. Muscular dystrophies, the cytoskeleton and cell adhesion. Bioessays. 2002;24:542–552. doi: 10.1002/bies.10098. [DOI] [PubMed] [Google Scholar]
- Stossel T. P., Condeelis J., Cooley L., Hartwig J. H., Noegel A., Schleicher M., Shapiro S. S. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- Thompson T. G., Chan Y. M., Hack A. A., Brosius M., Rajala M., Lidov H. G., McNally E. M., Watkins S., Kunkel L. M. Filamin 2 (FLN2): a muscle-specific sarcoglycan interacting protein. J. Cell Biol. 2000;148:115–126. doi: 10.1083/jcb.148.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y., Wu S., Shi X., Chen K., Wu C. Migfilin and Mig-2 link focal adhesions to filamin and the actin cytoskeleton and function in cell shape modulation. Cell. 2003;113:37–47. doi: 10.1016/s0092-8674(03)00163-6. [DOI] [PubMed] [Google Scholar]
- van der Flier A., Sonnenberg A. Structural and functional aspects of filamins. Biochim. Biophys. Acta. 2001;1538:99–117. doi: 10.1016/s0167-4889(01)00072-6. [DOI] [PubMed] [Google Scholar]
- van der Ven P. F., Obermann W. M., Lemke B., Gautel M., Weber K., Furst D. O. Characterization of muscle filamin isoforms suggests a possible role of gamma-filamin/ABP-L in sarcomeric Z-disc formation. Cell Motil. Cytoskeleton. 2000;45:149–162. doi: 10.1002/(SICI)1097-0169(200002)45:2<149::AID-CM6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Vorgerd M., et al. A mutation in the dimerization domain of filamin c causes a novel type of autosomal dominant myofibrillar myopathy. Am. J. Hum. Genet. 2005;77:297–304. doi: 10.1086/431959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. X., Zhang C. L., Yu R. T., Cho H. K., Nelson M. C., Bayuga-Ocampo C. R., Ham J., Kang H., Evans R. M. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2:e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Kimura A., Saltiel A. R. Cloning and characterization of Cbl-associated protein splicing isoforms. Mol. Med. 2003;9:18–25. [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Liu J., Cheng A., Deyoung S. M., Chen X., Dold L. H., Saltiel A. R. CAP interacts with cytoskeletal proteins and regulates adhesion-mediated ERK activation and motility. EMBO J. 2006;25:5284–5293. doi: 10.1038/sj.emboj.7601406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.