Abstract
Interactions between killer cell immunoglobulin-like receptors (KIRs) and human leukocyte antigen (HLA) class I ligands regulate the development and response of human natural killer (NK) cells. Natural selection drove an allele-level group A KIR haplotype and the HLA-C1 ligand to unusually high frequency in the Japanese, who provide a particularly informative population for investigating the mechanisms by which KIR and HLA polymorphism influence NK cell repertoire and function. HLA class I ligands increase the frequencies of NK cells expressing cognate KIR, an effect modified by gene dose, KIR polymorphism, and the presence of other cognate ligand–receptor pairs. The five common Japanese KIR3DLI allotypes have distinguishable inhibitory capacity, frequency of cellular expression, and level of cell surface expression as measured by antibody binding. Although KIR haplotypes encoding 3DL1*001 or 3DL1*005, the strongest inhibitors, have no activating KIR, the dominant haplotype encodes a moderate inhibitor, 3DL1*01502, plus functional forms of the activating receptors 2DL4 and 2DS4. In the population, certain combinations of KIR and HLA class I ligand are overrepresented or underrepresented in women, but not men, and thus influence female fitness and survival. These findings show how KIR–HLA interactions shape the genetic and phenotypic KIR repertoires for both individual humans and the population.
Human killer cell immunoglobulin-like receptors (KIRs) and murine lectin-like Ly49 receptors perform orthologous functions despite their striking difference in protein structure and evolutionary origin (1, 2). As receptors for polymorphic MHC class I, KIR and Ly49 regulate NK cell tolerance and response (3). Independently, the human KIR and murine Ly49 gene families acquired some remarkably similar features, including variegated expression (4–6), signaling pathways (7), and haplotypes varying in gene content, allelic polymorphism, and signaling potential (8). Two groups of human KIR haplotypes are defined (9, 10). Group A haplotypes have a fixed organization of seven genes, most with inhibitory potential; the group B haplotypes have a variable number of KIR genes, many with activating function. Clinical studies have correlated KIR gene content with infection, cancer, autoimmunity, pregnancy syndromes, and transplant outcome (11–16). Most of these associations are with activating KIR genes and group B haplotypes and favor success in reproduction and fighting infection, but also in increased autoimmunity (3).
Although the inhibitory receptors for HLA-A (KIR3DL2), -B (3DL1), -C (2DL1, 2 and 3), and the activating receptor for HLA-G (2DL4) are well studied (17–20), the functional effects of their polymorphism remain largely unexplored. Ligands for the inhibitory 2DL5 and the activating 2DS1, 2, 3, 4, 5, and 3DS1 are unknown or uncertain. The NK cell repertoire depends on both KIR and HLA polymorphisms. Thus, KIR and HLA identical siblings have similar NK cell repertoires, whereas siblings differing at KIR or HLA exhibit a range of phenotype difference (21). Uncertain from that analysis was whether HLA serves to make positive or negative selection. Complicating further investigation of KIR–HLA interactions has been the extent of their diversity. Population comparisons revealed ethnic differences in the ratio of group A and B KIR haplotypes. In East Asian populations, the simpler group A haplotypes dominate, suggesting their KIR diversity might be more simple and experimentally more tractable than for Caucasians (22–24). Here, in-depth analysis of KIR genotype, phenotype, and function in the Japanese shows this hypothesis to be true.
RESULTS
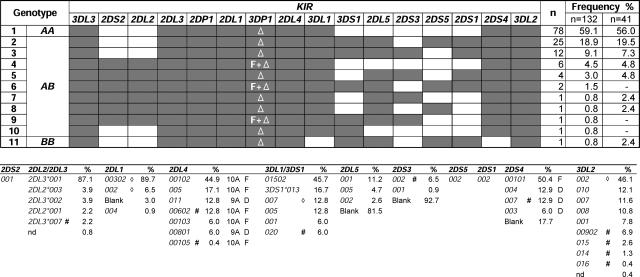
KIR gene content was determined for 132 unrelated Japanese donors. 11 KIR genotypes were distinguished (Fig. 1). With a frequency of 80%, the group A haplotypes dominated. These results extend the preliminary study of 41 donors (22), giving confidence that the results obtained here well represent the KIR system of the Japanese population. Full-length cDNA from KIR expressed in peripheral blood was cloned and sequenced from 53 panel members. In selecting donors and the KIR to be sequenced from each donor, we sampled the full range of the panel's genotypes (Fig. 1) and phenotypes (Fig. 2); 18–25% of donors positive for each KIR were analyzed by cDNA cloning and sequencing, predicting detection of all alleles with >6% frequency.
Figure 1.
KIR locus variability in the Japanese population. (top) KIR genotype and frequency in a panel of 132 donors. A shaded box indicates the presence of a gene; an unshaded box represents its absence. The deleted form of the 3DP1 pseudogene is indicated by “Δ,” the full-length form is indicated by “F.” The genotype frequencies are compared with previous analysis of 41 donors (reference 22). (bottom) Allele frequencies for the polymorphic genes are listed in order of decreasing frequency. Full-length (10A) and deleted (9A) forms of 2DL4 are designated “F” and “D,” respectively (this is also the case for full-length and deleted forms of 2DS4). KIR2DL2 and 2DL3 are considered alleles, as are 3DL1 and 3DS1. Alleles with novel changes in the coding region are denoted “#.” “⋄” denotes alleles defined by a novel change only in the untranslated region and are given preexisting allele names based on the coding region.
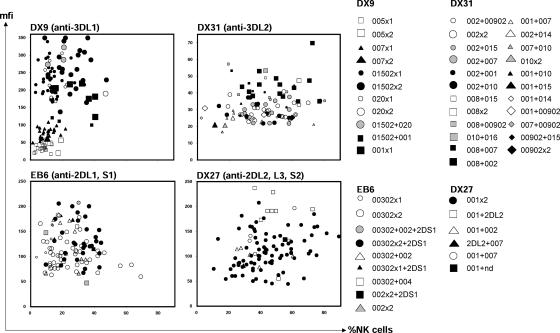
Figure 2.
Variability of KIR cell surface phenotype in the Japanese population. Each panel shows flow cytometry data obtained from NK cells stained with a different anti-KIR monoclonal antibody: EB6, anti-2DL1 and 2DS1; DX27, anti-2DL2/3 and 2DS2; DX9, anti-3DL1; and DX31, anti-3DL2. The percentage of NK cells that binds antibody (x axis) is plotted against the mean fluorescence intensity (mfi) (y axis), a measure of the amount of antibody bound. Each donor is represented by one data point in each of the four two-dimensional plots. The key gives allele-level genotypes that were subsequently assigned to each person. For donors giving bimodal patterns of DX9 binding, the high and low peaks are represented separately.
Sequencing >1,100 clones identified 41 distinct KIR, of which 14 were novel (Fig. 1). Polymorphism concentrated on the group A haplotype genes, particularly those in the telomeric part of the locus. Genes that distinguish group B haplotypes (2DS1, 2, 3, 5, 3DS1, 2DL2, 2DL5) were represented by only one or two variants. Methods that distinguish the KIR alleles were devised and used to type 116 panel members. Each polymorphic KIR gene has a dominant allele, with a frequency of 45–89% compared with 4–17% for the next most common allele (Fig. 1). For 2DL4 and 2DS4, nonfunctional alleles caused by frame-shifting deletions accounted for 19 and 38% of the total alleles, respectively. In contrast, the common nonfunctional 3DL1*004 allele of Caucasians (17%) is absent from the Japanese (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20051884/DC1).
Polymorphism modulates the level and frequency of KIR3D expression
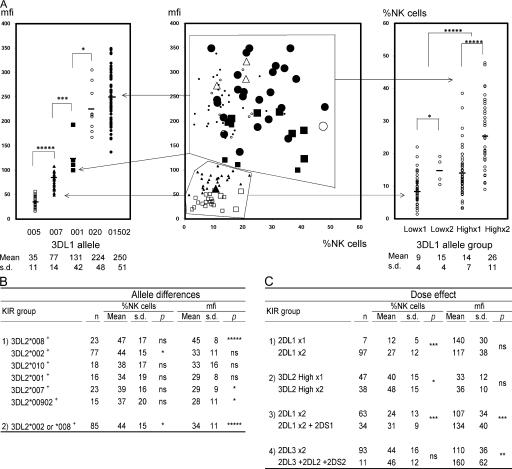
The variation in KIR phenotype of NK cells from 104 donors was assessed by flow cytometry using four anti-KIR antibodies and correlated with genotype (Fig. 2). The five KIR3DL1 allotypes had different levels of DX9 binding. The levels ranged over an order of magnitude, with the hierarchy 3DL1*005<*007<*001<*020<*01502 (Fig. 3 A, left). Comparison of different anti-KIR3DL1 antibodies and of DX9 binding kinetics showed the allotypic differences are the result of the abundance of cell surface 3DL1, not variable affinity for DX9 (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20051884/DC1, and references 25–27). Also correlating with binding level was the frequency of cellular expression so that high-binding allotypes (*001, *020, and *01502) were expressed by a larger proportion of NK cells than low-binding allotypes (*005 and *007) (Fig. 3 A, right). The frequency of NK cell expression also increased with gene dose: individuals having two low or high expressing 3DL1 allotypes had mean frequencies of expression that were 1.7- and 1.9-fold greater, respectively, than individuals having one.
Figure 3.
Allelic polymorphisms of KIR3DL genes determine different levels of cell surface expression. (A) The central panel shows flow cytometric analysis of NK cells stained with the DX9 antibody. The x and y axes are the same as in Fig. 2. Each large data point represents a single person; however, the high and low peaks in donors with bimodal DX9 binding patterns are each represented independently with small data points. 3DL1/3DS1 heterozygotes are represented as one small data point. The 3DL1 genotypes are represented by different symbols; ▪: 3DL1*001 × 1,  : *001+*01502, Δ: *01502+*020, □: *005 × 1,
: *001+*01502, Δ: *01502+*020, □: *005 × 1,  : *005 × 2, ▴: *007 × 1,
: *005 × 2, ▴: *007 × 1,  : *007 × 2, •: *01502 × 1,
: *007 × 2, •: *01502 × 1, 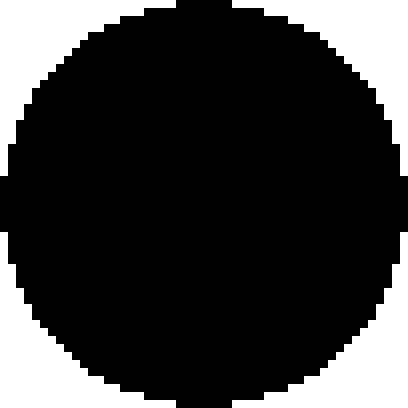 : *01502 × 2, ◯: *020 × 1,
: *01502 × 2, ◯: *020 × 1, 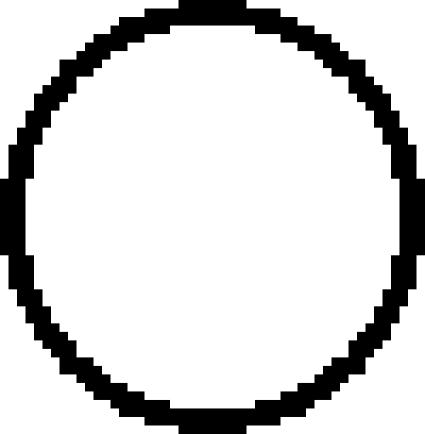 : *020 × 2. The staining as a result of the “low-binding allotypes” (3DL1*005 and *007) is distinguished from that of the “high-binding allotypes” (3DL1*001, *01502, and *020) by the gates. The left panel is a one-dimensional plot of mfi showing how each of the five 3DL1 allotypes corresponds to a different range and mean value of DX9 binding. The right panel is a one-dimensional plot of percentage of NK cells binding DX9 in which the panel members were divided into four groups according to 3DL1 genotype: one low-binding allotype, two low-binding allotypes, one high-binding allotype, and two high-binding allotypes. The frequency of NK cells expressing 3DL1 increases with gene dose. (B) Correlations between flow cytometric analysis (%NK cells and mfi) with DX31 and the different 3DL2 allotypes. Pairwise comparisons were made between phenotypes with and without each 3DL2 allotype in “1),” or with and without the allotype group comprising 3DL2*002 and *008 in “2).” (C) The effects of gene dose on %NK cells and mfi observed with EB6, DX31, and DX27. Pairwise comparisons were made in phenotypes between the two donor groups that differed in gene content as indicated from “1)” to “4).” One copy of a gene is shown by x1, two copies are shown by x2. *, P < 0.05; **, P < 0.01; ***, P < 0.005; and *****, P < 0.0005.
: *020 × 2. The staining as a result of the “low-binding allotypes” (3DL1*005 and *007) is distinguished from that of the “high-binding allotypes” (3DL1*001, *01502, and *020) by the gates. The left panel is a one-dimensional plot of mfi showing how each of the five 3DL1 allotypes corresponds to a different range and mean value of DX9 binding. The right panel is a one-dimensional plot of percentage of NK cells binding DX9 in which the panel members were divided into four groups according to 3DL1 genotype: one low-binding allotype, two low-binding allotypes, one high-binding allotype, and two high-binding allotypes. The frequency of NK cells expressing 3DL1 increases with gene dose. (B) Correlations between flow cytometric analysis (%NK cells and mfi) with DX31 and the different 3DL2 allotypes. Pairwise comparisons were made between phenotypes with and without each 3DL2 allotype in “1),” or with and without the allotype group comprising 3DL2*002 and *008 in “2).” (C) The effects of gene dose on %NK cells and mfi observed with EB6, DX31, and DX27. Pairwise comparisons were made in phenotypes between the two donor groups that differed in gene content as indicated from “1)” to “4).” One copy of a gene is shown by x1, two copies are shown by x2. *, P < 0.05; **, P < 0.01; ***, P < 0.005; and *****, P < 0.0005.
Polymorphism also affected the frequency and level of 3DL2 expression. Significant differences were observed between 3DL2*008 (high-binding) and 3DL2*007 and 3DL2*00902 (low-binding). Compared with other allotypes, 3DL2*008 and 3DL2*002 had a higher frequency of NK cell expression (Fig. 3 B). Like 3DL1, a gene dose effect was seen from comparison of individuals with one or two high-expressing allotypes (Fig. 3 C). Lower binding levels and smaller differences between allotypes limited 3DL2 analysis compared with 3DL1. KIR3DL2 heterozygotes did not give the readily interpreted bimodal distributions seen for 3DL1. Furthermore, there were nine 3DL2 allotypes, compared with five for 3DL1, which increased the number of heterozygous combinations and reduced the frequency of the more readily analyzed homozygotes.
For 2DL1/S1, the frequency of NK cells binding EB6 and their binding level both increased with the number of genes encoding KIR that bind the antibody (Fig. 3 C). Although no effects correlating with 2DL1 polymorphism were observed, the panel's high 2DL1*00302 frequency suggests that its high binding could have obscured the effects of low-binding allotypes in heterozygotes. Regarding the anti-2DL2/3+2DS2 antibody (DX27), panel members were either homozygous for 2DL3 or had one copy each of 2DL2, 2DL3, and 2DS2. Donors with the latter genotype had a similar frequency of antibody-binding NK cells, but a substantially increased level of binding (Fig. 3 C).
Cognate HLA class I increases the frequency of NK cells expressing inhibitory KIR
HLA-A, -B, -C types for the panel were determined at high resolution (Fig. S3 A, available at http://www.jem.org/cgi/content/full/jem.20051884/DC1). Both the alleles and their frequencies were consistent with previous studies (28, 29). Hardy-Weinberg equilibrium was observed for KIR and HLA (Fig. S3 B). Distinguishing Japanese from Caucasians are the frequencies of HLA-C ligands for KIR2DL. In Caucasians, 66% of HLA-C allotypes have C1 (the 2DL2/3 ligand) and 34% have C2 (the KIR2DL1 ligand); whereas in Japanese, 92% have C1 and 8% have C2. Thus, almost all Japanese use C1 as a self-ligand for inhibitory KIR, whereas a small minority use C2. The frequency of HLA-A3/11 ligands for 3DL2 is also lower in Japanese (8%) than in Caucasians (22%), whereas they have identical frequency for HLA-Bw4, the 3DL1 ligand (36%).
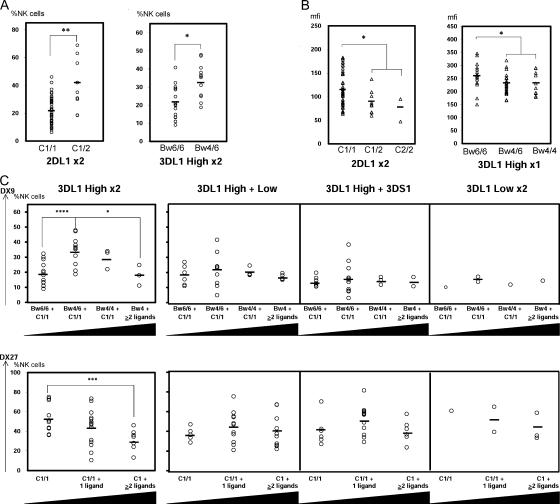
KIR phenotypes were compared for donors who have or lack a cognate HLA class I ligand for particular KIR. In several combinations, presence of cognate ligand increased the frequency of NK cells expressing the KIR (Fig. 4 A), while decreasing the amount of KIR-specific antibody that is bound (Fig. 4 B). In 2DL1 homozygotes, the presence of cognate C2 almost doubled the frequency of 2DL1+ NK cells while reducing the level of anti-2DL1 binding by 26%. Although the panel's high C1 frequency precluded comparable analysis of 2DL2/3, the two donors who lacked C1 showed decreased frequency of 2DL2/3-expressing cells and reduced levels of antibody binding (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20051884/DC1). C1 homozygotes had decreased binding of anti-2DL2/3 to NK cells compared with C1/C2 heterozygotes. Presence of Bw4 significantly increased the frequency of cells expressing 3DL1 for donors having two high-binding 3DL1 allotypes and decreased the level of anti-KIR3DL1 bound for donors having one high-binding 3DL1 allotype (Fig. 4, A and B). For donors having one high-binding 3DL1 allotype, paired with a low binding allotype or 3DS1, there was little effect on expression frequency, as also seen for donors having two low-binding allotypes (Fig. 4 C, top).
Figure 4.
The influence of cognate HLA class I on KIR expression by NK cells. (A) Influence of ligand on the frequency of cognate KIR expression. (left) For individuals who have two copies of 2DL1 and no 2DS1, the percent of NK cells expressing 2DL1 is compared between those who lack the C2 ligand (C1/1 genotype) and those who have one copy of it (C1/2 genotype). (right) For individuals who have two high-expressing 3DL1 allotypes, the percent of NK cells expressing 3DL1 is compared between those who lack the Bw4 ligand (Bw6/6 genotype) and those who have one copy of it (Bw4/6 genotype). (B) Influence of ligand on the antibody binding to cognate KIR. (left) For individuals who have two copies of 2DL1 and no 2DS1, the mean binding of anti-2DL1 to 2DL1-expressing NK cells is compared between those who have no C2 ligand, one C2 ligand, and two C2 ligands (C2/2 genotype). (right) For individuals who have one high-expressing 3DL1 allotype, the mean binding of anti-3DL1 to NK cells expressing 3DL1 is compared between those who have no Bw4 ligand, one Bw4 ligand, and two Bw4 ligands. (C) Influence of other ligand–receptor pairs on KIR expression. (top four panels) In each panel, the frequency of cells expressing 3DL1 is compared in four groups of individuals: those who lack Bw4 and are homozygous for C1 (Bw6/6+C1/1); those who are heterozygous for Bw4 and homozygous for C1 (Bw4/6+C1/1); those who are homozygous for Bw4 and C1 (Bw4/4+C1/1); and those who are either heterozygous or homozygous for Bw4 and have two or more other self-ligands for inhibitory KIR (Bw4 + ≥2 ligands). Each of the four panels corresponds to individuals with different 3DL1 genotypes: two high-expressing allotypes (far left), one high and one low-expressing allotype (center, left), one high-expressing allotype and 3DS1 (center, right), and two low-expressing allotypes (far right). Black triangles below each panel indicate increasing number of KIR ligands. (bottom) In each panel, the frequency of cells expressing 2DL2/3 and 2DS2 is compared in three groups of individuals: those who are homozygous for C1 (C1/1);, those who are homozygous for C1 and have one additional self-ligand for inhibitory KIR (C1/1 + 1 ligand); and those who are either homozygous for C1 and have two or more additional self-ligands for inhibitory KIR or are heterozygous for C1/C2 and have one or more additional self-ligands for inhibitory KIR (C1 + ≥2_ligands). The four panels correspond to the same groups based upon KIR3DL1/S1 genotype as the top panels. *, P < 0.05; **, P < 0.01; ***, P < 0.005; and ****, P < 0.001.
Cognate HLA class I decreases the frequency of NK cells expressing other inhibitory KIR
For donors with two high-binding 3DL1 allotypes, the increase in frequency of 3DL1+ NK cells is greater in Bw4 heterozygotes than in homozygotes (Fig. 4 C, top). The effect of cognate Bw4 to increase the frequency of 3DL1+ cells was also depressed by the presence of additional cognate HLA–KIR interactions, and the effects were additive. Thus, for individuals who had all four inhibitory ligands (C1, C2, Bw4, and A3/11), the frequencies of 3DL1+ cells are comparable to those in individuals lacking Bw4 (Fig. 4 C, top). Although similar trends were seen for individuals having one high-binding 3DL1 allotype, they did not reach statistical significance. A similar effect of other cognate HLA–KIR interactions was also seen from analysis of the frequency of NK cells expressing 2DL2/3 (Fig. 4 C, bottom).
Allelic polymorphism modulates the inhibitory function of KIR3DL1
Approximately one third of the Japanese population has C1-2DL3 as the only functional inhibitory KIR–HLA class I interaction, while one half of the population can also use the Bw4–3DL1 interaction. Although 2DL3 has little polymorphism in the Japanese, 3DL1 variation is substantial (Fig. 1). Consequently, the Bw4–3DL1 interaction is a potentially important source of functional NK cell diversification in the Japanese population. To address this question, the functions of the five 3DL1 allotypes present in the Japanese panel were compared.
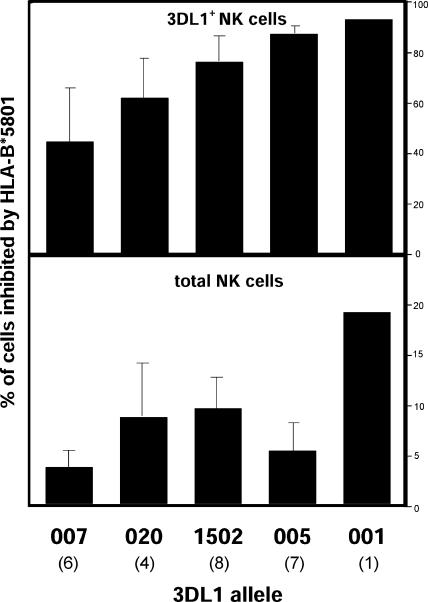
PBMCs from 13 donors, each having a high- and low-binding 3DL1 allotype, were challenged with class I–deficient 721.221 cells and with 221 cells transfected with HLA-B*5801, a 3DL1 ligand. In response to 221 cells, 3DL1+ NK cells produced IFN-γ as detected by intracellular staining. A diminished response was made to 221- B*5801, the inhibition varying 40–90% between the five 3DL1 allotypes (Fig. 5, top). The hierarchy for increasing inhibition was 3DL1*007<*020<*01502<*005<*001. That 3DL1 polymorphism modulates a major NK cell effector function demonstrates its functional importance. Of note, the two allotypes giving strongest inhibition are those expressed at intermediate (3DL1*001) and lowest levels (3DL1*005).
Figure 5.
KIR3DL1 allotypes have different inhibitory capacity. PBMCs from heterozygous donors expressing one high- and one low-expressing 3DL1 allotype were cultured with class I–deficient 721.221 cells or 221 cells transfected with HLA-B*5801, a 3DL1 ligand. The production of IFN-γ by CD3−3DL1+ cells (3DL1+NK cells) was determined by flow cytometry. Cells expressing the high and the low expressing allotypes formed distinct distributions and were analyzed separately. For each 3DL1 allotype, the number of cells producing IFN-γ in response to 221 and 221-B*5801 was compared and the inhibition mediated by B*5801 was calculated as the percentage of CD3−3DL1+ NK cells (top) or total NK cells (bottom), as defined by CD3−CD56+ PBMC in the KIR phenotype analysis of the donor panel. The number of donors tested for each allotype is shown in parenthesis below the allotype name. Significance values for pairwise comparisons are: *01502/*005: P < 0.02; *01502/*007: P < 0.005; *005/*007: P < 0.003; *020/*005: P < 0.03. Error bars represent the standard deviation of the results obtained with the number of donors shown (bottom).
Further modulating each 3DL1 allotype's capacity to influence the NK cell response is its expression frequency in the NK cell population. Consequently, the impact of an allotype such as 3DL1*005 could be restricted to a relatively small number of cells. By taking both factors (the frequencies of expression and inhibition) into account, we estimated the frequency of NK cells in peripheral blood that are inhibitable by B*5801 (Fig. 5, bottom). The difference between this hierarchy (3DL1*007<*005<*020<*01502<*001) and that in Fig. 5′s top half is the position of 3DL1*005, which shifts from the second strongest to second weakest allotype.
Positive and balancing selection of KIR
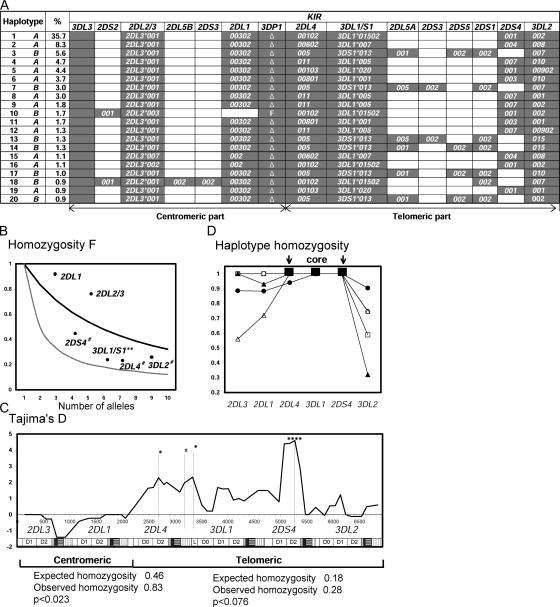
70 allele-level KIR genotypes were distinguished in the panel. From these genotypes and the sequences of prototypical group A (GenBank accession no. AC011501) and group B (GenBank accession no. AY320039) haplotypes, the structures of Japanese KIR haplotypes were inferred by two independent methods. The 20 most frequent haplotypes were identically ranked (Fig. 6 A). Each was identified in two or more donors and, collectively, they account for 83.4% of the panel's haplotypes. In both gene content and allelic polymorphism, the telomeric part of Japanese KIR haplotypes is more variable than the centromeric part. Together, the 12 deduced A haplotypes comprise 67.7% of total haplotypes compared with 15.7% for the 8 deduced B haplotypes. One A haplotype is markedly more common (35.7%) than any other haplotype (<9%) and, for each of its component KIR genes, this haplotype has the dominant allele.
Figure 6.
KIR haplotypes have been subject to balancing selection and positive directional selection. (A) Deduced allele-level KIR haplotypes. Shown are the 20 most common haplotypes, their frequencies, and their assignment to group A or B. These haplotypes were all identified by independent analytical methods: the EM-algorithm and the Bayesian phasing method. Independent analysis of three families confirmed the segregation of haplotypes 1, 2, 3, 5, 7, 13, and 15 (not depicted). (B) The Ewens-Watterson homozygosity statistic (F) was used to assess the departure from neutrality of the KIR allele frequencies in 116 Japanese donors. The thick line shows the neutral expectation. F values that are higher than the neutral expectation indicate purifying or directional selection; those that are lower than neutral expectation indicate balancing selection. The thin line shows the limit at maximum balancing selection. The 2DS4, 3DL1, 2DL4, and 3DL2 genes from the telomeric part of the KIR locus appear subject to balancing selection (**, P < 0.01 and #, P < 0.1). 2DL1 and 2DL3 genes in the centromeric part of the KIR locus show a trend toward purifying or directional selection. (C) Tajima's D statistic was applied to a composite of 182 group A coding-region haplotypes using a sliding window. The positions of the individual KIR in the composite are shown along the bottom of the panel. Positive values are indicative of balancing selection, negative values of purifying or directional selection. *, P < 0.05; ****, P < 0.001; and #, P < 0.1. Beneath the panel the results are shown for the Ewens-Watterson test on the centromeric and telomeric parts of the 182 group A haplotypes. Significance was assessed by Watterson's exact test. For both parts, the observed homozygosity was higher than expected. (D) The 182 group A haplotypes are all variants of five basic forms, which can be defined by the five KIR3DL1 alleles. For each of these five subgroups, the haplotype homozygosity is shown. The subgroups are as follows: •: 3DL1*01502, ▴: 3DL1*005, Δ: 3DL1*007, □: 3DL1*001, ◯: 3DL1*020. For each haplotype subgroup, there is a central region of high LD encompassing the 2DL4, 3DL1, and 2DS4 genes, which is flanked by regionsof reduced LD.
Application of the Ewens-Watterson test (30, 31) to the polymorphic KIR genes of the group A haplotypes shows that centromeric genes specifying HLA-C receptors (2DL1 and 3) have been subject to positive directional selection (Fig. 6 B), consistent with the high frequencies of the dominant alleles at these genes (Fig. 1). In contrast, the telomeric 2DL4, 3DL1, 2DS4, and 3DL2 genes, including those encoding HLA-A, -B, and -G receptors, appear subject to balancing selection, consistent with their wider range of allele frequencies. Tajima's D statistic (32) was also used to assess the mode and strength of selective pressure on polymorphic sites in group A haplotypes, using composite 7-kb haplotypes constructed from the coding sequences of KIR alleles in their order on the haplotype (Fig. 6 C, within the panel). Aside from confirming directional selection on the 2DL1 and 3 genes, this analysis revealed a strong signature for balancing selection in the telomeric region, one focusing on the ligand-binding domains of 2DL4, 3DL1, and 2DS4 and the signaling domain of 2DL4.
Linkage disequilibrium (LD) can indicate positive selection (33). Haplotype homozygosity revealed relatively high LD throughout the Japanese group A haplotypes; in particular, from 2DL4 through 3DL1 to 2DS4 (Fig. S5, available at http://www.jem.org/cgi/content/full/jem.20051884/DC1). Within this core region, the five 3DL1 alleles are linked with different combinations of alleles for the other two genes. These various cores are further diversified by combination with different 3DL2 alleles, the number depending on the core (Fig. 6 D). For example, 3DL1*005 pairs with four 3DL2 alleles, whereas the haplotype containing the dominant 3DL1*01502 allele is almost homogeneous, which is strong evidence for its positive selection, a selective sweep. As an independent assessment, the Ewens-Watterson homozygosity statistic was calculated for the centromeric and telomeric parts of the group A haplotypes and tested for departure from neutrality. In both parts of the KIR locus, signatures of positive selection were detected (Fig. 6 C, below the panel).
These analyses show that positive selection drove one group A haplotype to high frequency, such that its component alleles are now the dominant alleles of the polymorphic KIR genes. Positive selection also homogenized the centromeric part of the KIR locus. In contrast, balancing selection on telomeric KIR has maintained a variety of other haplotypes and their component alleles at intermediate frequencies. Because of these contrasting selections, a majority of Japanese (∼75%) have the same centromeric KIR genes, including the HLA-C–specific KIR2DL, but differ in the telomeric genes, including the HLA-A– and -B–specific KIR3D.
In addition to the 20 haplotypes shown in Fig. 6 A, 24 rarer haplotypes were deduced by assuming patterns of LD defined previously in other populations (34). Of the 44 haplotypes, 24 were group A and 20 group B. 30 of the 44 haplotypes share some alleles with the most common haplotype (no. 1) (Fig. S6, available at http://www.jem.org/cgi/content/full/jem.20051884/DC1). As well as a high frequency group A haplotype, the Japanese population has an abundance of low frequency A and B haplotypes with centromeric and telomeric motifs distinct from those present in the common haplotypes. These provide a pool of diverse functional potential for natural selection to operate on in the future.
Non-random associations of KIR–HLA in females
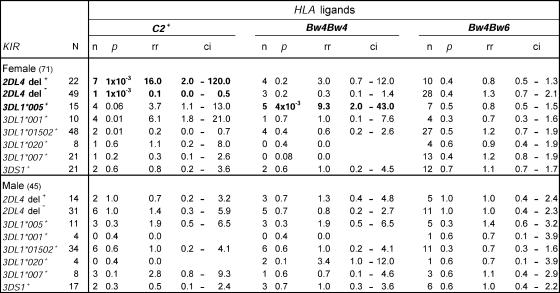
Systematic analysis revealed combinations of KIR and HLA genes in the donor panel at non-random frequencies. Stratification by sex showed that the effect was confined to females (Fig. 7). Deleted, nonfunctional forms of 2DL4 (35) preferentially associated with C2, whereas homozygosity for functional, full-length 2DL4 correlated negatively with C2. 3DL1*005 preferentially associated with Bw4, a combination also favoring inhibitory interaction. Although C2 and Bw4 are in strong LD in Caucasians, that is not so for Japanese, for whom only 40% of C2-carrying haplotypes also have HLA-Bw4 (36). Also, only 50% of donors carrying C2 have Bw4 (8 out of 16), a neutral distribution confirming the independence of C2 and Bw4 in the panel. Association with Bw4 is thus an independently associated factor and not a consequence of LD with C2. Weaker trends were HLA-C2 with 3DL1*005 or *001, the most potent inhibitors (Fig. 5), whereas 3DL1*01502, the dominant allotype and less potent inhibitor, correlated negatively with C2.
Figure 7.
Non-random KIR-HLA combinations in females. The observed frequencies of KIR-HLA combinations in the Japanese panel were compared with the frequencies predicted by random combination. This revealed a small number of deviations, which were then found to be present in females (top), but not males (bottom). Combinations with strong over-representation are shown in bold. As well as the p-value obtained from the chi-square test, relative risk (rr) was also calculated as a measure of deviation and is shown with 95% confidence intervals (ci).
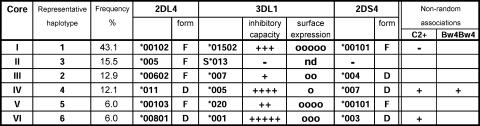
Six core haplotypes are defined by the five 3DL1 alleles and 3DS1 (Fig. 8). The cores defined by 3DL1*005 and *001 both encode deleted, nonfunctional forms of 2DL4 and 2DS4. Group A haplotypes with these cores lack activating KIR and have a strong inhibitory Bw4 receptor as well as inhibitory receptors for C1 and C2. Consequently, the overrepresented combination of 3DL1*005, nonfunctional 2DL4, Bw4 homozygosity, and C2 (usually accompanied by C1) is one that suggests some selection for strong inhibitory KIR–HLA interactions. Conversely, the underrepresented combinations of functional forms of 2DL4 with C2 suggest selection for less inhibition and more activation.
Figure 8.
Non-random KIR-HLA associations involve highly inhibitory KIR haplotypes. Although the centromeric part of the KIR locus lacks variability in the Japanese, the “core” region encompassing the 2DL4, 3DL1, and 2DS4 genes has six distinctive motifs (designated by Roman numerals I–VI) at significant frequency, each having a different 3DL1 allele or 3DS1. For 2DL4 and 2DS4, the full-length functional forms are denoted by “F” and the deleted, nonfunctional forms are indicated by “D.” Under “Non-random associations,” the overrepresented combinations of KIR with HLA described in Fig. 7 are indicated by “+,” the underrepresented combinations are indicated by “−.” The overrepresented combinations involve core haplotypes IV and VI that have no activating receptor and the most inhibitory 3DL1 allotypes. nd, not detected.
DISCUSSION
Selection of NK cell KIR repertoire by cognate HLA class I ligands
In mouse models, acquisition of inhibitory Ly49 receptors by NK cells is informed by interactions with cognate MHC class I ligands expressed on bone marrow stromal cells (37). The common observation is that cognate ligand, usually a transgene product, reduces the frequency of receptor-expressing cells. For humans, we find both increase and decrease in the frequency of receptor expression, which depends on the number of cognate KIR–HLA pairs contributing to NK cell self-tolerance. When acting alone, the cognate ligand increases the frequency of the KIR's expression, but the effect varies with receptor and ligand gene dose. Such increase is consistent with previous observation that NK cell self-tolerance is mediated by inhibitory HLA class I receptors (6). In the presence of other ligand–receptor pairs, the expression frequency for each receptor is reduced from that achieved when it acts alone, and to an extent inversely correlated with the number of functional ligand–receptor pairs.
For mice, it has been proposed that sequential or stochastic expression of inhibitory self-reactive receptors occurs until cumulative inhibition reaches some threshold, whereupon further receptor acquisition stops (38–40). Our results fit with this type of model. That the cognate ligand has not been observed to elevate receptor expression frequency in mice could reflect a difference in the mechanism between the two species or in the mode of analysis. For example, the presence in mice of multiple Ly49 products with overlapping ligand specificity could have obscured the positive effect, as we observed for humans with multiple KIR–HLA pairs.
KIR polymorphism modulates NK cell effector function
For ∼33% of the Japanese population, C1 is the only self-ligand for inhibitory KIR. Because the Japanese are practically monomorphic for KIR2D and polymorphic for KIR3DL1, they provided a good context in which to investigate the function of 3DL1 polymorphism. The five 3DL1 allotypes present in the Japanese differ in their capacity to inhibit NK cell function; they also differ in repertoire, which is the frequency and level of cell surface expression. Much evidence shows that DX9 binding measures the amount of cell surface KIR3DL1 (Fig. S10 and references 25–27), so the allotypic differences observed are not the result of varying affinity for DX9.
The allotypic differences in the frequency and level of KIR3DL1 expression are distinguished from those caused by cognate ligand and represent a ligand-independent influence of 3DL1 polymorphism on the primary NK cell repertoire. The effects of cognate 3DL1–Bw4 interactions are also modulated by KIR3DL1 polymorphism. Notably, the increase in frequency induced by the cognate ligand is greater for high than for low-expressing KIR3DL1 allotypes. The overall trend is for inhibitory capacity to increase with the level and frequency of 3DL1 expression, an exception being 3DL1*005, which combines low frequency and level of expression with strong inhibitory function.
KIR diversity is subject to positive and balancing selection
An outstanding question is whether the haplotypic diversity and allelic polymorphism of KIR genes arose from functional selection or neutral change. From functional assay and statistical analysis, we provide evidence that balancing selection maintains a pool of alleles and haplotypes with distinguishable functions upon which positive selection can act. In the Japanese, one group A KIR haplotype has risen to unusually high frequency, a selection that may have been targeted to the centromeric KIR genes, because the 2DL1*00302 and 2DL3*001 alleles of the selected haplotype are at frequencies approaching 90%. In contrast, in the telomeric parts of the locus, the balancing selection retained diversity as exemplified by the even frequency of other group A and group B haplotype motifs and the distribution of the six core group A haplotype structures (Fig. 8). In these regions, the selected haplotype encodes functional KIR2DL4 and KIR2DS4 forms and a KIR3DL1 allotype giving moderate inhibition. Within the set of Japanese A haplotypes, it is one that combines a full set of functional genes and good potential for both inhibition and activation. Because of this haplotype, the frequency of functional 2DL4 and 2DS4 alleles in the Japanese is much higher than in Caucasians (35, 41). One possible cause of the selective sweep is an epidemic of infectious disease where the selected haplotype enhanced the chance of surviving infection. Alternatively, women carrying this less inhibitory, more activating form of A haplotype could have above average reproductive success, as might be suggested from the results of Hiby et al. (15).
Evidence for selection of KIR–HLA combinations in female genotypes
An unexpected and thought-provoking discovery is the over- and underrepresentation of certain KIR–HLA class I combinations in Japanese females. Because KIR and HLA are on different chromosomes, the overrepresented combinations must confer some selective advantage, whereas the underrepresented combinations are disadvantageous. As only NK cells and T cells express KIR, the selective mechanism likely involves one or both of these cell types. Candidate mechanisms can be distinguished according to when they might act: before conception, during fetal life, or after birth.
Acting before conception and biasing progeny genotype is mate choice. Extensive literature points to MHC polymorphism influencing the choice of sexual partners (42–44), and it is plausible that this biology could extend to combinations of KIR and HLA polymorphisms. For example, preferential choice of Bw4+ partners by Bw4+, 3DL1*005+ individuals would lead to enrichment in the next generations of genotypes combining Bw4 homozygosity with 3DL1*005. For mate choice, a cause for the female bias is hard to envisage.
As only a fraction of human embryos survive implantation and fetal life (45), there is potential for improving survival. There are also sex differences in the mechanisms by which embryos fail to survive and the timing of these events (46). For example, for embryos with normal karyotype, the rate of spontaneous abortion during early pregnancy is greater for females than males, so any combination of genetic factors that reduced the effect would be specifically overrepresented in the female population (46). Candidate mechanisms involve KIR–HLA interactions within the growing fetus, which could contribute to tissue development or defense against infection. NK cells are detectable in fetuses aborted at 6–24 wk of gestation, pointing to their possible contribution (47). Also of possible relevance, KIR3DL3 transcripts are more abundant in the NK cells of females than males and, although the protein appears undetectable in adult cells, it may function during fetal development (48).
After birth, the favored KIR–HLA combinations could improve survival, relative to other combinations, through NK or T cell responses that eliminate infection or cancer, or prevent autoimmune disease. Female specificity could arise in a general way from the hormonal differences between men and women, and their influences on the immune system (49, 50), or more specifically to immune functions under circumstances experienced only by women, such as menstruation, pregnancy, nursing, cancer of breast, or ovary.
Precedent for KIR–HLA combinations influencing human reproduction and survival is the work of Hiby et al. (15). They found that inhibitory combinations of fetal HLA-C2 and maternal homozygosity for A KIR haplotypes predispose Caucasians to preeclampsia. These correlations fit well with the observed interactions of fetal trophoblast with maternal NK cells during placentation. Selection by preeclampsia can also explain the inverse correlation between C2 and A haplotype frequencies in human populations, as exemplified by the low C2 and high group A frequencies in the Japanese. In this context, it is important to note that the correlations reported here do not address the fitness of allogeneic combinations of maternal and fetal factors, but of the KIR and HLA genes brought together in the child's genotype. Consequently, the biology underlying the overrepresented and underrepresented KIR–HLA combinations we find may well be distinct from those described by Hiby et al. (15).
A signature of natural selection on KIR diversity
KIR-expressing NK cells participate in defense against infection and in reproduction, two functions that are essential for the survival of individuals, populations, and species. Both the KIR and their cognate HLA class I ligands are highly variable. In comparison with Caucasians, the Japanese population has a skewed distribution of KIR and HLA variants, reflecting a distinct history of directional and balancing selection. Functional and genetic study of a Japanese panel provides evidence here that the combination of KIR and HLA variation acts to diversify NK cell repertoire and effector function within the population and to facilitate human survival.
MATERIALS AND METHODS
Human subjects and blood samples.
Blood samples were from 132 healthy, unrelated Japanese individuals (54 males and 78 females, ages 22–73, mean 40 ± 12 yr [SD]) and from related donors in three pedigrees; informed consent was as approved by the Stanford University Institutional Review Board on Human Subjects. PBMCs were prepared with Ficoll-Paque PLUS (GE Healthcare). Genomic DNA and RNA were extracted with DNAzol (Molecular Research Center) and TRIzol (Invitrogen). Complementary DNA was synthesized from RNA, using oligo(dT) and SMART II oligonucleotide primers (SMART-RACE kit; CLONTECH Laboratories, Inc.).
KIR and HLA typing.
KIR genes were typed as described previously (22). 5′RACE used the Advantage 2 PCR kit (CLONTECH Laboratories, Inc.), with the universal primer (UPM) from the SMART-RACE kit or KIR-specific primers as the forward primer combined with KIR-specific reverse primers (Fig. S7, available at http://www.jem.org/cgi/content/full/jem.20051884/DC1). KIR transcripts were cloned into the pGEM T-easy vector (Promega) and sequenced using BigDye terminator 3.1 chemistry and a PRISM 377 DNA sequencer (Applied Biosystems). Sequence assembly and analysis used AlignIR version 2.0 (LICOR). Three or more clones from two or more independent PCR reactions were sequenced to define novel polymorphisms. KIR allele typing by sequence-specific polymorphism PCR used novel primers and those described previously (25, 34, 41, 51) (Figs. S8–S10, available at http://www.jem.org/cgi/content/full/jem.20051884/DC1). PCR used hot-start procedures using AmpliTaq FS on a PE9600 thermalcycler (Applied Biosystems). The juxtaposition of 2DL5, 2DS3, and 2DS5 to either 2DL2 or 3DS1 was determined by long-range PCR (52). Exon 1 was sequenced directly from the PCR products to distinguish 2DL5*002 from *005. In exon 6 of 2DL4, the cluster of 9 or 10 adenines (9A/10A) were typed by sequencing (35). HLA-A, B, C typing was by sequencing locus-specific PCR products (minimally exons 2 and 3, but also exon 4 for some samples) with adaptations from Dunn et al. (53) and PCR-SSOP (Sequence-Specific Oligonucleotide Probing), using LABType SSO Typing Tests (One Lambda).
Flow cytometry and NK cell secretion of IFN-γ.
KIR on CD56+CD3−NK cell surfaces was measured with monoclonal antibodies and flow cytometry (22). Phycoerythrin conjugates of anti-2DL1/2DS1 (EB6; Beckman Coulter), anti-2DL2/2DL3/2DS2 (DX27), and anti-3DL1(DX9) (BD Biosciences), anti-3DL2 (DX31, a gift from Drs. J. Phillips and L. Lanier, DNAX Research Institute, Palo Alto, CA) were used in combination with anti-CD3–PerCP (SK7) and anti-CD56–fluorescein (NCAM16.2) (BD Biosciences) to stain PBMCs. Three-color flow cytometry used a FACScan instrument and CELLQuest software (BD Biosciences). Parameters measured were the percent NK cells binding an antibody and the mean fluorescence intensity (mfi) for the positive cells.
PBMCs from donors heterozygous for high- and low-binding 3DL1 allotypes were cocultured with the class I–deficient 721.221 cell line or 221 transfected with HLA-B*5801. After gating on 3DL1+ NK cells, IFN-γ production was assessed by intracellular staining (54): PBMCs at 6 × 105 cells/well were cocultured with target cells at an optimized effector–target ratio of 1:1 for 9 h; fluorescent anti-CD3 and anti-CD85–PECy5 (GHI/75) (BD Biosciences) were added so that 3DL1+ T cells and the inhibitory effects of LIR1 could be excluded from analysis. Knowing the proportion of donor NK cells expressing each 3DL1 allotype, the inhibitory effect on total NK cells was calculated for each allotype: (% IFN-γ inhibition) × (% allotype-expressing NK cells).
Statistical methods.
Reconstruction of allele-level KIR haplotypes used the expectation maximization algorithm (Arlequin version 2.001; reference 55) and a Bayesian statistical method using hidden Monte Carlo Markov Chain modeling (PHASE version 2.1; reference 56). LD was calculated for pairs of KIR alleles (Arlequin version 2.001), D′ values >0.5 with P < 0.002 are shown in Fig. S5. The homozygosity statistic F of Ewens (30) and Watterson (31), calculated from KIR allele frequencies was compared with expected homozygosity statistics; significance levels were tested using Slatkin's exact test (57) (Arlequin version 2.001). Tajima's D (32) was calculated for 7-kb haplotypes constructed by combining the coding sequences of KIR alleles on group A haplotypes in order. 10 potential recombination points between haplotypes were first detected by the four-gamete test (58). D values were then calculated in a 300-bp sliding window along the composite haplotypes without extending over the recombination points, and visualized (DnaSP version 4.0; reference 59). For the 9A KIR2DL4 alleles and the 22-bp deletion 2DS4 alleles, each deleted nucleotide was considered an independent position. Haplotype homozygosity was calculated at each gene in group A haplotypes based on Sabeti et al.'s method (33).
Nonparametric tests of significance were used in the analyses described in Figs. 3, 4, 5 and Fig. S4 (two-tailed Mann-Whitney's U-test). Chi-squared tests were used in 2 × 2 contingency tables for pairwise comparisons of genetic associations in Fig. 7. P < 0.05 was considered statistically significant. Fisher's exact test was applied when expected numbers were small (n < 5). Tests used SPSS version 8 software (SPSS Inc). Significance values are as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001; and *****, P < 0.0005.
Online supplemental material.
Fig. S1 depicts comparison of KIR allele frequencies between Japanese and Caucasians. Fig. S2 shows kinetics of DX9 binding and phenotype comparison with Z27. Fig. S3 A contains comparison of HLA class I allele distributions between Japanese and Caucasians. Fig. S3 B shows heterozygosity of HLA class I and KIR. Fig. S4 depicts the influence of cognate HLA class I on KIR expression. Fig. S5 displays LD between KIR alleles. Fig. S6 shows the frequency of centromeric and telomeric parts in 44 haplotypes. Fig. S7 contains primers and PCR conditions for KIR cDNA cloning. Fig. S8 depicts primers for KIR allele typing. Fig. S9 shows PCR conditions for KIR allele typing. Fig. S10 depicts primer combinations for KIR allele typing. Online supplemental figures are available at http://www.jem.org/cgi/content/full/jem.20051884/DC1.
Supplemental Material
Acknowledgments
We thank all blood donors who participated in this study; we also thank M. Minami and his staff for their help in blood sample collection. DX31 was a gift from J. Phillips and L. Lanier. We thank T. Hastie for his advice on statistics.
This work was supported by National Institutes of Health (NIH) grant nos. AI-017892 and AI-022039 (to P. Parham) and a grant from the Leukemia Research Foundation (to M. Yawata).
The authors have no conflicting financial interests.
Abbreviations used: KIR, killer cell immunoglobulin-like receptor; LD, linkage disequilibrium; mfi, mean fluorescence intensity.
M. Yawata and N. Yawata contributed equally to this work.
References
- 1.Sun, P.D. 2003. Structure and function of natural-killer-cell receptors. Immunol. Res. 27:539–548. [DOI] [PubMed] [Google Scholar]
- 2.Abi-Rached, L., and P. Parham. 2005. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J. Exp. Med. 201:1319–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parham, P. 2005. MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 5:201–214. [DOI] [PubMed] [Google Scholar]
- 4.Tanamachi, D.M., D.C. Moniot, D. Cado, S.D. Liu, J.K. Hsia, and D.H. Raulet. 2004. Genomic Ly49A transgenes: basis of variegated Ly49A gene expression and identification of a critical regulatory element. J. Immunol. 172:1074–1082. [DOI] [PubMed] [Google Scholar]
- 5.Held, W., B. Kunz, V. Ioannidis, and B. Lowin-Kropf. 1999. Mono-allelic Ly49 NK cell receptor expression. Semin. Immunol. 11:349–355. [DOI] [PubMed] [Google Scholar]
- 6.Valiante, N.M., M. Uhrberg, H.G. Shilling, K. Lienert-Weidenbach, K.L. Arnett, A. D'Andrea, J.H. Phillips, L.L. Lanier, and P. Parham. 1997. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 7:739–751. [DOI] [PubMed] [Google Scholar]
- 7.McVicar, D.W., and D.N. Burshtyn. 2001. Intracellular signaling by the killer immunoglobulin-like receptors and Ly49. Sci. STKE. 2001:RE 1. [DOI] [PubMed]
- 8.Anderson, S.K., K. Dewar, M.L. Goulet, G. Leveque, and A.P. Makrigiannis. 2005. Complete elucidation of a minimal class I MHC natural killer cell receptor haplotype. Genes Immun. 6:481–492. [DOI] [PubMed] [Google Scholar]
- 9.Uhrberg, M., N.M. Valiante, B.P. Shum, H.G. Shilling, K. Lienert-Weidenbach, B. Corliss, D. Tyan, L.L. Lanier, and P. Parham. 1997. Human diversity in killer cell inhibitory receptor genes. Immunity. 7:753–763. [DOI] [PubMed] [Google Scholar]
- 10.Yawata, M., N. Yawata, L. Abi-Rached, and P. Parham. 2002. Variation within the human killer cell immunoglobulin-like receptor (KIR) gene family. Crit. Rev. Immunol. 22:463–482. [PubMed] [Google Scholar]
- 11.Martin, M.P., X. Gao, J.H. Lee, G.W. Nelson, R. Detels, J.J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, J. Trowsdale, et al. 2002. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 31:429–434. [DOI] [PubMed] [Google Scholar]
- 12.Khakoo, S.I., C.L. Thio, M.P. Martin, C.R. Brooks, X. Gao, J. Astemborski, J. Cheng, J.J. Goedert, D. Vlahov, M. Hilgartner, et al. 2004. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 305:872–874. [DOI] [PubMed] [Google Scholar]
- 13.Carrington, M., S. Wang, M.P. Martin, X. Gao, M. Schiffman, J. Cheng, R. Herrero, A.C. Rodriguez, R. Kurman, R. Mortel, et al. 2005. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J. Exp. Med. 201:1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson, G.W., M.P. Martin, D. Gladman, J. Wade, J. Trowsdale, and M. Carrington. 2004. Cutting edge: heterozygote advantage in autoimmune disease: hierarchy of protection/susceptibility conferred by HLA and killer Ig-like receptor combinations in psoriatic arthritis. J. Immunol. 173:4273–4276. [DOI] [PubMed] [Google Scholar]
- 15.Hiby, S.E., J.J. Walker, K.M. O'Shaughnessy, C.W. Redman, M. Carrington, J. Trowsdale, and A. Moffett. 2004. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J. Exp. Med. 200:957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shilling, H.G., K.L. McQueen, N.W. Cheng, J.A. Shizuru, R.S. Negrin, and P. Parham. 2003. Reconstitution of NK cell receptor repertoire following HLA-matched hematopoietic cell transplantation. Blood. 101:3730–3740. [DOI] [PubMed] [Google Scholar]
- 17.Pende, D., R. Biassoni, C. Cantoni, S. Verdiani, M. Falco, C. di Donato, L. Accame, C. Bottino, A. Moretta, and L. Moretta. 1996. The natural killer cell receptor specific for HLA-A allotypes: a novel member of the p58/p70 family of inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140-kD disulphide-linked dimer. J. Exp. Med. 184:505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Litwin, V., J. Gumperz, P. Parham, J.H. Phillips, and L.L. Lanier. 1994. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J. Exp. Med. 180:537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winter, C.C., J.E. Gumperz, P. Parham, E.O. Long, and N. Wagtmann. 1998. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J. Immunol. 161:571–577. [PubMed] [Google Scholar]
- 20.Rajagopalan, S., Y.T. Bryceson, S.P. Kuppusamy, D.E. Geraghty, A. van der Meer, I. Joosten, and E.O. Long. 2005. Activation of NK cells by an endocytosed receptor for soluble HLA-G. PLoS Biol. DOI: 10.1371/journal.pbio.0040009. [DOI] [PMC free article] [PubMed]
- 21.Shilling, H.G., N. Young, L.A. Guethlein, N.W. Cheng, C.M. Gardiner, D. Tyan, and P. Parham. 2002. Genetic control of human NK cell repertoire. J. Immunol. 169:239–247. [DOI] [PubMed] [Google Scholar]
- 22.Yawata, M., N. Yawata, K.L. McQueen, N.W. Cheng, L.A. Guethlein, R. Rajalingam, H.G. Shilling, and P. Parham. 2002. Predominance of group A KIR haplotypes in Japanese associated with diverse NK cell repertoires of KIR expression. Immunogenetics. 54:543–550. [DOI] [PubMed] [Google Scholar]
- 23.Whang, D.H., H. Park, J.A. Yoon, and M.H. Park. 2005. Haplotype analysis of killer cell immunoglobulin-like receptor genes in 77 Korean families. Hum. Immunol. 66:146–154. [DOI] [PubMed] [Google Scholar]
- 24.Jiang, K., F.M. Zhu, Q.F. Lv, and L.X. Yan. 2005. Distribution of killer cell immunoglobulin-like receptor genes in the Chinese Han population. Tissue Antigens. 65:556–563. [DOI] [PubMed] [Google Scholar]
- 25.Gardiner, C.M., L.A. Guethlein, H.G. Shilling, M. Pando, W.H. Carr, R. Rajalingam, C. Vilches, and P. Parham. 2001. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J. Immunol. 166:2992–3001. [DOI] [PubMed] [Google Scholar]
- 26.Gumperz, J.E., N.M. Valiante, P. Parham, L.L. Lanier, and D. Tyan. 1996. Heterogeneous phenotypes of expression of the NKB1 natural killer cell class I receptor among individuals of different human histocompatibility leukocyte antigens types appear genetically regulated, but not linked to major histocompatibililty complex haplotype. J. Exp. Med. 183:1817–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carr, W.H., M.J. Pando, and P. Parham. 2005. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J. Immunol. 175:5222–5229. [DOI] [PubMed] [Google Scholar]
- 28.Tokunaga, K., Y. Ishikawa, A. Ogawa, H. Wang, S. Mitsunaga, S. Moriyama, L. Lin, M. Bannai, Y. Watanabe, K. Kashiwase, et al. 1997. Sequence-based association analysis of HLA class I and II alleles in Japanese supports conservation of common haplotypes. Immunogenetics. 46:199–205. [DOI] [PubMed] [Google Scholar]
- 29.Itoh, Y., N. Mizuki, T. Shimada, F. Azuma, M. Itakura, K. Kashiwase, E. Kikkawa, J.K. Kulski, M. Satake, and H. Inoko. 2005. High-throughput DNA typing of HLA-A, -B, -C, and -DRB1 loci by a PCR-SSOP-Luminex method in the Japanese population. Immunogenetics. 57:717–729. [DOI] [PubMed] [Google Scholar]
- 30.Ewens, W.J. 1972. The sampling theory of selectively neutral alleles. Theor. Popul. Biol. 3:87–112. [DOI] [PubMed] [Google Scholar]
- 31.Watterson, G. 1978. The homozygosity test of neutrality. Genetics. 88:405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tajima, F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 123:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabeti, P.C., D.E. Reich, J.M. Higgins, H.Z. Levine, D.J. Richter, S.F. Schaffner, S.B. Gabriel, J.V. Platko, N.J. Patterson, G.J. McDonald, et al. 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature. 419:832–837. [DOI] [PubMed] [Google Scholar]
- 34.Shilling, H.G., L.A. Guethlein, N.W. Cheng, C.M. Gardiner, R. Rodriguez, D. Tyan, and P. Parham. 2002. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J. Immunol. 168:2307–2315. [DOI] [PubMed] [Google Scholar]
- 35.Witt, C.S., A. Martin, and F.T. Christiansen. 2000. Detection of KIR2DL4 alleles by sequencing and SSCP reveals a common allele with a shortened cytoplasmic tail. Tissue Antigens. 56:248–257. [DOI] [PubMed] [Google Scholar]
- 36.Nakajima, F., J. Nakamura, and T. Yokota. 2002. Analysis of HLA haplotypes in Japanese, using high resolution allele typing. MHC. 8:1–32. [Google Scholar]
- 37.Fahlen, L., U. Lendahl, and C.L. Sentman. 2001. MHC class I-Ly49 interactions shape the Ly49 repertoire on murine NK cells. J. Immunol. 166:6585–6592. [DOI] [PubMed] [Google Scholar]
- 38.Hanke, T., H. Takizawa, and D.H. Raulet. 2001. MHC-dependent shaping of the inhibitory Ly49 receptor repertoire on NK cells: evidence for a regulated sequential model. Eur. J. Immunol. 31:3370–3379. [DOI] [PubMed] [Google Scholar]
- 39.Dorfman, J.R., and D.H. Raulet. 1998. Acquisition of Ly49 receptor expression by developing natural killer cells. J. Exp. Med. 187:609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams, N.S., A. Kubota, M. Bennett, V. Kumar, and F. Takei. 2000. Clonal analysis of NK cell development from bone marrow progenitors in vitro: orderly acquisition of receptor gene expression. Eur. J. Immunol. 30:2074–2082. [DOI] [PubMed] [Google Scholar]
- 41.Norman, P.J., M.A. Cook, B.S. Carey, C.V. Carrington, D.H. Verity, K. Hameed, D.D. Ramdath, D. Chandanayingyong, M. Leppert, H.A. Stephens, and R.W. Vaughan. 2004. SNP haplotypes and allele frequencies show evidence for disruptive and balancing selection in the human leukocyte receptor complex. Immunogenetics. 56:225–237. [DOI] [PubMed] [Google Scholar]
- 42.Reusch, T.B., M.A. Haberli, P.B. Aeschlimann, and M. Milinski. 2001. Female sticklebacks count alleles in a strategy of sexual selection explaining MHC polymorphism. Nature. 414:300–302. [DOI] [PubMed] [Google Scholar]
- 43.Ziegler, A., H. Kentenich, and B. Uchanska-Ziegler. 2005. Female choice and the MHC. Trends Immunol. 26:496–502. [DOI] [PubMed] [Google Scholar]
- 44.Ober, C., L.R. Weitkamp, N. Cox, H. Dytch, D. Kostyu, and S. Elias. 1997. HLA and mate choice in humans. Am. J. Hum. Genet. 61:497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vatten, L.J., and R. Skjaerven. 2004. Offspring sex and pregnancy outcome by length of gestation. Early Hum. Dev. 76:47–54. [DOI] [PubMed] [Google Scholar]
- 46.Eiben, B., I. Bartels, S. Bahr-Porsch, S. Borgmann, G. Gatz, G. Gellert, R. Goebel, W. Hammans, M. Hentemann, R. Osmers, et al. 1990. Cytogenetic analysis of 750 spontaneous abortions with the direct-preparation method of chorionic villi and its implications for studying genetic causes of pregnancy wastage. Am. J. Hum. Genet. 47:656–663. [PMC free article] [PubMed] [Google Scholar]
- 47.Phillips, J.H., T. Hori, A. Nagler, N. Bhat, H. Spits, and L.L. Lanier. 1992. Ontogeny of human natural killer (NK) cells: fetal NK cells mediate cytolytic function and express cytoplasmic CD3 ɛ,δ proteins. J. Exp. Med. 175:1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trundley, A.E., S.E. Hiby, C. Chang, A.M. Sharkey, S. Santourlidis, M. Uhrberg, J. Trowsdale, and A. Moffett. 2006. Molecular characterization of KIR3DL3. Immunogenetics. 57:904–916. [DOI] [PubMed] [Google Scholar]
- 49.Da Silva, J.A. 1999. Sex hormones and glucocorticoids: interactions with the immune system. Ann. NY Acad. Sci. 876:102–118. [DOI] [PubMed] [Google Scholar]
- 50.Tanriverdi, F., L.F. Silveira, G.S. MacColl, and P.M. Bouloux. 2003. The hypothalamic-pituitary-gonadal axis: immune function and autoimmunity. J. Endocrinol. 176:293–304. [DOI] [PubMed] [Google Scholar]
- 51.Gomez-Lozano, N., and C. Vilches. 2002. Genotyping of human killer-cell immunoglobulin-like receptor genes by polymerase chain reaction with sequence-specific primers: an update. Tissue Antigens. 59:184–193. [DOI] [PubMed] [Google Scholar]
- 52.Vilches, C., C.M. Gardiner, and P. Parham. 2000. Gene structure and promoter variation of expressed and nonexpressed variants of the KIR2DL5 gene. J. Immunol. 165:6416–6421. [DOI] [PubMed] [Google Scholar]
- 53.Dunn, P.P., V. Carter, A. Dunn, S. Day, S.V. Fuggle, J. Ross, and G. Cavanagh. 2000. Identification of an HLA-B7 serological variant and its characterization by sequencing based typing. Tissue Antigens. 55:71–73. [DOI] [PubMed] [Google Scholar]
- 54.Draghi, M., N. Yawata, M. Gleimer, M. Yawata, N.M. Valiante, and P. Parham. 2005. Single-cell analysis of the human NK cell response to missing self and its inhibition by HLA class I. Blood. 105:2028–2035. [DOI] [PubMed] [Google Scholar]
- 55.Schneider, S., D. Roessli, and L. Excoffier. 2000. Arlequin ver. 2.000: A Software for Population Genetics Data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva. 111 pp.
- 56.Stephens, M., N.J. Smith, and P. Donnelly. 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68:978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slatkin, M. 1996. A correction to the exact test based on the Ewens sampling distribution. Genet. Res. 68:259–260. [DOI] [PubMed] [Google Scholar]
- 58.Kaplan, N., and R.R. Hudson. 1985. The use of sample genealogies for studying a selectively neutral m-loci model with recombination. Theor. Popul. Biol. 28:382–396. [DOI] [PubMed] [Google Scholar]
- 59.Rozas, J., J.C. Sanchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 19:2496–2497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.