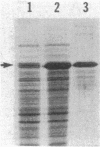
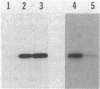
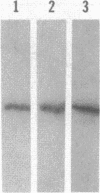
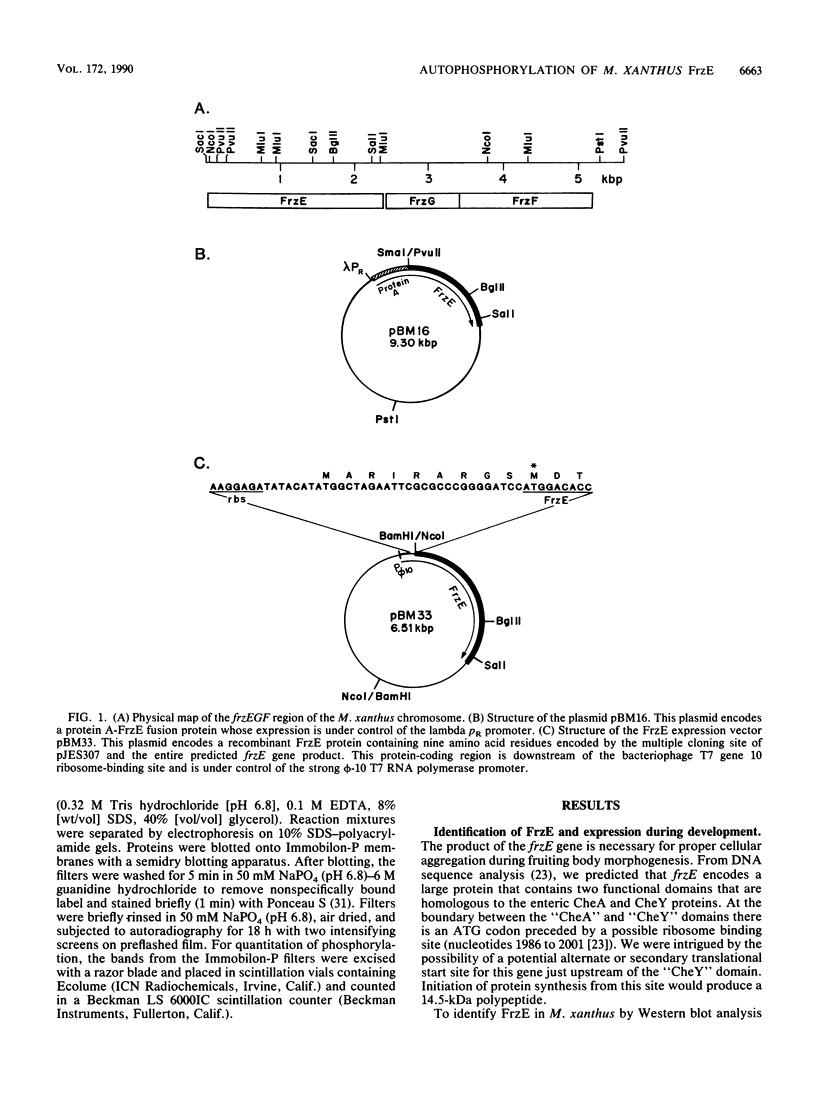
Abstract
Myxococcus xanthus exhibits multicellular interactions during vegetative growth and fruiting body formation. Gliding motility is needed for these interactions. The frizzy (frz) genes are required to control directed motility. FrzE is homologous to both CheA and CheY from Salmonella typhimurium. We used polyclonal antiserum raised against a fusion protein to detect FrzE in M. xanthus extracts by Western immunoblot analysis. FrzE was clearly present during vegetative growth and at much lower levels during development. A recombinant FrzE protein was overproduced in Escherichia coli, purified from inclusion bodies, and renatured. FrzE was autophosphorylated when it was incubated in the presence of [gamma-32P]ATP and MnCl2. Chemical analyses of the phosphorylated FrzE protein indicated that it contained an acylphosphate; probably phosphoaspartate. FrzE was phosphorylated in an intramolecular reaction. Based on these observations, we propose a model of the mechanism of FrzE phosphorylation in which autophosphorylation initially occurs at a conserved histidine residue within the "CheA" domain and then, via an intramolecular transphosphorylation, is transferred to a conserved aspartate residue within the "CheY" domain.
Full text
PDF


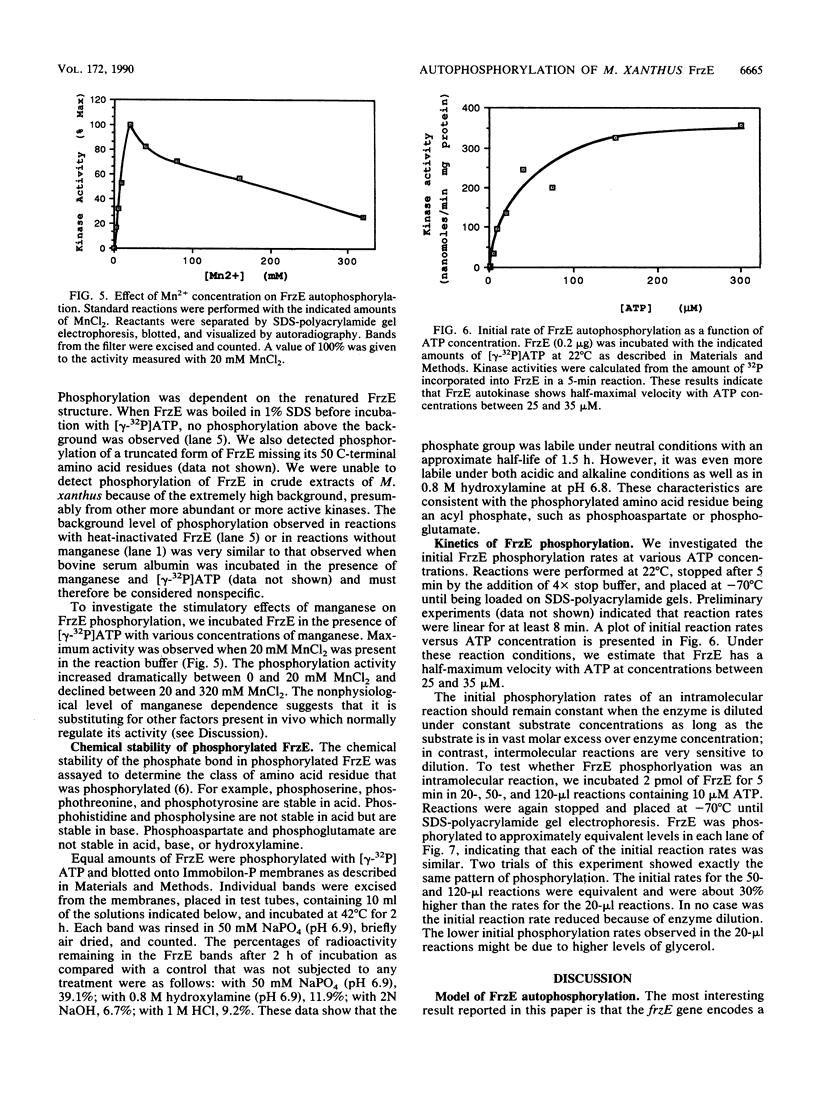
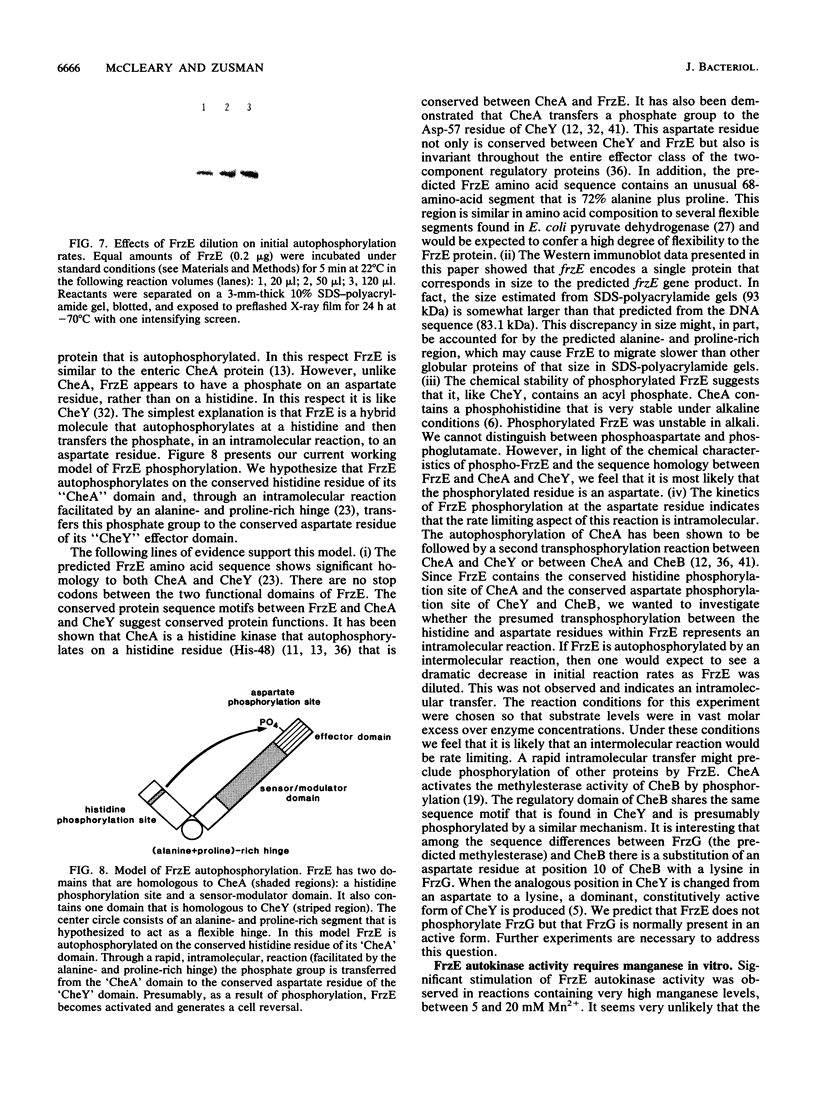


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blackhart B. D., Zusman D. R. "Frizzy" genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackhart B. D., Zusman D. R. Analysis of the products of the Myxococcus xanthus frz genes. J Bacteriol. 1986 May;166(2):673–678. doi: 10.1128/jb.166.2.673-678.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackhart B. D., Zusman D. R. Cloning and complementation analysis of the "Frizzy" genes of Myxococcus xanthus. Mol Gen Genet. 1985;198(2):243–254. doi: 10.1007/BF00383002. [DOI] [PubMed] [Google Scholar]
- Borkovich K. A., Kaplan N., Hess J. F., Simon M. I. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret R. B., Hess J. F., Simon M. I. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci U S A. 1990 Jan;87(1):41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss J. E., Stull J. T. Measurement of chemical phosphate in proteins. Methods Enzymol. 1983;99:7–14. doi: 10.1016/0076-6879(83)99035-3. [DOI] [PubMed] [Google Scholar]
- Campos J. M., Geisselsoder J., Zusman D. R. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978 Feb 25;119(2):167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- DWORKIN M. Nutritional requirements for vegetative growth of Myxococcus xanthus. J Bacteriol. 1962 Aug;84:250–257. doi: 10.1128/jb.84.2.250-257.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forst S., Delgado J., Inouye M. Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6052–6056. doi: 10.1073/pnas.86.16.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamood A. N., Iglewski B. H. Expression of the Pseudomonas aeruginosa toxA positive regulatory gene (regA) in Escherichia coli. J Bacteriol. 1990 Feb;172(2):589–594. doi: 10.1128/jb.172.2.589-594.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J. F., Bourret R. B., Simon M. I. Histidine phosphorylation and phosphoryl group transfer in bacterial chemotaxis. Nature. 1988 Nov 10;336(6195):139–143. doi: 10.1038/336139a0. [DOI] [PubMed] [Google Scholar]
- Hess J. F., Oosawa K., Kaplan N., Simon M. I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988 Apr 8;53(1):79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- Hess J. F., Oosawa K., Matsumura P., Simon M. I. Protein phosphorylation is involved in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7609–7613. doi: 10.1073/pnas.84.21.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Roitsch T., Ankenbauer R. G., Gordon M. P., Nester E. W. The VirA protein of Agrobacterium tumefaciens is autophosphorylated and is essential for vir gene regulation. J Bacteriol. 1990 Feb;172(2):525–530. doi: 10.1128/jb.172.2.525-530.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J., Kustu S. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc Natl Acad Sci U S A. 1988 Jul;85(14):4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D. E., Jr Biochemistry of sensing and adaptation in a simple bacterial system. Annu Rev Biochem. 1981;50:765–782. doi: 10.1146/annurev.bi.50.070181.004001. [DOI] [PubMed] [Google Scholar]
- Koshland D. E., Jr, Sanders D. A., Weis R. M. Roles of methylation and phosphorylation in the bacterial sensing system. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 1):11–17. doi: 10.1101/sqb.1988.053.01.004. [DOI] [PubMed] [Google Scholar]
- Liu J. D., Parkinson J. S. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8703–8707. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Stock J. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J Biol Chem. 1989 Oct 15;264(29):17337–17342. [PubMed] [Google Scholar]
- McBride M. J., Weinberg R. A., Zusman D. R. "Frizzy" aggregation genes of the gliding bacterium Myxococcus xanthus show sequence similarities to the chemotaxis genes of enteric bacteria. Proc Natl Acad Sci U S A. 1989 Jan;86(2):424–428. doi: 10.1073/pnas.86.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary W. R., McBride M. J., Zusman D. R. Developmental sensory transduction in Myxococcus xanthus involves methylation and demethylation of FrzCD. J Bacteriol. 1990 Sep;172(9):4877–4887. doi: 10.1128/jb.172.9.4877-4887.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary W. R., Zusman D. R. FrzE of Myxococcus xanthus is homologous to both CheA and CheY of Salmonella typhimurium. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5898–5902. doi: 10.1073/pnas.87.15.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Parker S. R., Talbert P. B., Houts S. E. Interactions between chemotaxis genes and flagellar genes in Escherichia coli. J Bacteriol. 1983 Jul;155(1):265–274. doi: 10.1128/jb.155.1.265-274.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. Protein phosphorylation in bacterial chemotaxis. Cell. 1988 Apr 8;53(1):1–2. doi: 10.1016/0092-8674(88)90478-3. [DOI] [PubMed] [Google Scholar]
- Radford S. E., Laue E. D., Perham R. N., Martin S. R., Appella E. Conformational flexibility and folding of synthetic peptides representing an interdomain segment of polypeptide chain in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. J Biol Chem. 1989 Jan 15;264(2):767–775. [PubMed] [Google Scholar]
- Ravid S., Matsumura P., Eisenbach M. Restoration of flagellar clockwise rotation in bacterial envelopes by insertion of the chemotaxis protein CheY. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7157–7161. doi: 10.1073/pnas.83.19.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronson C. W., Nixon B. T., Ausubel F. M. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987 Jun 5;49(5):579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- Salinovich O., Montelaro R. C. Reversible staining and peptide mapping of proteins transferred to nitrocellulose after separation by sodium dodecylsulfate-polyacrylamide gel electrophoresis. Anal Biochem. 1986 Aug 1;156(2):341–347. doi: 10.1016/0003-2697(86)90263-0. [DOI] [PubMed] [Google Scholar]
- Sanders D. A., Gillece-Castro B. L., Stock A. M., Burlingame A. L., Koshland D. E., Jr Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J Biol Chem. 1989 Dec 25;264(36):21770–21778. [PubMed] [Google Scholar]
- Sanders D. A., Mendez B., Koshland D. E., Jr Role of the CheW protein in bacterial chemotaxis: overexpression is equivalent to absence. J Bacteriol. 1989 Nov;171(11):6271–6278. doi: 10.1128/jb.171.11.6271-6278.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock A., Chen T., Welsh D., Stock J. CheA protein, a central regulator of bacterial chemotaxis, belongs to a family of proteins that control gene expression in response to changing environmental conditions. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1403–1407. doi: 10.1073/pnas.85.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Weinberg R. A., Zusman D. R. Evidence that the Myxococcus xanthus frz genes are developmentally regulated. J Bacteriol. 1989 Nov;171(11):6174–6186. doi: 10.1128/jb.171.11.6174-6186.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente S. R., Villalba M., Schramm V. L., Rosen O. M. Mn2(+)-binding properties of a recombinant protein-tyrosine kinase derived from the human insulin receptor. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2805–2809. doi: 10.1073/pnas.87.7.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie D., Stock A., Wong C. Y., Stock J. Sensory transduction in bacterial chemotaxis involves phosphotransfer between Che proteins. Biochem Biophys Res Commun. 1988 Mar 15;151(2):891–896. doi: 10.1016/s0006-291x(88)80365-6. [DOI] [PubMed] [Google Scholar]
- Zusman D. R. "Frizzy" mutants: a new class of aggregation-defective developmental mutants of Myxococcus xanthus. J Bacteriol. 1982 Jun;150(3):1430–1437. doi: 10.1128/jb.150.3.1430-1437.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]