Abstract
Background
Chronic obstructive pulmonary disease (COPD) is an inflammatory lung disease associated with significant systemic consequences. Recognition of the systemic manifestations has stimulated interest in identifying circulating biomarkers in these patients. A systematic analysis was undertaken of multiple protein analytes in the serum of well characterised patients with COPD and matched controls using novel protein microarray platform (PMP) technology.
Methods
Forty‐eight patients (65% men) with COPD (forced expiratory volume in 1 s <55%) and 48 matched controls were studied. Anthropometric parameters, pulmonary function tests, 6‐minute walk distance, the BODE index and the number of exacerbations were measured and the association of these outcomes with the baseline levels of 143 serum biomarkers measured by PMP was explored.
Results
Thirty biomarker clusters were identified and ranked by computing the predictive value of each cluster for COPD (partial least squares discriminant analysis). From the 19 best predictive clusters, 2–3 biomarkers were selected based on their pathophysiological profile (chemoattractants, inflammation, tissue destruction and repair) and the statistical significance of their relationship with clinically important end points was tested. The selected panel of 24 biomarkers correlated (p<0.01) with forced expiratory volume in 1 s, carbon monoxide transfer factor, 6‐minute walk distance, BODE index and exacerbation frequency.
Conclusion
PMP technology can be useful in identifying potential biomarkers in patients with COPD. Panels of selected serum markers are associated with important clinical predictors of outcome in these patients.
Chronic obstructive pulmonary disease (COPD) is projected to be the third leading cause of death in the world by the year 2020.1,2 Despite the well‐documented role of cigarette smoking in the genesis of COPD, it is unclear what steps are involved in its pathogenesis.3 Most, if not all, patients with COPD develop a combination of lung emphysema with its characteristic pattern of alveolar destruction and abnormal repair as well as small airway inflammation that persists even years after smoking cessation.4
The current pathogenetic theories for the development of COPD include an imbalance between the protease and antiprotease system, dysregulation of oxidant‐antioxidant activity and chronic airway inflammation, processes that lead to the progressive destruction and abnormal repair of the lung connective tissue matrix.5 Recent studies have suggested that increased apoptosis of the alveolar wall accounts in part for the loss of lung tissue that characterises emphysema.6,7 Transgenic and null mutant mouse studies have identified a number of genes and pathways that, when altered, result in the morphological changes of emphysema.8,9,10
Although COPD primarily affects the lungs, it is associated with important systemic consequences which include malnutrition with a low body mass index (BMI)11 and impaired peripheral muscle function.12 These clinically relevant expressions of the disease have been associated with detectable systemic changes including evidence of increased oxidative stress, activation of circulating inflammatory cells and increased levels of proinflammatory cytokines.13,14 The multidimensional expression of COPD can be expressed by a clinical score including BMI, degree of obstruction (O), perception of dyspnoea (D) and exercise capacity (E) by the 6‐minute walk distance known as the BODE index.15 This index predicts mortality better than the forced expiratory volume in 1 s (FEV1).
We reasoned that the pathobiological processes that occur in the lungs and possibly in systemic tissues such as the peripheral muscles of patients with COPD could be associated with systemic biomarker levels detectable in the systemic circulation. Despite the many studies aimed at identifying the pathogenesis of COPD, to our knowledge only one study16 has explored the potential value of high‐density microarray technology to systematically define the serum protein expression profile in patients with COPD. Using a novel protein microarray platform (PMP) technology, we compared the serum proteomic profile of 143 serum biomarkers in patients with COPD with that of age and sex‐matched controls. We also explored the relationship between a selected subset of 24 biomarkers with clinically important outcome variables in COPD including lung function, the BODE index and its components and the frequency of exacerbations.
Methods
Patient recruitment
This is a matched case‐control study of 48 patients with severe COPD (FEV1 <55% predicted), 8 of whom were current smokers. We then matched 8 control smokers and 40 subjects who had smoked <5 pack years and had stopped at least 20 years previously or who had always been non‐smokers. All controls had a ratio of FEV1 to forced vital capacity (FVC) of >0.7 and FEV1 >70% predicted. Participants were >35 years of age and patients with COPD had to be clinically stable and without exacerbations for at least 3 months. Subjects with a history of asthma or atopy, conditions precluding performance of the tests, and a systemic infection or an inflammatory process that could be associated with abnormal biomarker profile were excluded. All patients were followed for 1 year and were stratified according to smoking history into ex‐smokers (never smoked or ex‐smokers for >15 years and <20 pack‐years) and active smokers. The controls were frequency matched according to sex, age and smoking history (table 1).
Table 1 Cases and controls stratified by exacerbation frequency and smoking status.
| Never/ex‐smoker | Active smoker | Total | |
|---|---|---|---|
| COPD | |||
| Group 1 (no exacerbations) | 12 | 4 | 16 |
| Group 2 (⩽2 exacerbations/ year) | 12 | 4 | 16 |
| Group 3 (>2 exacerbations/ year) | 15 | 0 | 15 |
| Controls | 40 | 8 | 48 |
The pulmonary function tests were measured according to ATS standards17 and the BODE index was calculated as previously reported.15 Exacerbations were defined as episodes of increased dyspnoea, sputum or cough lasting >24 h and requiring treatment with antibiotics and/or corticosteroids.18 After follow‐up for 1 year, patients were stratified into no exacerbations (n = 12), <2 exacerbations (n = 12) and ⩾2 exacerbations (n = 15).
Specimen collection
Blood samples were drawn, centrifuged and the serum frozen at –80°C. Rolling cell amplification (RCA) immunoassay was performed by Molecular Staging Inc (MSI, New Haven, Connecticut, USA) using a protein microarray platform that measured levels of 143 analytes (see table S1 available online at http://thorax.bmj.com/supplemental) on five separate arrays.19,20 After incubating and washing the serum samples on microarrays, the captured proteins were detected by specific biotinylated second antibodies and a universal anti‐biotin antibody was bound to the secondary antibodies. The anti‐biotin antibody contained an oligonucleotide DNA primer used for amplification. During the process, a circular DNA hybridises to the oligonucleotide DNA primer in the presence of DNA polymerase and fluorescent nucleotides to generate a signal. Following RCA, the slides were scanned (L200 scan, TECAN, Durham, North Carolina, USA) using a proprietary software. The fluorescence intensity of microarray spots was analysed and the resulting mean intensity values were measured. Dose‐response curves for the biomarkers were determined with increasing intensity indicating increasing analyte concentration.
Data analysis
A more complete discussion of the analysis used in this study is available in the online supplement at http://thorax.bmj.com/supplemental. In summary, two independent statistical approaches were used: (1) we tested the distribution of biomarkers for an association with COPD by univariate analysis adjusting for multiple comparisons using false discovery rate analysis;21 and (2) we used a variable clustering (VARCLUS) tool which divides the biomarkers into non‐overlapping unidimensional groups or clusters,22 a process similar to factor analysis. Each cluster's predictive value was determined by computing the partial regression coefficient of individual cluster centroids with COPD using partial least squares discriminant analysis (PLS‐DA). After the initial analysis, we selected a group of 24 biomarkers from those clusters that showed a significant association with the diagnosis of COPD (clinical history and presence of airflow limitation). The biomarkers were chosen to reflect a variety of pathobiological mechanisms relevant to the disease process. The resultant panel of biomarkers was then tested for strength of association with variables known to predict outcome in COPD, including transfer factor for carbon monoxide (Tlco),23 6‐minute walk distance (6MWD), the BODE index and exacerbation frequency.
Results
Study population
The characteristics of the patients and the controls are summarised in table 2. As expected, the patients had higher smoking exposure (pack‐years), significant airflow limitation, higher lung volumes, worse BODE scores and health‐related quality of life than controls. Patients and controls were of similar age, sex and BMI.
Table 2 Basic characteristics of patients with COPD and controls*.
| Variable | COPD (n = 47) M = 28/F = 19 | Controls (n = 48) M = 29/F = 19 | T test for independent samples (p value) |
|---|---|---|---|
| Age | 64 (8) | 64 (7) | NS |
| BMI (kg/m2) | 26.6 (5.2) | 27.6 (4.8) | NS |
| Smoking history (pack‐years) | 70 (40) | 23 (3) | <0.001 |
| Pre‐bronchodilator FEV1 (l) | 0.87 (0.33) | 2.46 (0.61) | <0.001 |
| Pre‐bronchodilator FEV1 (% predicted) | 32 (10) | 90 (15) | <0.001 |
| Post‐bronchodilator FEV1 (l) | 0.94 (0.36) | NA | |
| Post‐bronchodilator FEV1 (% predicted) | 35 (11) | NA | |
| TLC (l) | 7.00 (1.57) | 5.33 (1.48) | <0.001 |
| TLC (% predicted) | 126 (2) | 95 (19) | <0.001 |
| RV (l) | 4.46 (1.36) | 2.07 (0.81) | <0.001 |
| RV (% predicted) | 211 (60) | 96 (33) | <0.001 |
| Tlco (ml/min/mmHg) | 9.9 (3.8) | 19.5 (6) | <0.001 |
| Tlco (% predicted) | 48 (5) | 92 (24) | <0.001 |
| MRC dyspnoea | 2.5 (0.8) | 0.08 (0.3)* | <0.001 |
| 6MWD (m) | 365 (109) | 540 (95) | <0.001 |
| SGRQ composite score | 53 (18) | 4.8 (2.6)* | <0.001 |
| BODE index | 4.7 (1.8) | 2.2 (1.8) | <0.001 |
NA, not available; NS, not significant; BMI, body mass index; FEV1, forced expiratory volume in 1 s; TLC, total lung capacity; RV, residual volume; Tlco, carbon monoxide transfer factor; MRC, Medical Research Council; 6MWD, 6‐minute walk distance; SGRQ, St George's Respiratory Disease Questionnaire; BODE, body mass index (B), obstruction (O), dyspnoea (D) and exercise endurance (E).
Data presented as mean (SD).
*Log transformation was performed on variables with non‐normal distribution.
Biomarkers that distinguish between patients with COPD and controls
In the univariate analysis, 43 biomarkers were identified that differed between patients and controls. To adjust for multiple analysis, these were filtered by false discovery rate adjusted p value (FDR_p) of <0.015 (table 3).
Table 3 Statistical comparison of biomarkers in patients with COPD and controls.
| Sequence | Protein ID | Controls | COPD patients | T ratio | FDR_p | p Value | ||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||||
| 1 | MMP‐9 | 4129.05 | 1524.12 | 6298.61 | 2437.93 | 5.07 | 0.00028 | 0.00000 |
| 2 | Eotaxin‐2 | 3821.17 | 2786.26 | 6897.35 | 4303.97 | 4.37 | 0.00078 | 0.00003 |
| 3 | MMP‐7 | 860.72 | 340.62 | 1431.51 | 1021.53 | 4.37 | 0.00078 | 0.00003 |
| 4 | MMP‐10 | 1111.17 | 445.91 | 1813.39 | 1424.64 | 4.54 | 0.00078 | 0.00002 |
| 5 | TARC | 178.84 | 124.23 | 386.29 | 541.28 | 4.44 | 0.00078 | 0.00002 |
| 6 | TNF‐α | 83.24 | 19.97 | 112.99 | 48.21 | 4.40 | 0.00078 | 0.00003 |
| 7 | BDNF | 293.34 | 347.45 | 992.07 | 1348.16 | 4.17 | 0.00106 | 0.00007 |
| 8 | GCP‐2 | 100.41 | 26.19 | 131.26 | 45.91 | 4.16 | 0.00106 | 0.00007 |
| 9 | MIF | 702.08 | 165.77 | 1062.19 | 931.81 | 4.15 | 0.00106 | 0.00007 |
| 10 | NE | 3951.64 | 925.28 | 4917.56 | 1284.20 | 4.18 | 0.00106 | 0.00006 |
| 11 | IL‐1‐sR1 | 173.18 | 89.25 | 264.36 | 157.92 | 4.04 | 0.00144 | 0.00011 |
| 12 | MMP‐8 | 7479.60 | 5112.69 | 14511.88 | 9305.13 | 4.00 | 0.00151 | 0.00013 |
| 13 | TIMP‐1 | 2885.88 | 1477.78 | 4677.85 | 2424.25 | 3.91 | 0.00197 | 0.00018 |
| 14 | AR | 117.76 | 30.93 | 146.85 | 47.46 | 3.77 | 0.00253 | 0.00029 |
| 15 | IL‐10rβ | 689.21 | 200.70 | 874.60 | 287.25 | 3.70 | 0.00253 | 0.00037 |
| 16 | IL‐12p40 | 130.59 | 41.13 | 162.29 | 54.10 | 3.69 | 0.00253 | 0.00037 |
| 17 | IL‐1rα | 176.72 | 53.05 | 221.43 | 61.74 | 3.80 | 0.00253 | 0.00026 |
| 18 | IL‐2rβ | 275.40 | 92.33 | 366.01 | 155.99 | 3.73 | 0.00253 | 0.00033 |
| 19 | IL‐8 | 209.52 | 80.41 | 278.40 | 101.25 | 3.79 | 0.00253 | 0.00027 |
| 20 | I‐TAC | 86.84 | 51.75 | 138.41 | 95.26 | 3.71 | 0.00253 | 0.00036 |
| 21 | VEGF | 127.21 | 40.98 | 168.05 | 65.40 | 3.70 | 0.00253 | 0.00036 |
| 22 | VEGF‐D | 263.56 | 142.27 | 361.02 | 186.09 | 3.56 | 0.00377 | 0.00058 |
| 23 | CD30 | 158.85 | 69.99 | 210.88 | 102.48 | 3.48 | 0.00457 | 0.00077 |
| 24 | Eotaxin | 235.07 | 80.48 | 282.00 | 65.23 | 3.49 | 0.00457 | 0.00075 |
| 25 | MCP‐1 | 633.02 | 303.92 | 936.95 | 584.67 | 3.44 | 0.00496 | 0.00087 |
| 26 | IL‐1α | 91.98 | 31.06 | 113.74 | 38.44 | 3.41 | 0.00508 | 0.00096 |
| 27 | Prolactin | 925.25 | 244.88 | 1274.07 | 838.37 | 3.42 | 0.00508 | 0.00093 |
| 28 | TNF‐R1 | 525.09 | 455.95 | 800.09 | 616.36 | 3.35 | 0.00597 | 0.00117 |
| 29 | IL‐1β | 98.21 | 34.65 | 124.21 | 53.06 | 3.31 | 0.00662 | 0.00134 |
| 30 | BLC | 192.76 | 89.61 | 294.84 | 240.72 | 3.29 | 0.00675 | 0.00142 |
| 31 | FGF‐4 | 347.42 | 95.24 | 434.21 | 169.61 | 3.26 | 0.00719 | 0.00156 |
| 32 | MPIF‐1 | 624.64 | 331.03 | 825.75 | 398.73 | 3.17 | 0.00932 | 0.00209 |
| 33 | IGFBP‐4 | 6118.33 | 1188.73 | 7136.63 | 2703.80 | 3.15 | 0.00962 | 0.00222 |
| 34 | CTACK | 3480.02 | 1367.85 | 4745.23 | 3090.78 | 3.12 | 0.01005 | 0.00239 |
| 35 | HCC‐1 | 1201.77 | 352.24 | 1549.81 | 870.31 | 3.07 | 0.01130 | 0.00276 |
| 36 | Eotaxin‐3 | 229.35 | 114.73 | 609.19 | 2006.73 | 3.06 | 0.01138 | 0.00291 |
| 37 | IL‐17 | 343.13 | 69.30 | 390.90 | 84.96 | 3.05 | 0.01138 | 0.00302 |
| 38 | TGF‐α | 210.31 | 107.82 | 595.09 | 2168.58 | 3.05 | 0.01138 | 0.00299 |
| 39 | MIP‐1δ | 2094.72 | 836.45 | 2878.11 | 1762.62 | 2.99 | 0.01326 | 0.00362 |
| 40 | EGF | 158.28 | 64.86 | 216.59 | 164.16 | 2.93 | 0.01458 | 0.00428 |
| 41 | M‐CSF | 78.34 | 24.86 | 96.89 | 35.82 | 2.93 | 0.01458 | 0.00422 |
| 42 | Protein C | 8112.46 | 1788.08 | 9087.41 | 1603.96 | 2.94 | 0.01458 | 0.00412 |
| 43 | HCC‐4 | 8572.51 | 2501.97 | 10071.78 | 2581.08 | 2.90 | 0.01498 | 0.00461 |
The protein markers were filtered by false discovery adjusted p‐value (FDR_p) of 0.015. The FDR adjusted p value (sometimes called just FDR) is a practical way of dealing with multiple testing issues and can be interpreted as estimated proportion of false positives in the list. The T ratio was computed from common log scale. The names of all the analytes are given in the online supplement available at http://thorax.bmj.com/supplemental.
The second approach (variable cluster analysis) resulted in 30 different clusters, 19 of which correlated significantly with the diagnosis of COPD. We selected biomarkers from among these 19 clusters to reflect a variety of pathophysiological mechanisms considered relevant to COPD. In order to enrich the exploratory value of the panel, two biomarkers—prolactin and plasminogen activator inhibitor type 2 (PAI‐II)—were included despite lack of an obvious disease association. The selected panel biomarkers are shown in table 4 and their full description is given in the online supplement available at http://thorax.bmj.com/supplemental.
Table 4 Biomarkers selected for analysis.
| Pathobiological function | |
|---|---|
| Chemoattractants | I‐TAC, eotaxin‐2, MPIF‐1, MCP‐1, MIP‐1β, IL‐8, PARC |
| Inflammation | IL‐15, IL‐1ra, IL‐17, TNFα, TNF R1, IFNγ, IL‐12 p40, IL‐2Rγ |
| Destruction and repair | TGFα, VEGF, AR, BDNF, βNGF, MMP‐9, TIMP‐1 |
| Novel markers | PAI‐II, prolactin |
The biomarkers were selected from clusters statistically associated with the diagnosis of COPD and thought to have known or potential significance in the pathobiology of COPD.
AR, amphiregulin; BDNF, brain‐derived neurotrophic factor; βNGF, β‐nerve growth factor; IFNγ, interferon γ; IL, interleukin; IL‐1ra, interleukin 1 receptor antagonist; IL‐2Rγ, interleukin 2 receptor gamma; I‐TAC, interferon γ‐inducible T cell α chemoattractant; MCP‐1, monocyte chemotactic protein 1; MIP‐1β, macrophage inflammatory protein 1β; MMP‐9, matrix metalloproteinase 9; MPIF‐1, myeloid progenitor inhibitory factor 1; PAI‐II, plasminogen activator inhibitor II; TGFα, transforming growth factor α; TIMP‐1, tissue inhibitors of metalloproteinases 1; TNFα, tumour necrosis factor α; TNF R1, tumour necrosis factor receptor I; VEGF, vascular endothelial growth factor.
Associations of the biomarker panel with FEV1, Tlco, 6MWD, BODE index, BMI and exacerbation rate in patients with COPD
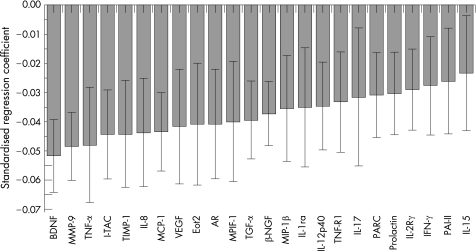
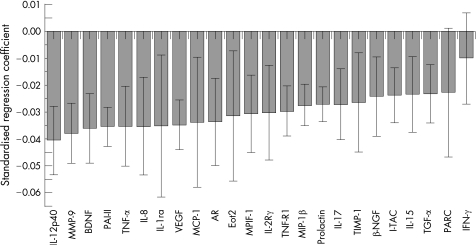
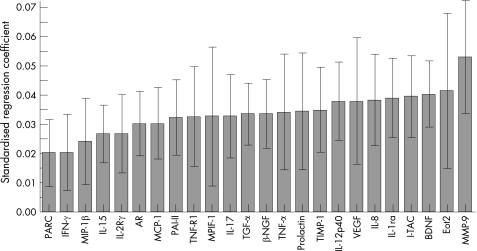
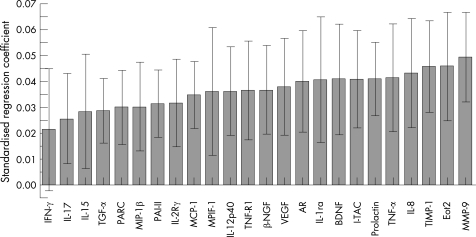
In the patients with COPD, the selected biomarkers tested in the panel correlated significantly with FEV1 (fig 1). The findings were replicated for the Tlco (fig 2), the BODE index (fig 3) and the exacerbation rate (fig 4).
Figure 1 Correlation of the selected biomarker panel with forced expiratory volume in 1 s (FEV1) in patients with COPD. The size of the bar in the graph indicates the magnitude of the regression coefficients and the 95% confidence interval is also indicated for each bar. If the confidence interval includes zero, the associated biomarker is “not significant”. The overall regression model was significant by a permutation test (p<0.01). The standardised coefficients for this and for figs 2, 3 and 4 are for scaled and centred markers and scaled response. These coefficients can be used to interpret the influence of the markers on the clinical response. The standardised regression coefficient for each marker measures the effect of the marker on the clinical response adjusted for all other markers in the regression (partial correlation). The coefficients can also be compared across the clinical responses since they are scaled. Definitions of the biomarkers are given in the footnote to table 4.
Figure 2 Correlation of the selected biomarker panel with carbon monoxide transfer factor (Tlco) in patients with COPD. The size of the bar in the graph indicates the magnitude of the regression coefficients and the 95% confidence interval is also indicated for each bar. If the confidence interval includes zero, the associated biomarker is “not significant”. The overall regression model was significant by a permutation test (p<0.01). Definitions of the biomarkers are given in the footnote to table 4.
Figure 3 Correlation of the selected biomarker panel with the BODE index in patients with COPD. The size of the bar in the graph indicates the magnitude of the regression coefficients and the 95% confidence interval is also indicated for each bar. If the confidence interval includes zero, the associated biomarker is “not significant”. The overall regression model was significant by a permutation test (p<0.01). Definitions of the biomarkers are given in the footnote to table 4.
Figure 4 Correlation of the selected biomarker panel with the exacerbation rate in patients with COPD. The size of the bar in the graph indicates the magnitude of the regression coefficients and the 95% confidence interval is also indicated for each bar. If the confidence interval includes zero, the associated biomarker is “not significant”. The overall regression model was significant by a permutation test (p<0.01). Definitions of the biomarkers are given in the footnote to table 4.
We also observed a correlation with the 6MWD while there was no correlation with BMI (not shown). The same selected biomarkers are shown for each analysis. Most of the markers were associated with all of the physiological indicators of disease, but the strength of the association differed from outcome to outcome as did the rank order of each biomarker.
Discussion
This study had two important findings: (1) that PMP technology can be useful in identifying potential biomarkers in patients with COPD; and (2) that a pattern of systemic biomarkers identified in these patients can be associated with different clinical variables known to predict disease outcome including degree of airflow limitation, lung transfer factor, functional capacity, the BODE index and exacerbation frequency.
Several groups have shown an increase in a number of circulating inflammatory biomarkers in COPD,24,25,26 suggesting that it might be possible to characterise patients with COPD using systemic biomarkers. To address this question we used a novel technology that simultaneously evaluated analytes covering diverse potential processes including inflammation, chemo‐attraction, cell activation, tissue destruction and repair. Based on a collaborative effort of statistical results and scientific plausibility, a subset of 24 biomarkers was identified and selected for subsequent testing against a variety of clinically important parameters. Many studies have been published on the association between a specific marker and COPD disease status, with both positive and negative results being reported.24,25,26,27,28,29,30,31 The disagreement in results can be attributed to different factors, including the heterogeneity of COPD phenotypes, low sample size, or the use of different methodologies and assays. The development of a panel of biomarkers addressing preconceived multiple pathophysiological pathways may provide a more specific tool to serve as an intermediate end point reflecting the natural history of the disease.
One obvious limitation of this preliminary dataset is that the biomarkers identified are limited by the pool of analytes that were available for the primary assessment. Clearly, use of an “open” proteomic platform would give information about a much broader range of proteins and might provide additional insights into biomarker selection and disease processes. However, a recent report16 using a panel developed from the one reported here shows that the panel as developed is valid and capable of reflecting changes induced by exacerbations. Recognising the fact that the discussion is valid only for the analytes explored, our findings may help to shed light on the underlying pathogenetic processes involved in this disease.
It has been proposed that various proteases break down lung connective tissue components to cause emphysema,5,6 leading to aberrant remodelling and/or degradation of the extracellular matrix. In our study, several proteins (table 3) related to the protease‐antiprotease mechanism were clearly different between patients with COPD and controls. Thus, metalloproteinases 7, 8, 9 and 10 (MMP‐7, MMP‐8, MMP‐9 and MMP‐10) were among the proteins with large differences between groups. Of these, MMP‐9 showed the strongest association with FEV1 and Tlco, which is interesting because MMP‐9 has been implicated in the experimental genesis of emphysema.32,33 The tissue inhibitor of metalloproteinase 1 (TIMP‐1), a collagenase inhibitor, was also different between patients and controls, providing evidence that the final expression of the disease may rest upon the appropriate balance of the system.33
Differences were also found in enzymes other than the metalloproteinases that are related to tissue destruction, as well as proteins related to repair, that deserve some comments. While the fold increase of neutrophil elastase in COPD was not as great as that found for the metalloproteinases, the difference was still statistically significant. Previous studies of experimental emphysema produced by pancreatic or neutrophil elastase showed that increased levels of elastase enzymes lead to the degradation of connective tissue components and, thus, enlargement of distal airspaces.34 While both elastin and collagen are rapidly re‐synthesised in these animal models and mRNA levels for both are increased, the connective tissue remodelling process is ineffective and lung mechanical properties remain abnormal.35
The differences in tissue growth factor alpha (TGFα), amphiregulin (AR), brain‐derived neurotropic factor (BDNF) and nerve growth factor β (βNGF) and their association with low FEV1 and Tlco (figs 1 and 2) suggest that connective tissue remodelling continues even in severe advanced COPD in humans, but the process fails effectively to restore the mechanical properties of the diseased lung. The role of TGFα is something of a mystery. Mice genetically manipulated to overexpress TGFα develop emphysema postnatally,36 yet an in vitro model of alveolar re‐epithelialisation showed that TGFα induced faster wound repair.37 The presence of significant associations between BDNF and lung function and the BODE index (fig 3) is particularly interesting. Recent evidence indicates that BDNF decreases conversion from oxygen to hydrogen peroxide in experimental cell cultures,38 suggesting a role in the modulation of oxidative stress, and makes this an interesting marker to study. Furthermore, similar to results seen with AR, exogenous BDNF can protect cells from serum deprivation‐induced cell death.39
It has been suggested that angiogenesis and apoptosis of the alveolar wall may have a role in emphysema. While little is known about the role of the EGF family member AR in the aetiology of COPD, one study has found that AR can inhibit apoptosis of non‐small cell lung cancer cell line.40 Blockade of vascular endothelial growth factor R2 (VEGF‐R2) receptor in rats induces apoptosis of the alveolar cell wall and results in an emphysema‐like pathology.41,42 Several studies have found decreased expression of VEGF in induced sputum or bronchoalveolar lavage (BAL) fluid from patients with obstructive lung disease in comparison with normal subjects.43,44 These studies have also shown a direct association between the reduction in VEGF and FEV1. While our study showed an increase in VEGF serum content that was inversely associated with FEV1, this difference could be due to differential expression of VEGF in lung tissue and serum. Studies of VEGF expression in human lung tissue by immunohistochemistry have shown increased VEGF in pulmonary and airway smooth muscle in subjects with COPD that correlated with decreased FEV1.45 Furthermore, patients with cystic fibrosis show an inverse relationship in the level of VEGF in serum and BAL fluid compartments. These patients had a higher level of VEGF in serum and a lower level of VEGF in BAL fluid compared with controls.46 The role of apoptosis and its relationship to inflammation and repair seem supported by our findings.
Current thinking places inflammation at the centre of the pathogenetic mechanisms of COPD. The inflammation is characterised by increased numbers of alveolar macrophages, neutrophils and T lymphocytes, together with the release of multiple inflammatory mediators that result in a high level of oxidative stress. Multiple proteins related to inflammation were detected in the serum of patients with COPD (table 4). These included interleukin (IL)‐12, IL‐15, IL‐17, IL‐1 receptor antagonist (IL‐1ra), tumour necrosis factor α (TNFα), tumour necrosis factor receptor 1 (TNF R1), interferon γ (IFNγ), IL‐12p40 and IL‐2Rγ. There is experimental evidence for the participation of all of these proteins in the inflammation that characterises COPD, and raises the possibility that the systemic manifestations of COPD may be intimately related to this process. Indeed, the association between inflammatory markers and exacerbation rate (fig 4) suggests that this manifestation of the disease could be modulated by amplification of the inflammatory cascade. In this regard, eotaxin‐2—which had one of the strongest associations with the exacerbation rate in our patients—is a strong chemotactic cytokine for eosinophils,47 cells that have been found to be increased in airway biopsy tissue from patients with COPD exacerbations.48 Indeed, although the inflammatory pathways of COPD appear to be more related to lymphocytes expressing a T helper 1 (Th1) bias,49 a high level of Th2 chemokines have been reported in experimental models of emphysema induced by cigarette smoking.10
There were several novel proteins that differed between patients with COPD and controls. We selected two of them—plasminogen activator inhibitor type 2 (PAI‐II) and prolactin—because of their presence in one of the eight clusters with the strongest association with COPD. PAI‐II belongs to the serpine class of protease inhibitors and is involved in the thrombogenic cascade. Known to be produced by activated monocytes in the peripheral blood,50 this protein (together with PAI‐I) may have a role in tissue remodelling in airways disease.51 These data warrant further investigation to explore the possible role of serpines in COPD.52 Prolactin upregulation presents an enigma. Prolactin receptor has recently been reported to be upregulated in the lungs of mice exposed to lipopolysaccharide,53 and prolactin can activate the inflammatory natural killer (NF)‐κβ cascade in pulmonary fibroblasts.54 It is therefore plausible that prolactin may play a role in the inflammatory environment in COPD.
There are a number of important limitations to our study. Not all of the possible proteins that participate in the complex mechanism of COPD were tested. Absent were some with a known relationship to COPD such as C‐reactive protein and fibrinogen, and some of potential importance such as MMP‐12. The reason for their omission was not any preconceived mechanistic bias. Our study was designed as a proof of principle rather than a totally comprehensive evaluation of all of the markers that could potentially be explored. Many complex diseases have components related to inflammation, tissue remodelling, apoptosis and chemoattraction of specific cell types. This observation suggests that a panel of analytes might provide insight into the pathobiology of the disease under study in the absence of, or in conjunction with, novel “disease‐specific” biomarkers. We also acknowledge that not all phenotypic expressions of COPD were analysed; for example, it would have been interesting to have related the biomarkers to changes in the CT scan of patients with emphysema, but unfortunately the technique needed to quantitatively express CT changes was not available. However, the Tlco does relate to the phenotypic expression of emphysema. We believe that this study represents a proof of concept and opens a window for hypothesis testing and perhaps the discovery of yet to be described pathway interactions and targets.
For the correlation analyses we attempted to address the issue of many proteins representing the same pathophysiological mechanism by empirically grouping them according to their statistical strength and their presumed pathobiological role. We acknowledge the latter to be empirical, but it is based on the data currently available and aimed at simplifying the prospective testing. Furthermore, the inclusion of too many proteins may be intellectually desirable but may cause important cross‐correlative noise that may actually cloud the interpretation of the results. We also acknowledge that the patients included in the study do not represent the large population of patients with COPD since all of them had severe disease. However, the patients included represent those likely to be seen by clinicians and to benefit from new therapeutic strategies. On the other hand, this study is unique in that patients and controls were phenotypically well characterised and matched by age, sex and—very importantly—by smoking habits to minimise the hypothetical influence of these confounding factors. Indeed, the inclusion or exclusion of smokers in each of the groups did not affect the results. In addition, the evaluation of important associations of the panel markers with clinical markers of COPD such as the BODE index and its individual components offers a more comprehensive picture of the value of the technique. The association with exacerbation frequency is particularly interesting because exacerbations constitute an extremely important outcome and one where elucidation of the factors that may help prevent their occurrence would prove extremely useful. Finally, we also acknowledge that the stability of biomarker levels in serum samples is not well characterised and that we did not repeat the tests at different times. However, the recent report by Hurat and colleagues16 using the panel derived from this study independently validated our findings.
In summary, using a serum PMP, we have identified a biomarker profile whose expression levels can distinguish patients with COPD from smokers and non‐smokers without COPD. We have also found an association between the level of selected biomarkers and lung function, the degree of airflow limitation and Tlco, a marker of lung tissue destruction. Furthermore, we documented an association between the expression of the serum biomarkers and the integrated local and systemic manifestations of the disease as represented by the functional capacity and the BODE index. The expression of biomarkers was also associated with the exacerbation rate, crucial events in the natural course of the disease. The ease of sampling of peripheral blood and the continuing improvement and availability of multiplexed immunoassay technology should provide us with a new tool for research in this deadly disease.
Further information is given in the online supplement available at http://thorax.bmj.com/supplemental.
Supplementary Material
Abbreviations
AR - amphiregulin
BAL - bronchoalveolar lavage
BDNF - brain‐derived neurotropic factor
BMI - body mass index
BODE index - body mass index (B), degree of obstruction (O), perception of dyspnoea (D) and exercise capacity (E)
COPD - chronic obstructive pulmonary disease
FDR_p - false discovery adjusted p value
FEV1 - forced expiratory volume in 1 s
IFNγ - interferon γ
IL - interleukin
MMP - metalloproteinase
6MWD - 6‐minute walk distance
βNGF - nerve growth factor β
PLS‐DA - partial least squares discriminant analysis
PMP - protein microarray platform
RCA - rolling cell amplification
TGFα - tissue growth factor α
TIMP‐1 - tissue inhibitor of metalloproteinase 1
Tlco - carbon monoxide transfer factor
TNFα - tumour necrosis factor α
VEGF - vascular endothelial growth factor
Footnotes
This work was supported by an unrestricted grant from GlaxoSmithKline and the Thoracic and Overholt Foundation. No funds were received from the tobacco industry.
Competing interests: JT, KL, DP, JB, HM, MMDS and RV are all full time employees of GlaxoSmithKline.
Further information is given in the online supplement available at http://thorax.bmj.com/supplemental.
References
- 1.Ezzati M, Lopez A D. Estimates of global mortality attributable to smoking in 2000. Lancet 2003362847–852. [DOI] [PubMed] [Google Scholar]
- 2.Mannino D M, Homa D M, Akinbami L J.et al Chronic obstructive pulmonary disease surveillance – United States, 1971–2000. MMWR 2002511–16. [PubMed] [Google Scholar]
- 3.Celli B R, MacNee W. Standards for the diagnosis and treatment of COPD. Eur Respir J 200423932–946. [DOI] [PubMed] [Google Scholar]
- 4.Hogg J C, Chu F, Utokaparach S.et al The nature of small‐airway obstruction in chronic obstructive pulmonary disease. N Engl J Med 20043502645–2653. [DOI] [PubMed] [Google Scholar]
- 5.Barnes P J, Shapiro S D, Pauwels R A. Chronic obstructive pulmonary disease: molecular and cellular mechanisms. Eur Respir J 200322672–688. [DOI] [PubMed] [Google Scholar]
- 6.Tuder R M, Zhen C Y L. Cho L, et al. Oxidative stress and apoptosis interact and cause emphysema due to vascular endothelial growth factor receptor blockade. Am J Respir Cell Mol Biol 20032988–97. [DOI] [PubMed] [Google Scholar]
- 7.Aoshiba K, Yokohori N, Nagai A. Alveolar wall apoptosis causes lung destruction and emphysematous changes. Am J Respir Cell Mol Biol 200328555–562. [DOI] [PubMed] [Google Scholar]
- 8.Fehrenbach H. Animal models of chronic obstructive pulmonary disease: some critical remarks. Pathobiology 200270277–283. [DOI] [PubMed] [Google Scholar]
- 9.Mahadeva R, Shapiro S D. Chronic obstructive pulmonary disease: experimental animal models of pulmonary emphysema. Thorax 200257908–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elias J. The relationship between asthma and COPD. Lessons from transgenic mice. Chest 2004126111–6S. [DOI] [PubMed] [Google Scholar]
- 11.Schols A M, Slangen J, Volovics L.et al Weight loss is a reversible factor in the prognosis of chronic obstructive pulmonary disease. Am J Respir Crit Care Med 19981571791–1797. [DOI] [PubMed] [Google Scholar]
- 12.Maltais F, Simard A A, Simard C.et al Oxidative capacity of the skeletal muscle and lactic acid kinetics during exercise in normal subjects and in patients with COPD. Am J Respir Crit Care Med 1996153288–293. [DOI] [PubMed] [Google Scholar]
- 13.Agustí A G, Noguera A, Sauleda J.et al Systemic effects of chronic obstructive pulmonary disease. Eur Respir J 200321347–360. [DOI] [PubMed] [Google Scholar]
- 14.Rahman I, Morrison D, Donaldson K.et al Systemic oxidative stress in asthma, COPD, and smokers. Am J Respir Crit Care Med 19961541055–1060. [DOI] [PubMed] [Google Scholar]
- 15.Celli B R, Cote C G, Marin J M.et al The body mass index, airflow obstruction, dyspnea and exercise capacity index in chronic obstructive pulmonary disease. N Engl J Med 20043501005–1012. [DOI] [PubMed] [Google Scholar]
- 16.Hurst J R, Donaldson G C, Perea W R.et al Utility of plasma biomarkers at exacerbation of chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2006174867–874. [DOI] [PubMed] [Google Scholar]
- 17.American Thoracic Society Lung function testing: selection of reference values and interpretative strategies. Am Rev Respir Dis 19911441202–1218. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez‐Roisin R. Toward a consensus definition for COPD exacerbations. Chest 2000117398–401S. [DOI] [PubMed] [Google Scholar]
- 19.Schweitzer B, Wiltshire S, Lambert J.et al Immunoassays with rolling circle DNA amplification: a versatile platform for ultrasensitive antigen detection. Proc Natl Acad Sci USA 20009710113–10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perlee L, Christiansen J, Dondero R.et al Development and standardization of multiplexed antibody microarrays for use in quantitative proteomics. Proteome Sci 200429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradley E. Bayesians, frequentists and scientists. J Am Stat Assoc 20051001–5. [Google Scholar]
- 22.SAS/STAT User's Guide. Version 8. Chapter 68. The VARCLUS procedure. 3593–620
- 23.Nakano Y, Muro S, Sakai H.et al Computed tomographic measurements of airway dimensions and emphysema in smokers. Correlation with lung function. Am J Respir Crit Care Med 20001621102–1108. [DOI] [PubMed] [Google Scholar]
- 24.Gan W Q, Man S F, Senthilselvan A.et al Association between chronic obstructive pulmonary disease and systemic inflammation: a systematic review and a meta‐analysis. Thorax 200459574–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vernooy J H, Kucukaycan M, Jacobs J A.et al Local and systemic inflammation in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 20021661218–1224. [DOI] [PubMed] [Google Scholar]
- 26.Schols A M W J, Buurman W A, Staal‐van den Brekel A J.et al Evidence for a relation between metabolic derangement and increased levels of inflammatory mediators in a subgroup of patients with chronic obstructive pulmonary disease. Thorax 199651819–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aaron S D, Angel J B, Lunau M.et al Granulocyte inflammatory markers and airway infection during acute exacerbation of chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2001163349–355. [DOI] [PubMed] [Google Scholar]
- 28.Noguera A, Busquets X, Sauleda J.et al Expression of adhesion molecules and G proteins in circulating neutrophils in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 19981581664–1668. [DOI] [PubMed] [Google Scholar]
- 29.Malo O, Sauleda J, Busquets X.et al Systemic inflammation during exacerbations of chronic obstructive pulmonary disease. Arch Bronconeumol 200238172–176. [PubMed] [Google Scholar]
- 30.Broekhuizen R, Wouters E F, Creutzberg E C.et al Raised CRP levels mark metabolic and functional impairment in advanced COPD. Thorax 20066117–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinto‐Plata V, Mullerova H, Toso J.et al C‐reactive protein in patients with COPD, control smokers and non‐smokers. Thorax 20066123–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Culpitt S V, Rogers D F, Traves S L.et al Sputum matrix metalloproteases: comparison between chronic obstructive pulmonary disease and asthma. Respir Med 200599703–710. [DOI] [PubMed] [Google Scholar]
- 33.Higashimoto Y, Yamagaga Y, Iwata T.et al Increased serum concentrations of tissue inhibitor of metalloproteinase‐1 in COPD patients. Eur Respir J 200525885–890. [DOI] [PubMed] [Google Scholar]
- 34.Churg A, Wright J L. Proteases and emphysema. Curr Opin Pulm Med 200511153–159. [DOI] [PubMed] [Google Scholar]
- 35.Shapiro S D, Goldstein N M, Houghton A M.et al Neutrophil elastase contributes to cigarette smoke‐induced emphysema in mice. Am J Pathol 20031632329–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardie W D, Piljan‐Gentle A, Dunlavy M R.et al Dose‐dependent lung remodeling in transgenic mice expressing transforming growth factor‐alpha. Am J Physiol Lung Cell Mol Physiol 20012811088–1094. [DOI] [PubMed] [Google Scholar]
- 37.Kheradmand F, Folkesson H G, Shum L.et al Transforming growth factor‐alpha enhances alveolar epithelial cell repair in a new in vitro model. Am J Physiol 1994267728–738. [DOI] [PubMed] [Google Scholar]
- 38.Yamagata T, Satoh T, Ishikawa Y.et al Brain‐derived neurotropic factor prevents superoxide anion‐induced death of PC12h cells stably expressing TrkB receptor via modulation of reactive oxygen species. Neurosci Res 1999359–17. [DOI] [PubMed] [Google Scholar]
- 39.Zheng W H, Quirion R. Comparative signaling pathways of insulin‐like growth factor‐1 and brain‐derived neurotrophic factor in hippocampal neurons and the role of the PI3 kinase pathway in cell survival. J Neurochem 200489844–852. [DOI] [PubMed] [Google Scholar]
- 40.Hurbin A, Dubrez L, Coll J L.et al Inhibition of apoptosis by amphiregulin via an insulin‐like growth factor‐1 receptor‐dependent pathway in non‐small cell lung cancer cell lines. J Biol Chem 200227749127–49133. [DOI] [PubMed] [Google Scholar]
- 41.Voelkel N F, Cool C D. Pulmonary vascular involvement in chronic obstructive pulmonary disease. Eur Respir J 200346(Suppl)28–32s. [DOI] [PubMed] [Google Scholar]
- 42.Kasahara Y, Tuder R M, Taraseviciene‐Stewart L.et al Inhibition of VEGF receptors causes lung cell apoptosis and emphysema. J Clin Invest 20001061311–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanazawa H, Hirata K, Yoshikawa J. Imbalance between vascular endothelial growth factor and endostatin in emphysema. Eur Respir J 200322609–612. [DOI] [PubMed] [Google Scholar]
- 44.Kanazawa H, Asai K, Hirata K.et al Possible effects of vascular endothelial growth factor in the pathogenesis of chronic obstructive pulmonary disease. Am J Med 2003114354–358. [DOI] [PubMed] [Google Scholar]
- 45.Kranenburg A R, de Boer W I, Alagappan V K.et al Enhanced bronchial expression of vascular endothelial growth factor and receptors (Flk‐1 and Flt‐1) in patients with chronic obstructive pulmonary disease. Thorax 200560106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer K C, Cardoni A, Xiang Z Z. Vascular endothelial growth factor in bronchoalveolar lavage from normal subjects and patients with diffuse parenchymal lung disease. Lab Clin Med 2000135332–338. [DOI] [PubMed] [Google Scholar]
- 47.Tachimoto H, Kikuchi M, Hudson S A.et al Eotaxin‐2 alters eosinophil integrin function via mitogen‐activated protein kinases. Am J Respir Cell Mol Biol 200226645–649. [DOI] [PubMed] [Google Scholar]
- 48.Zhu J, Qui Y S, Majumdar S.et al Exacerbations of bronchitis: bronchial eosinophilia and gene expression for interleukin‐4, interleukin‐5, and eosinophil chemoattractants. Am J Respir Crit Care Med 2001164109–116. [DOI] [PubMed] [Google Scholar]
- 49.Grumelli S, Corry D B, Song L Z.et al An immune basis for lung parenchymal destruction in chronic obstructive pulmonary disease and emphysema. PLoS Med 20041e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ritchie H, Booth N A. The distribution of the secreted and intracellular forms of plasminogen activator inhibitor 2 (PAI‐2) in human peripheral blood monocytes is modulated by serum. Thromb Haemost 199879813–817. [PubMed] [Google Scholar]
- 51.Kucharewicz I, Kowal K, Buczko W.et al The plasmin system in airway remodeling. Thromb Res 20031121–7. [DOI] [PubMed] [Google Scholar]
- 52.DeMeo D L, Mariani T J, Lange C.et al The SERPINE2 gene is associated with chronic obstructive pulmonary disease. Am J Hum Genet 200678253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corbacho A M, Valacchi G, Kubala L.et al Tissue‐specific gene expression of prolactin receptor in the acute‐phase response induced by lipopolysaccharides. Am J Physiol Endocrinol Metab 2004287750–757. [DOI] [PubMed] [Google Scholar]
- 54.Macotela Y, Mendoza C, Corbacho A M.et al 16K prolactin induces NF‐kappaB activation in pulmonary fibroblasts. J Endocrinol 200217513–18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.