Abstract
The chronic graft-versus-host (cGvH) reaction is a model of induced lupus caused by alloreactive CD4+ T cells from a Bm-12 mouse in a C57BL/6 recipient. We used this cGvH reaction in C57BL/6 anti-DNA H chain transgenic mice, 56R/B6, to understand the structure, specificity, and origin of the induced autoantibodies (auto-Abs). We found anti-DNA Abs that reacted to several different antigens, such as phosphatidylserine, myelin basic protein, thyroglobulin, histone, insulin, cytochrome C, and β-galactosidase. This polyreactivity was found for Abs from B cells that expressed the 56R H chain transgene with “editor” L chains that did not completely veto autoreactivity. We suggest that such incomplete editing results in polyreactivity and that incompletely edited polyreactive B cells influence the subsequent expression of pathogenic auto-Abs in disease. We also found B cells that coexpress κ and λ L chain. These B cells contributed to the autoimmune response and are possibly in the marginal zone of the spleen.
Systemic lupus erythematosus patients produce a broad spectrum of autoantibodies (auto-Abs), including anti–double-stranded (ds)DNA (1, 2). In healthy individuals these auto-Abs are regulated at multiple checkpoints during B cell development (3, 4), and defects, such as loss of T cell tolerance (5), late-stage B cell deregulation (6), and clearance defects of nuclear and cytoplasmic antigens, can lead to autoimmunity.
A reproducible appearance of auto-Abs can be achieved by induction of a systemic lupus erythematosus–like syndrome by a chronic graft-versus-host (cGvH) reaction (7). To initiate cGvH, lymphocytes from a Bm-12 mouse are transferred into a C57BL/6 (B6) recipient (8, 9). The Bm-12 and the B6 mouse differ at the MHC class II protein by three amino acids; hence, the cGvH is thought to be mediated by Bm-12 CD4+ T cells (10). cGvH leads to B cell hyperactivation followed by the production of a variety of auto-Abs directed to chromatin, Sm, DNA, and other nuclear antigens. In addition, the mice develop glomerulonephritis (8, 11). The cGvH is thought to require B cell receptor cross-linking (12). The spectrum of auto-Abs induced by cGvH is narrowed in an anti-DNA transgenic mouse, referred to as 56R/B6 (13–15). This 56R H chain site-directed knock-in mouse was generated using the VH from an anti-DNA Ab isolated from a diseased MRL/lpr mouse (16). The DNA reactivity of the 56R H chain is due to positive charges of arginines (Args) in the complementarity-determining regions (CDRs) (17).
The 3H9 and 56R transgenic models have been useful for studying the mechanism of receptor editing (18, 19). L chain rearrangement in the 56R model seems to persist until the self-reactivity of the transgenic 56R H chain is altered. Such B cells bearing the 56R transgenic H chain are allowed to migrate into the periphery when paired with so called “editor” L chains. Such editor L chains have low isoelectric points due to a high frequency of aspartates (Asps) in their CDRs, and we think they block the interaction between H chain Arg and DNA (17).
The 56R H chain on a BALB/c background pairs mainly with the κ L chain Vκ21D, an efficient editor of anti-DNA activity (17). On a B6 background, however, less efficient editors Vκ38C and Vκ20 L chains are used (17, 20, and unpublished data). Among λ chains, λx has been shown to modify DNA binding of the 56R H chain (21, 22) and may result in altered self-reactivity. In 56R/BALB/c mice, a new population of edited 56R H chain transgenic B cells has been identified. In this mouse, B cells express two receptors of which one is κ and the other is λ1 (17), and these “partially edited” B cells are thought to reside in the marginal zone (MZ) (23, 24).
Using cGvH, we asked whether incompletely and partially edited B cells were activated and secreted auto-Abs in cGvH. We find that the induction of anti-DNA Abs is accompanied by the induction of other auto-specificities. Hybridoma panels derived from cGvH-induced mice were analyzed and showed that auto-Abs arose from B cells that were polyreactive and bound to self-antigens and proteins, such as dsDNA, phosphatidylserine (PS), myelin basic protein (MBP), thyroglobulin, histone, insulin, cytochrome C, and β-galactosidase. We found that the observed polyreactivity was a feature of the 56R H chain in combination with the editor L chains Vκ20 and Vκ38C. Consistent with the expression of both the κ and λ L chain in the serum after cGvH disease, hybridoma panels derived from cGvH-induced mice included partially edited B cells with λ1 L chain that coexpressed κ L chain. The B cells with the λ and κ L chain bound to dsDNA, MBP, and PS and could account for some of the auto-Ab expression in cGvH. In addition, those double positive B cells may be located in the MZ.
RESULTS
Induction of auto-Abs in cGvH
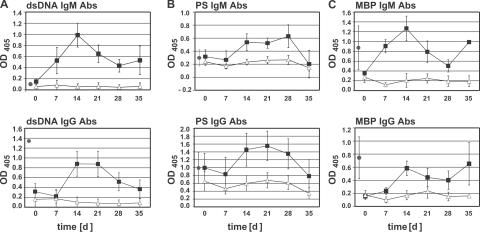
The cGvH reaction induces a lupus-like syndrome that includes activated lymphocytes and the production of auto-Abs (8, 9). In previous studies, cGvH was induced by the transfer of splenocytes (7–11, 14, 15, 25, 26) or partially purified CD4+ T cells (27) from Bm-12 mice to B6 recipients. The cGvH induces a variety of auto-Abs in B6 mice (8, 10), but in the case of 56R/B6, the auto-Ab repertoire is enriched in anti-DNA specificities as has been observed in studies on 56R Abs from unmanipulated and cGvH 56R/B6 mice (13–15, 28). To rule out possible donor B cell response in the cGvH, we used purified Bm-12 CD4+ T cells (CD4+CD3+ > 96%; not depicted). Under these conditions, we found that cGvH caused an increase of anti-dsDNA Abs (Fig. 1 A), accompanied by an increase of anti-PS Abs (Fig. 1 B), but we also find induction of anti-MBP Abs (Fig. 1 C).
Figure 1.
cGvH leads to the concomitant induction of anti-dsDNA, anti-PS, and anti-MBP Abs in 56R H chain transgenic mice. For induction of cGvH, Bm12 CD4+ T cells were injected into 56R/B6 (▪). As a control, B6 CD4+ T cells were injected into 56R/B6 (▵). Anti-DNA, anti-PS, and anti-MBP Abs in sera were tested at the indicated time points by ELISA at a serum dilution of 1:180. IgM and IgG Abs to dsDNA (n = 6 for control group and n = 7 for cGvH group) (A), to PS (n = 5 for control and cGvH group) (B), and to MBP (n = 3 for control group and n = 6 for cGvH group) (C) were tested by ELISA. MRL/lpr sera were tested at the same concentration for comparison (•; n = 4). Results represent means ± SEM.
The cGvH also activated peripheral B cells in the 56R/B6 recipient as seen by the transient expression of CD69 and CD86 as well as by the increased levels of MHC class II and Fas (CD95). A slight decrease of CD24 was also observed (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20060075/DC1). We conclude that cGvH induced by purified CD4+ T cells leads to secretion of anti-DNA Ab and activation of B cells in the 56R/B6 recipient. The induction of different auto-Ab specificities in 56R transgenic mice raised the following questions: Do these specificities come from different nontransgenic B cell populations, or can these specificities be attributed to polyreactive Abs expressed by 56R H chain with editor L chains as has been found to be the case for Vκ20 and Vκ38C? The 56R transgene in B6 mice both accelerates the onset and increases the titers of anti-dsDNA Abs in cGvH as reported previously (14, 15). To study the role of 56R in auto-Ab induction, we tested the expression of 56R H chain by flow cytometry and in hybridoma panels.
Role of 56R H chain in auto-Ab induction
To test whether the anti-DNA, anti-PS, and anti-MBP Ab specificities (Fig. 1) were associated with the 56R H chain, we used allotype-specific Abs that distinguish the transgenic allele Igha (16, 29) and the nontransgenic endogenous allele Ighb of the 56R/B6 mouse. In the control mice, 45–48% of the B220+ splenic B cells were IgMa+ and IgDa+ at day 60. An almost identical number (44–45%) was found in the cGvH group. Of the B220+ B cells, 8–10% expressed endogenous H chain according to allotype IgMb and IgDb expression. This may result from aberrant VH replacement followed by rearrangement of the untargeted allele. The IgMb and IgDb fraction did not change after cGvH (Fig. S2 A, available at http://www.jem.org/cgi/content/full/jem.20060075/DC1). From the correlation of allotype expression and auto-Ab production, we conclude that 56R H chain is mainly associated with the cGvH-induced auto-Abs.
mAbs isolated after cGvH
To test whether κ-edited 56R Abs are responsible for the different autoreactivities found in the serum (Fig. 1), we studied mAbs secreted by the hybridomas in two independent fusion panels generated 4 and 10 wk after cGvH. We tested the hybridomas for several characteristics: binding to multiple self-antigens, the H chain isotype, and the type of L chains.
Specificity and isotype.
In the 4-wk fusion, 53 out of 78 hybridomas secreted IgM or IgG (Table I. A and Table S1, which is available at http://www.jem.org/cgi/content/full/jem.20060075/DC1). Similar results were obtained in the 10-wk fusion, as 42 out of 139 hybridomas secreted IgM (Table S2). Both fusions yielded a large number of clones that were IgM−IgG−. These “nonsecretors” were not analyzed further. Supernatants from both fusions were tested for binding to dsDNA, PS, and MBP. Most of the mAbs in the 4-wk fusion bound dsDNA as well as PS. In addition, these mAbs bound MBP (Table I A). Polyreactive mAbs that bound to dsDNA, PS, and MBP were also found in the 10-wk fusion (see below; Table II).
Table I.
B cell hybridomas generated from 56R/B6 splenocytes 4 wk after cGvH reaction
| A | |||||
|---|---|---|---|---|---|
| Total clones (78) |
56R tg+
(33) |
||||
| IgM+ (29) | 24 | ||||
| Reactivity to | |||||
| dsDNA | 23 | ||||
| PS | 18 | ||||
| MBP | 21 | ||||
| IgG+ (24) | 9 | ||||
| Reactivity to | |||||
| dsDNA | 3 | ||||
| PS | 2 | ||||
| MBP | 3 | ||||
| B | |||||
| Total clones (36) |
Vκ12-13 (3) |
Vκ20 (10) |
Vκ21D (4) |
Vκ38C (8) |
Vκ(S) (17) |
| 56R tg+ (25) | 1(J2)a 2(J5)a | 3(J2)a 5(J4)a | 4(J2)a | 1(J2) 1(J4) 4(J5)a | 9 |
| 56R tg− b (11) | 2(J5)a | 1(J2)a 1(J4) | 8 |
56R+ clones from fusion 4 wk after cGvH (Bm-12 into 56R/B6) are polyreactive (A). κ L chain usage in 56R+ and 56R− clones from fusion 4 wk after cGvH (Bm-12 into 56R/B6). Only clones with H and L chain secretion are listed and were tested for rearrangement of κ L chains using specific primers for Vκ12-13, Vκ20, Vκ21D, and Vκ38C, and a degenerate primer Vκ(S) for other κ rearrangements (B). For more details see Table S1.
Groups that contain clones with two L chain rearrangements.
56R H chain transgenic−/CDR3JHCH+.
J, Jκ gene in rearrangement.
Table II.
Binding of MBP and PS of 56R+IgM+ B cell hybridomas with different L chains
| A | ||
|---|---|---|
| Vκ-gene | MBP/dsDNA binding | PS/dsDNA binding |
| Vκ20/J2 | 0.77 ± 0.10 (n = 7) | 5.55 ± 1.78 (n = 3) |
| Vκ20/J4 | 0.58 ± 0.05 (n = 14) | 3.62 ± 1.02 (n = 6) |
| Vκ38c/J2 | 0.61 ± 0.09 (n = 7) | 0.40 ± 0.12 (n = 3) |
| Vκ38c/J4 | 0.92 ± 0.06 (n = 7) | 0.51 ± 0.27 (n = 3) |
| Vκ38c/J5 | 0.35 ± 0.03 (n = 11) | 1.06 ± 0.24 (n = 4) |
| Vκ(S)/J1 | 1.03 ± 0.07 (n = 7) | 0.25 ± 0.02 (n = 3) |
| B | ||
| Vκ-gene | MBP/dsDNA binding | PS/dsDNA binding |
| Vκ20/J2 | 1.46 ± 0.05 (n = 3) | 14.26 ± 0.85 (n = 3) |
| Vκ20/J4 | 0.85 ± 0.08 (n = 7) | 10.68 ± 2.41 (n = 7) |
| Vκ38c/J5 | 1.36 ± 0.14 (n = 3) | 1.25 ± 0.06 (n = 3) |
56R+ IgM hybridomas are compared from two fusions. Clones were polyreactive to at least two antigens in ELISA assays testing for dsDNA, PS, and MBP. The mean of the MBP/dsDNA and PS/dsDNA was calculated from at least three clones, and ± indicates the error (SEM). The values show the binding to PS and MBP relative to DNA binding. Fusion panels are from 4 (A) and 10 wk (B) after GvH. For more details see Table S2.
H chain gene usage in polyreactive Abs.
To determine whether these polyreactive mAbs were associated with 56R, we tested whether the hybridomas have an intact 56R gene. PCR products from primers complementary to the 3H9/56R (56R) H chain leader sequence and the CDR3 of the 56R transgene (16) indicate the presence of the complete 56R transgene. In the 4-wk fusion, we found 33 clones out of 53 secretors to be 56R+ (Table I A). The remaining 20 clones did not amplify the 56R transgene by this assay and are classified as 56R−. 11 of these 20 56R− clones did amplify a product with a 56R CDR3 reverse primer and the JHCH primer. This PCR product represents 56R VH genes that have been truncated or VH replaced (13). No PCR product was detected with either of the 56R PCRs in 9 out of 20 56R− clones, and they were not analyzed further (Table S1).
Most of the clones that bound dsDNA, PS, and MBP were 56R+ (Table I A). A few 56R− clones bound to PS, dsDNA, and MBP, but their affinity for these antigens relative to the 56R+ mAbs was low (Table S1).
L chain gene usage.
We determined the types of L chains of these polyreactive mAbs. The Vκ genes from both the 56R+ group and the 11 56R− mAbs that have truncated or VH replaced the 56R gene were identified by PCR using primers specific for the editors Vκ12−13, Vκ20, Vκ21D, and Vκ38C (Table I B) (17). Other Vκ rearrangements were detected using a degenerate Vκ primer, Vκ(S) or the L5 primer. Jκ was also determined. Most clones with the 56R H chain had rearranged the L chain editors Vκ12–13, Vκ20, Vκ21D, and Vκ38C to Jκ4 or Jκ5 (15 rearrangements) and slightly less to Jκ1 or Jκ2 (10 rearrangements). Some clones in the 56R+ and 56R− group had more than one editor rearrangement (marked with an asterisk in Table I B). Editor L chains were more abundant in the 56R+ group, and Vk20 and Vk38C were rearranged more often than Vk21D as has been shown for 56R/B6 (20 and unpublished data). This is a different distribution than in 56R/BALB/c, where Vκ21D is the predominant editor (17, 20). We think this may be related to the susceptibility to autoimmunity of 56R/B6 mice (30–34).
Contribution of L chain to polyreactivity
The 56R+ mAbs had distinguishable reactivities depending on the Vκ and Vκ/Jκ combination. Vκ21D vetoed binding to dsDNA and the other antigens: PS, MBP, thyroglobulin, cytochrome C, histone, β-galactosidase, and insulin. Both Vκ20 and Vκ38C Abs were polyreactive, but their binding pattern was different (Fig. 2 A). 56R+ clones with Vκ20 or Vκ38C bound dsDNA with higher relative affinity than Vκ21D. In addition, Vκ20 and Vκ38C rearranged to J2 bound dsDNA with a higher relative affinity than the same V regions rearranged to J4 or J5 (Fig. 2 A). We studied the binding to PS, MBP, and the other proteins relative to their binding to dsDNA. We showed that binding to PS was favored over binding to dsDNA in clones with the L chain Vκ20, whereas Vκ38C L chains bound to dsDNA and PS with similar affinity. Affinity for MBP, thyroglobulin, cytochrome C, histone, and β-galactosidase was higher in Vκ38C as compared with Vκ20, and no difference was found between Vκ20 and Vκ38C for insulin (Fig. 2 B).
Figure 2.
Subclones from selected 56R+ hybridomas from the fusion 4 wk after cGvH induction were tested for binding to dsDNA, PS, and the proteins MBP, thyroglobulin, cytochrome C, histone, β-galactosidase, and insulin by ELISA. The OD405 of the binding assays was normalized based on the Ig concentration of 1 μg/ml for each supernatant. All clones were IgM+ and expressed the indicated editor L chain. The following groups were tested: Vκ20/J2: n = 2 (clones 50.1 and 50.2 from one original clone); Vκ20/J4: n = 4 (clones 71.1, 71.5, 73.2, and 73.4 from two original clones); Vκ21D/J2: n = 2 (clones 79.1 and 79.5 from one original clone); Vκ38C/J2: n = 2 (clones 160.1 and 160.5 from one original clone); Vκ38C/J4: n = 2 (clones 222.4 and 222.6 from one original clone); Vκ38C/J5: n = 3 (clones 89.4, 89.6 and 105.5 from two original clones); and Vκ(S)/J1: n = 2 (clones 206.1 and 206.5 from one original clone). All clones tested were sequenced using specific primers for Vκ20, Vκ21D, and Vκ38C and were unmutated. All tested binding specificities are shown in comparison for one representative result for each group. SEM from triple values in the ELISA assay (A). To analyze the binding of the subclones (to PS and the proteins MBP, thyroglobulin, histone, insulin, cytochrome C, and β-galactosidase) relative to dsDNA binding, dsDNA binding of each subclone was set to 1; therefore, only clones with considerable binding to dsDNA were included in this representation. Relative binding to the tested antigens is shown for clones using the indicated L chain. Numbers show p-values determined by the Anova Single Factor Test (B).
Relative binding to PS and MBP was also assessed by calculating the ratios of PS to DNA and MBP to DNA binding of the mAbs that expressed the 56R transgene from clones in both fusions. Again, PS/dsDNA binding relative to MBP/dsDNA binding was greater in clones that expressed the Vκ20 as compared with the Vκ38C L chain, whereas binding to PS/dsDNA and to MBP/dsDNA was similar in clones that expressed the Vκ38C L chain (Table II).
Plasma cell differentiation in cGvH
We searched for the origin of polyreactivity by flow cytometry. After cGvH induction, there was an increase in B220+IgG+ splenocytes expressing the plasma cell marker CD138 (2.57 and 4.05% of the B220+ cells by days 28 and 60 after cGvH, respectively). We considered these surface Ig–expressing B cells to be pre-plasma cells (Fig. 3 A). We also found an increase of B220+IgMa+ B cells expressing CD138 (0.37 and 0.75% of the B220+ cells by days 28 and 60 after cGvH, respectively), indicating that B cells with the H chain transgenic allele Igha of the 56R/B6 mouse differentiated into plasma cells, and were giving rise to anti-DNA Abs. B220+IgMa– B cells expressing CD138 (2.29 and 2.95% of the B220+ cells by days 28 and 60 after cGvH, respectively) were also increased and most likely to be of the IgG class (Fig. S2 B).
Figure 3.
cGvH induces differentiation of plasma cells and the redistribution of MZ and follicular B cells in the spleen of 56R/B6 mice. The plasma cell marker CD138 was increased on a small population of B220+ splenocytes in the IgG+ B cell population 28 and 60 d after cGvH. 56R/B6 mice were injected with Bm-12 CD4+ T cells (cGvH mice) or injected with B6 CD4+ T cells (control mice). Results are representative of four experiments (A). The percentage of MZ B cells (CD21high and CD23low) was decreased after cGvH, and the percentages of CD21−CD23− B cells and the follicular B cells (CD21intCD23high) were increased (dot plots). CD21−CD23− B cells show an increase of CD138 by days 14 and 60 after cGvH (black line, histograms) as compared with control recipients (gray line, histograms). One representative experiment out of six is shown (B). B cells expressing κ and λ L chain were found among B220+ B cells in the spleen. The decreasing percentage of κ/λ double positive B cells may be due to the increase in total lymphocyte numbers in cGvH. One experiment out of three is shown (C).
Potentially pathogenic auto-Abs are thought to home to the MZ (35, 36); therefore, we tested whether MZ B cells were affected by cGvH in the 56R/B6. This is relevant because it has been shown that 56R/BALB/c has an enlarged MZ B cell compartment. Here we show that in the 56R/B6, the MZ B cell population (CD21high and CD23low) comprised as much as 50% of the B220+ splenocytes. The percentage of the MZ B cell population decreased after cGvH, whereas the CD21−CD23− B cell population increased. The percentage of follicular B cells (CD21intCD23high) slightly increased (Fig. 3 B, dot plots). The majority of the CD138+ B cells was found in a subpopulation of CD21−CD23− B cells (Fig. 3 B, histograms). Yet, during cGvH, the number of lymphocytes in the spleen increased on average 2.5-fold and the spleen weight increased 3.2-fold (not depicted); therefore, we cannot conclude that the absolute numbers of MZ B cells in cGvH were decreasing. However, we observed a decrease of CD21 expression by the mean fluorescence intensity on MZ B cells (not depicted). This suggests that MZ B cells were activated during cGvH. The concommitant increase of plasma cell differentiation and the decrease of CD21 expression in MZ B cells suggest that potentially autoreactive B cells from the MZ were affected by the cGvH reaction and may have differentiated to become auto-Ab–producing B cells.
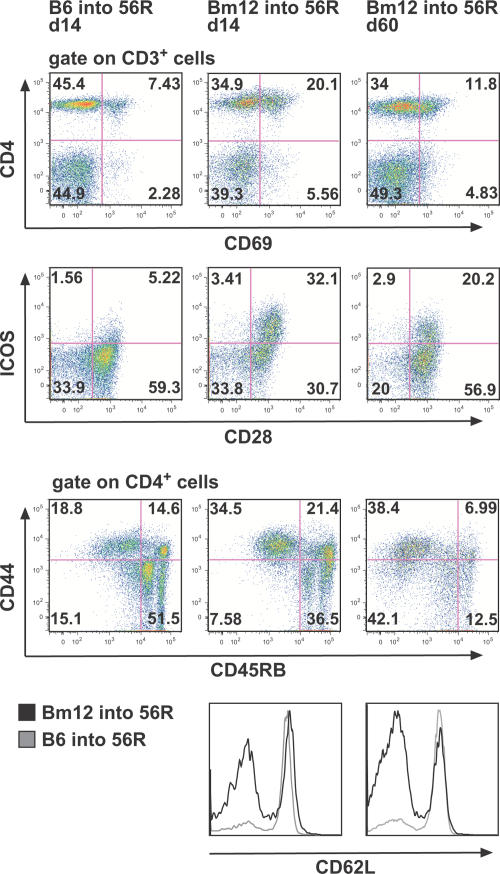
T cells in the spleen of 56R/B6 recipients were also activated as seen by the down-regulation of CD3 (not depicted) and CD4 as well as by the increased expression of CD69 (tested at day 14; Fig. 4). The expression of the T cell costimulatory molecules CD28 and ICOS also increased, and CD4+ T cells displayed a memory phenotype as shown by the expression of CD44high, CD45RBlow, and CD62Llow (Fig. 4). We could not distinguish between donor and recipient CD4+ T cells in these studies, but the activation of T cells as late as day 60, by which time we would not expect Bm-12 donor T cells, indicates a role of endogenous T cells in antigen activation. We conclude from these data that cGvH induced by purified Bm-12 CD4+ T cells leads to plasma cell differentiation (Fig. 3) and a sustained activation profile of T cells in the 56R/B6 recipient (Fig. 4). The role of endogenous T cells in the cGvH has been established by studies showing that CD4 knockout mice are unresponsive to cGvH (37).
Figure 4.
cGvH induces long-lasting activation of T cells in splenocytes of 56R mice. Expression of CD4, CD69, and the T cell costimulatory molecules CD28 and ICOS is shown on CD3+ T cells, and the expression of CD44 and CD45RB is shown on CD4+ T cells (dot plots). The levels of CD62L on CD4+ T cells (histograms) were decreased after cGvH (black line) as compared to the control (gray line) at days 14 and 60. One representative experiment out of five is shown.
Expression of λ L chain in cGvH
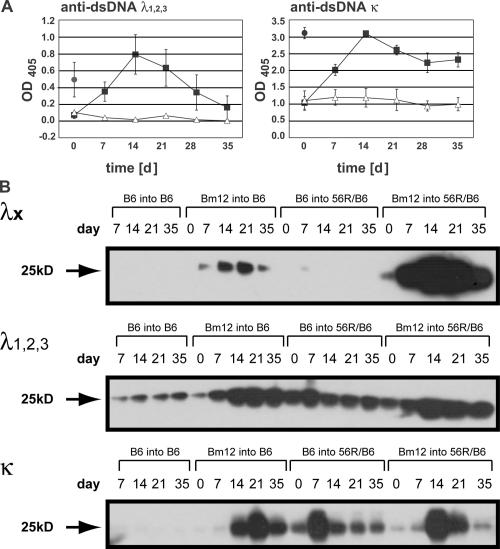
We tested anti-DNA Abs for the usage of κ and λ L chain in the serum after cGvH and developed our anti-dsDNA assay with anti-λ and anti-κ reagents. We found λ- and κ-containing anti-dsDNA Abs (Fig. 5 A). The strong induction of λ anti-dsDNA Abs led us to ask whether these Abs come from isotypically included B cells that express λ1 and κ L chain (18). We also asked whether these isotypically included B cells are activated by cGvH and differentiate into anti-dsDNA–producing plasma cells. This is relevant because it has been shown that MZ B cells in the 56R/BALB/c mouse can coexpress a κ and a λ L chain. The λ1 L chain with 56R H chain binds DNA (38), but the κ L chain is an editor (17). These isotypically included B cells are thought to be sequestered in the MZ (23, 24). We analyzed the composition of B cells in the MZ of a 56R/B6 mouse using a single cell PCR technique and tested for B cells with both λ1 and κ L chain (Table III, A and B). L chain rearrangements were identified using nested PCRs on cDNA from single cells. Specific primers were used for Vλ1/2 and Cλ1/2, Vκ21D and Cκ, and Vκ38C and Cκ (20). Other κ L chains were amplified using the degenerate primer Vκ(S) (39). We found that all B cells with λ L chain also had a κ L chain rearrangement. Sequence analysis showed that B cells with in-frame λ1 L chain had rearranged the editor L chain Vk21D and Vk38C. Two examples with in-frame λ1 L chain had, in addition to the κ editor, another κ rearrangement. Other λ L chain PCR products had large deletions in the V or V/J regions. Rearrangement of λ L chain in-frame or out of frame was always accompanied by an in-frame κ L chain that was often the editor Vκ38C or Vκ20 (Table III B). Thus, the MZ contains a population of B cells that seems to have rearranged excessively and introduced λ deletions.
Figure 5.
λ and κ L chain usage in anti-dsDNA Abs in 56R/B6 after cGvH. For induction of cGvH, Bm-12 CD4+ T cells were injected into 56R/B6 (▪). As a control, B6 CD4+ T cells were injected into 56R/B6 (▵). Anti-DNA Abs were detected by ELISA with an anti-λ (n = 6 for cGvH group and n = 3 for control group) and anti-κ (n = 4 for cGvH group and n = 3 for control group) detection Ab by ELISA at a serum dilution of 1:180. Results represent means ± SEM. MRL/lpr sera were tested for comparison (•; n = 4) (A). Detection of L chain expression during cGvH in serum was analyzed by Western blot. Sera from mice after cGvH (Bm-12 CD4+ T cells injected into B6 or 56R/B6 recipients) or sera from control mice (B6 CD4+ T cells were injected into B6 or 56R/B6 recipients) were run under reducing conditions. λx was detected using a polyclonal anti-Vλx Ab (reference 22). Serum samples were diluted at 1:100 for λ and at 1:2,000 for κ L chain detection. The expression of λx after exposure for 5 min, λ1,2,3 after exposure for 20 min, and κ after exposure for 30 s is shown. Results are representative of seven experiments. λx, λ1,2,3 and κ in the serum are shown from one mouse (B).
Table III.
Single cell analysis of B cells from the MZ of a 56R/B6 mouse
| A | |||||
|---|---|---|---|---|---|
| Single cells (179) | 56R tg+ (124) | Vκ(S) (124) | Vκ38C (100) | Vκ21D | Vλ (26) |
| Plate I | 67 | 80 | 30 | 11 | 15 |
| Plate II | 57 | 44 | 70 | n.d. | 11 |
| 56R tg+ (124) | Vκ (S) (87) | Vκ38C (64) | Vκ21D | Vλ (17) | |
| Plate I | 58 | 29 | 10 | 9 | |
| Plate II | 29 | 35 | n.d. | 8 |
| B | |||||
|---|---|---|---|---|---|
| Cell no. | 56R | Vκ(S)/Cκ | Vκ38C/Cκ | Vκ21D/Cκ | Vλ1/Cλ |
| Plate I | |||||
| 10 | + | +a | |||
| 18 | +a | ||||
| 20 | + (Vκ21-4 (98%)/J2 (100%) IF) | +b (IF) | |||
| 22 | + | + | +a | ||
| 40 | + | + | |||
| 43 | + | + | + | ||
| 53 | + | + (Vκkk4 (91%)/J5 (95%) IF) | + (Vκ38C/J5 IF) | + (Vλ1/Jλ1/Cλ1 IF) | |
| 59 | + | + | +a | ||
| 62 | + | + | + (Vκ38C/J4 IF) | + (Vλ1/Jλ3/Cλ2 OF) | |
| 63 | + | + (Vκ21-4 (98%)/J2 (100%) IF) | + (Vκ38C/J5 IF) | + (Vλ1/Jλ1/Cλ1 OF) | |
| 74 | + | + (Vλ2/Jλ2/Cλ2 OF) | |||
| 75 | + (Vκ21-4 (98%)/J1 (100%) IF) | + | + (n.d.) | ||
| 76 | + | + | +a | ||
| 95 | + | + | + (Vλ1/Jλ1/Cλ1 OF) | ||
| Plate II | |||||
| 13 | + (Vκ21-4 (98%)/J2 (100%) IF) | n.d. | + (Vλ1/Jλ1/Cλ1 IF) | ||
| 14 | + | + (Vκ19-23 (98%)/J5 (100%) IF) | n.d. | + (Vλ1/Jλ1/Cλ1 OF) | |
| 29 | + | + (Vκ21-4%) (98)/J2 (97%) IF) | n.d. | + (Vλ1/Jλ3/Cλ? OF) | |
| 33 | + | + (Vκ19-15 (92%)/J5 (100%) OF) | + (Vκ38C/J4 IF) | n.d. | + (Vλ1/Jλ3/Cλ2 OF) |
| 34 | + | + (Vκ38C/J4 IF) | n.d. | + | |
| 49 | + | + (Vκ38C/J5 IF) | n.d. | + (Vλ1/Jλ1/Cλ1 OF) | |
| 52 | + | + (Vκ19-23 (98%)/J2 (100%) IF) | n.d. | + (Vλ1/Jλ1/Cλ1 OF) | |
| 59 | + | + (Vκ21-4 (98%)/J2 (96%) IF) | + (Vκ38C/J4 IF) | n.d. | +a (Vλ1) |
| 60 | + (Vκkk4 (99%)/J5 (100%) IF) | + (Vκ38C/J4 IF) | n.d. | + (Vλ1/Jλ1/Cλ1 IF) | |
| 71 | + | + (Vκ19-23 (97%)/J2 (100%) IF) | + (Vκ38C/J4 IF) | n.d. | + |
Single cells from the MZ (B220+CD21high and CD23low) in a 56R/B6 mouse were analyzed by PCR for Igκ, Igλ, and 56R H chain. All PCR product-positive cells were included in the table (A). B cells contained Igλ+ cells. B cells with in-frame λ L chain show message for one or two in-frame κ L chain(s) (in bold) (B). For further details see Table S3. Numbers in parentheses indicate sequence similarity to the germline gene in percentages.
n.d., not done.
IF, in frame.
OF, out of frame.
%, sequence similarity in percent, determined for κ L chains amplified with the degenerate primer V κ (S).
Deleterious events in V and J.
Deleterious events in V.
We tested whether κ/λ1 56R B cells were activated by cGvH by analysis of a fusion panel 10 wk after cGvH. We found isotypically “included” B cells that coexpressed a λ1 and a κ L chain. 5 out of 139 clones were λ1, and 4 of these expressed the 56R H chain. One λ1 clone did not express the 56R H chain and was negative for IgM and IgG. All of the 56R/λ1-expressing clones coexpressed a κ L chain by ELISA, and in three of those examples, we found rearrangement of λx by PCR. In addition, we found one clone that expressed the 56R H chain with λ2. All clones that expressed the 56R H chain with λ1/κ or λ2 L chain bound to dsDNA, PS, and MBP. None of the clones expressed only λ1, indicating that B cells that express the 56R H chain with λ1 alone are eliminated from the repertoire (Table IV). Thus, partially edited B cells coexpressing λ and κ L chain were activated in cGvH and could be fused. We also found κ/λ-expressing B cells from cGvH-induced mice by flow cytometry (Fig. 3 C); however, the number of isotypically included B cells was higher as found by single cell PCR analysis (Table III). We are considering that this phenomenon is due to the slight cross-reactivity of anti-λ and anti-κ Abs used in flow cytometry analysis (Fig. 3 C).
Table IV.
κ/λ L chain double-expressing clones from fusion 10 wk after cGvH (Bm-12 into 56R/B6)
| Clone no. | 56R PCR | Vκ(S)/J5 | λ1 PCR | λ2 PCR | λx PCR | IgM ELISA | κ ELISA | λ1,2,3 ELISA | dsDNA ELISA [OD] |
MBP ELISA [OD] |
PS ELISA [OD] |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 30 | + | J5 | + | + | + | + | + | 1.284 | 1.919 | 0.274 | |
| 35 | + | J5 | + | + | + | + | + | 1.669 | 2.154 | 0.536 | |
| 41 | + | J5 | + | + | + | + | 1.741 | 2.006 | 0.768 | ||
| 124 | + | J5 | + | + | + | + | + | 1.361 | 1.846 | 0.842 | |
| 57 | + | + | + | + | 0.979 | 0.36 | 0.904 | ||||
| 59 | - | + | + | + | n.d. | n.d. | n.d. |
n.d., not done.
The editor L chain, λx, is also induced by cGvH. λx is known to bind MBP both as free L chain and in association with H chains (40, 41), including 56R (unpublished data). We tested the possibility that cGvH might induce λx-associated Abs by a Western blot of sera developed with a λx antiserum (Fig. 5 B). We were unable to detect λx in the serum of 56R/B6 mice or in nontransgenic B6 mice, but after initiation of the cGvH, we found that λx levels in both 56R/B6 and nontransgenic B6 mice rose rapidly and were maintained until day 60 (Fig. 5 B; not depicted for day 60). Expression of λx after cGvH was significantly higher in the 56R transgenic than in the nontransgenic B6, suggesting that this L chain is associated with the 56R H chain. Consistent with the ELISA data (Fig. 5 A), we also found increased levels of λ and κ; however, in 56R/B6, expression of those L chains did not increase to the same extent as λx (Fig. 5 B).
DISCUSSION
The 56R H chain transgene enhances the anti-DNA production in cGvH (14, 15) and in the quasi-autoimmune B6 (30–34 and unpublished data). This is unexpected because Abs with the 56R H chain in BALB/c are edited. DNA binding is either vetoed by L chains, such as Vκ21D (17) and λx (21, 22), or DNA binding on the cell surface is diluted because more than one receptor is expressed (17, 23, 24). In this study, we show that editing can also be incomplete (Fig. 2). Editors were originally defined in BALB/c transgenics, a strain that does not convert to autoimmunity even with an anti-DNA transgene. Most of the B cells from this mouse express the 56R H chain with Vκ21D, an L chain that vetoes DNA binding completely (17). In the B6 mouse, the L chain repertoire is reversed, very few 56R/Vκ21D combinations are found, and the majority of B cells express 56R H chain with incomplete editors, such as Vκ20 and Vκ38C (reference 20, unpublished data, and Table III). We think that the difference lies in the tolerance threshold of the strain: BALB/c, the prototypic “healthy” strain, tolerates only completely edited Abs, whereas B6, a lupus-susceptible strain (30–34), is more permissive. We show that the cGvH-induced autoimmunity in 56R/B6 mice activates B cells using the incomplete editors Vκ20 and Vκ38C (Fig. 2 and Table I B). Thus, incomplete editing contributes to autoimmunity by determining an available repertoire that includes autoreactive B cells. In support of this idea is the finding that 56R/B6 and the congenic 56R/Sle2/B6 (expressing the additional lupus susceptibility gene Sle2) spontaneously produce anti-DNAs that include 56R/Vκ20 Abs (unpublished data). Evidence for such a “lupus repertoire” is also found in lupus-affected individuals (3, 4).
Incomplete editing is inevitable. The ability of certain L chains to edit is correlated with certain structural features of these L chains, in particular, a high frequency of Asp residues in or near the CDRs (17). Hence, we think editing may be the result of an interaction between the VH Arg residues and the VL Asp residues to interfere with the VH Arg interaction with DNA (38, 42, 43). However, editor Asps may not neutralize every Arg and might account for residual anti-DNA activity (44). Similarly, not all Asps need to interact with Args and might be available for interactions with basic proteins such as MBP, as in the 56R/λx.
The incompletely edited Abs, such as 56R/Vκ20, retain activity for DNA and cardiolipin. This is thought to be a cross-reaction based on binding to a phosphate-containing moiety shared by both (45). But a novel source of polyreactivity comes from binding activities that some editors confer upon an anti-DNA VH. In this case, polyreactivity is not because of cross-reactivity but because of alternate binding sites. A relevant example has been described in which the 3H9 H chain gives different patterns of antinuclear antigens depending on the L chain with which it is associated (46). We conclude from these results that editor L chains are correlated with anti-self activity. J gene usage also seemed to play a role in binding some proteins, namely MBP, thyroglobulin, cytochrome C, and histone, as shown for its relevance for Vκ20 and Vκ38C in binding to dsDNA (Fig. 2, A and B). Although polyreactivity can come about by the creation of different combining sites, it can also be due to enhanced cross-reactivity, as in the case of Vκ20.
Receptor editing acts in B cells with autoreactive receptors; hence, rearrangement persists and can produce isotypically included B cells as we have described in the 56R/BALB/c transgenic mouse. One of the two receptors (56R/λ1) is anti-DNA (38), and the other is a 56R/κ editor (24). Such isotypically included κ/λ double-expressing B cells in 56R/ BALB/c are found in the MZ of the spleen (24) and in hybridomas from LPS-activated fusions. Fusion panels generated after cGvH include clones that coexpressed λ1 and κ L chain and secrete anti-DNA Abs. In this study, none of the clones (Table IV) expressed λ1 L chain only. Therefore, we think the κ/λ B cell may represent a state of tolerance. In this sense, a B cell may have a self-reactive receptor and expression of two L chains may alter or reduce the affinity of the B cell to an antigen. Other examples of dual expression come from recent studies in 56R mice in which the κ locus has been knocked out. Hybridoma panels from these mice showed that the vast majority of the 56R/λ1 B cells coexpressed λx L chain, an efficient editor, and only very few have rearranged λ1 alone (47 and unpublished data), but these 56R/λ1 clones bound DNA even when λx L chain was coexpressed. cGvH appeared to activate these κ/λ double-expressing B cells because we find anti-DNA Abs with λ1 in the serum in cGvH (Fig. 5 A). By single cell analysis we tested whether these κ/λ B cells may be in the MZ of 56R/B6 mice and confirmed the existence of κ/λ-included B cells (Table III). Single cell analysis of MZ B cells also revealed interesting properties that may act on the λ locus. All λ L chain rearrangements, whether out-of-frame or in-frame, were accompanied by a κ L chain, which was (except in one case) in-frame (Table III).
As discussed above, we think that DNA and/or MBP binding can result from incomplete editing. For example, the Asp-rich λx editor in association with H chains including 56R binds MBP (40, 41, and unpublished data). The acidic property of λx to which we attribute the MBP activity is also a feature of κ editors, and these L chains might also confer MBP activity to 56R. And, as we show here, 56R with Vκ38C and Vκ20 also bound MBP as well as DNA and PS (Fig. 2, A and B) (28). Therefore, an incompletely edited Ab may have both Arg(s) and Asp(s) available for binding both to DNA and proteins.
During the cGvH reaction there was a change in the percentages of MZ B cells, follicular B cells, and CD21−CD23− B cells (Fig. 3 B). MZ B cells may have activated by alloreactive Bm-12 T cells, which in turn may have activated endogenous T cells of the 56R/B6 recipient. These T cells could give help to the differentiation of plasma cells. This idea is consistent with recent findings that showed that MZ B cells, not follicular B cells, are potent activators of naive CD4+ T cells. MZ B cells migrate to T cell areas and differentiate upon interaction with T cells into plasma cells (48). This could be the case in the cGvH reaction, and plasma cells found in the CD21−CD23− population might be a source of auto-Abs to DNA (Fig. 3 B, histograms). Support for this idea comes from features of 56R/Sle2/B6. Sle2 further increases the tendency to autoimmunity of 56R/B6, and in 56R/Sle2/B6, the MZ and the CD21−CD23− B cell populations are increased as compared with 56R/B6. These CD21−CD23− B cells secreted auto-Abs of the IgMa allotype, the transgene-encoded allele, in culture (unpublished data). We also find Abs reactive to DNA expressing the H chain of the Iga allotype (Table I). Assuming that both Sle2 and cGvH enhance autoimmunity, we think that the increase of the CD21−CD23− cells in 56R/Sle2/B6 is analogous to the increase of CD21−CD23− B cells in cGvH-induced autoimmunity. The question remains whether partially edited κ and λ double positive B cells are able to move out of MZ to become plasma cells.
Conclusions
Autoreactive and polyreactive Abs.
Here we present a detailed description of the auto-Abs induced by cGvH. The study reveals the structural basis for autoreactivity of certain Abs and the origin of these auto-Abs.
Origin.
Natural auto-Abs are found in abundance in both healthy and autoimmune individuals, and there is a vast and confusing literature on their origin, rationale, and implications for autoimmunity and immune protection (49). Our study answers these questions by showing that at least some if not all “natural” auto-Abs result from receptor editing. Of course, the abundance of these autoAbs in our model is in part because of the anti-DNA transgene. This produces an immature B cell population, most of which must be edited to survive. But even nontransgenics must have a substantial repertoire that is shaped by editing. Based on the mechanisms that generate the immature VH repertoire, we estimate that at least 50% of this repertoire is anti-DNA (50, 51). Thus, editing of this population will have a profound influence on the frequency of natural auto-Abs in the peripheral B cell repertoire.
Relevance.
A question raised by natural auto-Abs is how to explain their presence in a self-tolerant individual. This dilemma is usually explained by invoking affinity differences; i.e., natural auto-Abs are low affinity anti-self and are ignored by tolerance mechanisms (52). But the question remains: Why is there a large abundance of these auto-Abs? The only explanation is positive selection (53) of this population, and several roles for natural auto-Abs have been proposed. These roles range from housekeeping Abs to stimulators of the repertoire to anti-virus Abs (49). If indeed natural auto-Abs are important in normal immunity, their existence in the repertoire can be understood. It has been shown that the B cell repertoire that is induced by the cGvH is not random but a selective process and requires B cell receptor cross-linking (12) and is supported by the involvement of endogenous T cells (as seen in this study [Fig. 4] and by others [37]). In addition, certain edited auto-Abs appear to be selected as suggested by differences in the repertoire of editors between strains; for example, the distribution of editors in B6 as compared with BALB/c.
Implications.
We show here that the peripheral repertoire is shaped by receptor editing and selection. Negative selection can continue to act on edited B cells if they retain self-reactivity after the initial editing event. In this sense, a healthy BALB/c mouse may only tolerate perfectly edited, nonautoreactive B cells. Other strains may be more permissive and allow incompletely edited B cells to enter the periphery. The ability to regulate this population may determine lupus susceptibility by supplying the peripheral B cell population with the precursors to pathogenic auto-Abs. That these B cells are involved in the onset of disease is demonstrated by these findings.
MATERIALS AND METHODS
Mice.
The generation of site-directed 3H9/56R knock-in mice has been described previously (13, 17). The 3H9/56R was backcrossed onto the C57/BL6 background for 14 generations to engender 3H9/56R/B6. The 3H9/56R transgene was determined by PCR amplification of tail DNA (16, 17). B6 and coisogenic Bm-12 (B6.C-H2bm12/KhEg) mice were obtained from The Jackson Laboratory. All mice were maintained in our mouse colony at the University of Chicago. All animal care and procedures were conducted in accordance with the Animal Welfare Act.
cGvH.
Mice were 2–8 mo old at time of cGvH initiation. All cGvH experiments for flow cytometry, serum data, and the fusion experiment at 10 wk after cGvH were performed with 107 Bm-12 (for cGvH) or B6 (for controls) CD4+ T cells. For the fusion at 4 wk, cGvH was performed as described previously (9). In brief, recipient mice were injected i.p. with Bm-12 (for cGvH) or B6 (for controls) splenocytes prepared using standard techniques, or with 107 CD4+ T cells (purity >96%) purified using the CD4 (L3T4) MicroBead MACS system (Miltenyi Biotec). Experimental mice were bled before onset of cGvH and weekly thereafter.
Flow cytometry.
Before staining, cells were preincubated with excess mouse anti–Fcγ III/II R (2.4G2; BD Biosciences) to block Fc receptors. Dead cells were excluded by propidium iodide staining (0.33 μg/ml). The following mAbs were used for staining: RA3-6B2-APC (anti-B220), 145-2C11-FITC (anti-CD3), 145-2C11-APC (anti-CD3), 30-F1-biotin (anti-CD24), L3T4-PE (anti-CD4), Jo2-PE (anti-CD95), H1.2F3-FITC (anti-CD69), AF6-78-PE (anti-IgM allotype b), AMS9.1-FITC (anti-IgD allotype a), 217-170-biotin (anti-IgD allotype b), R26-46-FITC (anti-λ1,2,3), H139.52.1-PE (anti-κ), 7G6-FITC (anti-CD21), B3B4-PE (anti-CD23), 281-2-PE (anti-CD138), and 281-2-biotin (anti-CD138, used in Fig. 3 B only; all from BD Biosciences); GL1-FITC (anti-CD86), M5/114.15.2-FITC (anti–MHC II), 7E.17G9-PE (anti-ICOS), 37.51-APC (anti-CD28), IM7-APC (anti-CD44), C363.16A-PE (anti-CD45RB), and MEL-14-FITC (anti-CD62L; all from eBioscience); and RS3.1-Alexa 488 (anti-IgM allotype a, own hybridoma, conjugated to Alexa 488; Invitrogen) and anti–IgM-FITC (SouthernBiotech). Detection of biotinylated Abs was done using streptavidin-FITC, streptavidin-PE, or streptavidin-PE-Cy5 (all from BD Biosciences). Data were generated on a FACSCanto (BD Biosciences) and analyzed using FlowJo software.
Hybridomas.
3H9/56R/B6 mice were injected with Bm-12 splenocytes (fusion 4 wk after injection as described previously [14, 15]) or with Bm-12 CD4+ T cells (fusion 10 wk after injection). Hybridomas were generated in the Monoclonal Antibody Facility of the University of Chicago as described previously (21). In brief, spleen cells from cGvH mice were fused without further manipulation to Sp2/0 myeloma cells. Hybridomas were plated at limiting dilution, wells bearing single colonies on 96-well plates were expanded, and hybridomas were grown 90% confluent in six-well plates for ELISA assays.
Antigens.
dsDNA (Vector Laboratories) was biotinylated and diluted 1 μg/ml PBS. PS was dissolved in chloroform (90%) and methanol (10%; 10 μg/ml). Human MBP from postmortal brain (RDI), total histone (Roche), β-galactosidase from Escherichia coli, cyochrome C from bovine heart, thyroglobulin from bovine thyroid, and human insulin (all Sigma-Aldrich) were dissolved in 10 μg/ml PBS.
Auto-Abs in serum.
Unless otherwise indicated, sera were diluted at 1:180 in the appropriate blocking buffer of the ELISA assay. Immulon 2 HBX was used for PS, and Immulon 4 HBX ELISA plates were used for all other ELISA assays (Thermo-Lab Systems).
DsDNA.
Anti-dsDNA Abs were detected as described previously (19). Plates were coated with 10 μg avidin D (Vector Laboratories) in PBS at 4°C, blocked in PBS with 1% BSA at 37°C for 2 h, and washed with PBS with 0.05% Tween (PBS-T). Biotinylated dsDNA was bound to avidin-coated plates at 37°C for 1.5 h. Sera were applied, plates were washed three times, and DNA serum complexes were detected with anti–mouse IgM-Ap, IgG-AP, κ-AP, or λ-AP (all from SouthernBiotech). After absorption for 60 min, plates were washed and the retaining anti-dsDNA IgM, IgG, κ, or λ Abs were quantified using AP substrate (Sigma-Aldrich). The OD was determined at 450 nm.
PS.
Anti-PS Abs were detected as described previously (45). Plates were coated with PS (Sigma-Aldrich) in 100% EtOH, dried, and blocked in 0.1% gelatin/1% BSA at 37°C for 2 h. Plates were washed with PBS. Sera were applied, plates were washed, and PS serum complexes were detected as described for dsDNA binding.
MBP.
Anti-MBP Abs were detected as described previously (40). Plates were coated with 10 μg human MBP in PBS at 4°C overnight and blocked in 0.5% casein at 37°C for 2 h. Plates were washed with PBS-T. Diluted sera were applied, plates were washed, and MBP serum complexes were detected as described for dsDNA binding.
Auto-Abs in hybridoma supernatants.
Supernatants were tested for dsDNA and PS binding as described above for serum. Blocking buffer for MBP and all other protein binding assays contained 8% casein in PBS. Supernatants from fusions were tested for binding to dsDNA, PS, and to proteins MBP, thyroglobuline, cytochrome C, histone, β-galactosidase, and insulin. The OD405 of the binding assays was normalized based on the Ig concentration of each supernatant.
Isotype and Ig concentration in hybridoma supernatants.
Isotype and concentration in culture supernatants were determined as described previously (47). In brief, plates were coated with unconjugated anti–mouse Ig, supernatants from hybridomas were added, and binding was detected with goat anti–IgM-AP or anti–IgG-AP and developed with AP substrate. The Ig concentration was determined by comparing samples to a standard curve generated by an isotype-matched Ab (Sigma-Aldrich).
PCRs on hybridoma DNA.
Genomic DNA was purified from individual hybrids. 150 pg DNA was used in each reaction. Primers and conditions for H and L chain PCR assays have been detailed previously (13, 47, 54). The presence of 3H9/56R H chain transgene was identified by PCR using primers complementary to the 3H9 H chain leader exon and the CDR3 sequence (16), or with a 56R CDR3 reverse primer (the complement of the 56R/CDR3 primer described above) and the JHCH primer. The JHCH primer binds to a site between JH4 and CH (55). This primer amplifies truncated or replaced 3H9 VH genes because both occur at the embedded heptamer 5′ of CDR3.
For typing of κ L chain rearrangements, Vκ-specific forward primers (Vκ12-13, Vκ20, Vκ21D, and Vκ38C) and Vκ(S) (39) or L5 (56) forward primers were used under conditions described previously (17). The Vκ(S) PCR primer should amplify 70% and the L5 primer should amplify ∼60% of Vκ genes. Using Jκ2 or Jκ5 reverse Jκ primers (57), the size of the PCR product corresponds to the Jκ segment participating in the rearrangement event as described previously (54). To determine editor rearrangements, PCRs were performed in 1× buffer II (PerkinElmer) with a final concentration of 200 μM of each dNTP (Boehringer), 50 pmol of each primer, 1.5 mM MgCl2, and 1 U of AmpliTaq Gold (PerkinElmer). Selected PCR products were sequenced using primers for Vκ20, Vκ21D, and Vκ38C as indicated in Results.
Single cell sorting, cDNA synthesis, and PCR.
Single cell PCR was performed as described previously (3). MZ B cells B220+CD21intCD23high from one 56R/B6 mouse were sorted on a FACSVantage (Becton Dickinson) into 96-well PCR plates containing 4 μl lysis solution (0.5× PBS containing 10 mM dithiothreitol, 8 U RNAsin [Promega], and 0.4 U 5′-3′ Prime RNase Inhibitor [Eppendorf]) and were immediately frozen on dry ice. All samples were stored at −70°C. The cDNA was synthesized in a total volume of 14 μl in the original 96-well PCR plate. RNA from single cells was reverse transcribed at 37°C for 55 min with 150 ng random hexamer primer (pd(N)6; GE Healthcare), 0.5 μl dNTP mix (10 mM each), 1 μl of 0.1 M dithiothreitol, 0.5% (vol/vol) Nonidet P-40, RNase inhibitors (4 U RNasin and 6 U Prime RNase inhibitor), and 50 U Superscript III reverse transcriptase (Invitrogen). Ig gene rearrangement was tested using the degenerate primer Vκ(S) (39) with nested Cκ reverse primers, specific nested primers for Vκ21D and Vκ38C with nested Cκ reverse primers (for primers see reference 20), primers for the 56R H chain transgene (20), and primers for Vλ (designed as follows: Vλ1/2 ext, 5′caggctgttgtgactcaggaatct3′; Vλ1/2int, 5′cagtcacactcacttgtcgctcaa3′; Cλext, 5′gcacgggacaaactcttctccaca3′; Cλint, 5′gagctcttcagaggaaggtggaaa-3′). Products were amplified by nested PCR (for Vκ(S)Cκ “semi-nested PCR”) in 40-μl reactions containing 20 pM primers and 1.2 U HotStar Taq DNA polymerase (QIAGEN). The PCR conditions were as follows: Denaturation for 4 min at 94°C, followed by 30 s at 94°C and 30 s at 55°C (for Vκ21D, Vκ38C, and 56R H chain), or at 50°C (for Vκ(S) and Vλ1/2) and 55 s (first PCR) or 45 s (nested PCR) at 72°C for 50 cycles. Elongation was allowed for 10 min at 72°C. As a template, 2 μl cDNA was used for the first PCR and 3 μl of the first PCR product was used for the nested PCR. PCR products were sequenced using primers for Vκ20, Vκ21D, Vκ38C, Vκ(S) and Cκ, and Vλ1/2 and Cλ as indicated in Results.
Western blot.
Serum samples were diluted to 1:100 for detection of λ and 1:2,000 for detection of κ L chain in Tris- and SDS-containing buffer. Proteins were denatured with β-ME and boiled for 5 min. Proteins were resolved on a 12% SDS page and transferred to a PVDF membrane (Bio-Rad Laboratories) at 100 mV, 500 mA. After blocking for 2 h with 4% milk in PBS-T, the blot was probed at 4°C overnight with a rabbit anti–mouse Vλx polyclonal Ab (22) or with a horseradish peroxidase–labeled goat anti–mouse Vκ or Vλ1,2,3 polyclonal Abs (both SouthernBiotech) in 1% milk in PBS-T. Membranes were washed three times at room temperature with PBS-T. For λx, membrane was incubated with horseradish peroxidase–conjugated goat anti–rabbit IgG (Bio-Rad Laboratories) for 2 h in 1% milk in PBS-T. Membranes for λx, λ1, and k were developed using ECL reagent.
Statistics.
Statistical significance was determined using Anova Single Factor Test for analysis of ELISAs. Anova Single Factor Test is appropriate for multiple statistical comparisons on a single dataset. Numbers in figures give the actual p-value determined by this test (Fig. 2 B). For all other ELISA assays on single clones, the SEM was determined for triple repeats (Fig. 2 A) and on serum from the indicated number of mice in each group (Figs. 1 and 5 A).
Online supplemental material.
Online supplemental material gives greater details about fusion 4 wk after cGvH, fusion 10 wk after cGvH, and single cell analysis of MZ B cells isolated from a 56R/B6 mouse. Figs. S1 and S2 and Tables S1–S3 are available at http://www.jem.org/cgi/content/full/jem.20060075/DC1 .
Supplemental Material
Acknowledgments
We thank Azusa Tanaka for technical assistance, Natan Dotan for help with statistical analyses, the Flow Facility, the Sequencing Facility, and the Monoclonal Antibody Facility of the University of Chicago for single cell sorting, sequencing, and the generation of fusion panels. We thank Dr. Marko Radic for helpful discussions.
This work was funded by a grant from the Lupus Research foundation and by National Institutes of Health grants R21 A105 9897 (Receptor Editing in Autoimmunity and Free L Chain Production) and R01GM 20964 (Genetics and Regulation of Autoantibodies), and supported by the Gwen Knapp Center for Autoimmunity and Research.
The authors have no conflicting financial interests.
Abbreviations used: Ab, antibody; Arg, arginine; Asp, aspartate; CDR, complementarity-determining region; cGvH, chronic graft-versus-host; ds, double-stranded; MBP, myelin basic protein; MZ, marginal zone; PS, phosphatidylserine.
References
- 1.Hardin, J.A. 1986. The lupus autoantigens and the pathogenesis of systemic lupus erythematosus. Arthritis Rheum. 29:457–460. [DOI] [PubMed] [Google Scholar]
- 2.Hardin, J.A., and J.O. Thomas. 1983. Antibodies to histones in systemic lupus erythematosus: localization of prominent autoantigens on histones H1 and H2B. Proc. Natl. Acad. Sci. USA. 80:7410–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wardemann, H., S. Yurasov, A. Schaefer, J.W. Young, E. Meffre, and M.C. Nussenzweig. 2003. Predominant autoantibody production by early human B cell precursors. Science. 301:1374–1377. [DOI] [PubMed] [Google Scholar]
- 4.Yurasov, S., H. Wardemann, J. Hammersen, M. Tsuiji, E. Meffre, V. Pascual, and M.C. Nussenzweig. 2005. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J. Exp. Med. 201:703–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kronenberg, M., and A. Rudensky. 2005. Regulation of immunity by self-reactive T cells. Nature. 435:598–604. [DOI] [PubMed] [Google Scholar]
- 6.Ishigatsubo, Y., A.D. Steinberg, and D.M. Klinman. 1988. Autoantibody production is associated with polyclonal B cell activation in autoimmune mice which express the lpr or gld genes. Eur. J. Immunol. 18:1089–1093. [DOI] [PubMed] [Google Scholar]
- 7.Gleichmann, E., E.H. Van Elven, and J.P. Van der Veen. 1982. A systemic lupus erythematosus (SLE)-like disease in mice induced by abnormal T-B cell cooperation. Preferential formation of autoantibodies characteristic of SLE. Eur. J. Immunol. 12:152–159. [DOI] [PubMed] [Google Scholar]
- 8.van Rappard-van der Veen, F.M., A.G. Rolink, and E. Gleichmann. 1982. Diseases caused by reactions of T lymphocytes towards incompatible structures of the major histocompatibility complex. VI. Autoantibodies characteristic of systemic lupus erythematosus induced by abnormal T–B cell cooperation across I-E. J. Exp. Med. 155:1555–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris, S.C., P.L. Cohen, and R.A. Eisenberg. 1990. Experimental induction of systemic lupus erythematosus by recognition of foreign Ia. Clin. Immunol. Immunopathol. 57:263–273. [DOI] [PubMed] [Google Scholar]
- 10.Morris, S.C., R.L. Cheek, P.L. Cohen, and R.A. Eisenberg. 1990. Autoantibodies in chronic graft versus host result from cognate T–B interactions. J. Exp. Med. 171:503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rolink, A.G., H. Gleichmann, and E. Gleichmann. 1983. Diseases caused by reactions of T lymphocytes to incompatible structures of the major histocompatibility complex. VII. Immune-complex glomerulonephritis. J. Immunol. 130:209–215. [PubMed] [Google Scholar]
- 12.van Rappard-van der Veen, F.M., U. Kiesel, L. Poels, W. Schuler, C.J. Melief, J. Landegent, and E. Gleichmann. 1984. Further evidence against random polyclonal antibody formation in mice with lupus-like graft-vs-host disease. J. Immunol. 132:1814–1820. [PubMed] [Google Scholar]
- 13.Chen, C., Z. Nagy, E.L. Prak, and M. Weigert. 1995. Immunoglobulin heavy chain gene replacement: a mechanism of receptor editing. Immunity. 3:747–755. [DOI] [PubMed] [Google Scholar]
- 14.Sekiguchi, D.R., S.M. Jainandunsing, M.L. Fields, M.A. Maldonado, M.P. Madaio, J. Erikson, M. Weigert, and R.A. Eisenberg. 2002. Chronic graft-versus-host in Ig knockin transgenic mice abrogates B cell tolerance in anti-double-stranded DNA B cells. J. Immunol. 168:4142–4153. [DOI] [PubMed] [Google Scholar]
- 15.Sekiguchi, D.R., R.A. Eisenberg, and M. Weigert. 2003. Secondary heavy chain rearrangement: a mechanism for generating anti–double-stranded DNA B cells. J. Exp. Med. 197:27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erikson, J., M.Z. Radic, S.A. Camper, R.R. Hardy, C. Carmack, and M. Weigert. 1991. Expression of anti-DNA immunoglobulin transgenes in non-autoimmune mice. Nature. 349:331–334. [DOI] [PubMed] [Google Scholar]
- 17.Li, H., Y. Jiang, E.L. Prak, M. Radic, and M. Weigert. 2001. Editors and editing of anti-DNA receptors. Immunity. 15:947–957. [DOI] [PubMed] [Google Scholar]
- 18.Gay, D., T. Saunders, S. Camper, and M. Weigert. 1993. Receptor editing: an approach by autoreactive B cells to escape tolerance. J. Exp. Med. 177:999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radic, M.Z., J. Erikson, S. Litwin, and M. Weigert. 1993. B lymphocytes may escape tolerance by revising their antigen receptors. J. Exp. Med. 177:1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukuyama, H., F. Nimmerjahn, and J.V. Ravetch. 2005. The inhibitory Fcgamma receptor modulates autoimmunity by limiting the accumulation of immunoglobulin G+ anti-DNA plasma cells. Nat. Immunol. 6:99–106. [DOI] [PubMed] [Google Scholar]
- 21.Chen, C., M.Z. Radic, J. Erikson, S.A. Camper, S. Litwin, R.R. Hardy, and M. Weigert. 1994. Deletion and editing of B cells that express antibodies to DNA. J. Immunol. 152:1970–1982. [PubMed] [Google Scholar]
- 22.Li, Y., Y. Louzoun, and M. Weigert. 2004. Editing anti-DNA B cells by Vlambdax. J. Exp. Med. 199:337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Y., H. Li, D. Ni, and M. Weigert. 2002. Anti-DNA B cells in MRL/lpr mice show altered differentiation and editing pattern. J. Exp. Med. 196:1543–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Y., H. Li, and M. Weigert. 2002. Autoreactive B cells in the marginal zone that express dual receptors. J. Exp. Med. 195:181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris, S.C., R.L. Cheek, P.L. Cohen, and R.A. Eisenberg. 1990. Allotype-specific immunoregulation of autoantibody production by host B cells in chronic graft-versus host disease. J. Immunol. 144:916–922. [PubMed] [Google Scholar]
- 26.Choudhury, A., M.A. Maldonado, P.L. Cohen, and R.A. Eisenberg. 2005. The role of host CD4 T cells in the pathogenesis of the chronic graft-versus-host model of systemic lupus erythematosus. J. Immunol. 174:7600–7609. [DOI] [PubMed] [Google Scholar]
- 27.Busser, B.W., B.S. Adair, J. Erikson, and T.M. Laufer. 2003. Activation of diverse repertoires of autoreactive T cells enhances the loss of anti-dsDNA B cell tolerance. J. Clin. Invest. 112:1361–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, H., Y. Jiang, H. Cao, M. Radic, E.L. Prak, and M. Weigert. 2003. Regulation of anti-phosphatidylserine antibodies. Immunity. 18:185–192. [DOI] [PubMed] [Google Scholar]
- 29.Shlomchik, M.J., A.H. Aucoin, D.S. Pisetsky, and M.G. Weigert. 1987. Structure and function of anti-DNA autoantibodies derived from a single autoimmune mouse. Proc. Natl. Acad. Sci. USA. 84:9150–9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rozzo, S.J., T.J. Vyse, C.G. Drake, and B.L. Kotzin. 1996. Effect of genetic background on the contribution of New Zealand black loci to autoimmune lupus nephritis. Proc. Natl. Acad. Sci. USA. 93:15164–15168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vidal, S., D.H. Kono, and A.N. Theofilopoulos. 1998. Loci predisposing to autoimmunity in MRL-Fas lpr and C57BL/6-Faslpr mice. J. Clin. Invest. 101:696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santiago, M.L., C. Mary, D. Parzy, C. Jacquet, X. Montagutelli, R.M. Parkhouse, R. Lemoine, S. Izui, and L. Reininger. 1998. Linkage of a major quantitative trait locus to Yaa gene-induced lupus-like nephritis in (NZW x C57BL/6)F1 mice. Eur. J. Immunol. 28:4257–4267. [DOI] [PubMed] [Google Scholar]
- 33.Morel, L., X.H. Tian, B.P. Croker, and E.K. Wakeland. 1999. Epistatic modifiers of autoimmunity in a murine model of lupus nephritis. Immunity. 11:131–139. [DOI] [PubMed] [Google Scholar]
- 34.Bolland, S., Y.S. Yim, K. Tus, E.K. Wakeland, and J.V. Ravetch. 2002. Genetic modifiers of systemic lupus erythematosus in FcγRIIB−/− mice. J. Exp. Med. 195:1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen, X., F. Martin, K.A. Forbush, R.M. Perlmutter, and J.F. Kearney. 1997. Evidence for selection of a population of multi-reactive B cells into the splenic marginal zone. Int. Immunol. 9:27–41. [DOI] [PubMed] [Google Scholar]
- 36.Qian, Y., H. Wang, and S.H. Clarke. 2004. Impaired clearance of apoptotic cells induces the activation of autoreactive anti-Sm marginal zone and B-1 B cells. J. Immunol. 172:625–635. [DOI] [PubMed] [Google Scholar]
- 37.Chen, F., M.A. Maldonado, M. Madaio, and R.A. Eisenberg. 1998. The role of host (endogenous) T cells in chronic graft-versus-host autoimmune disease. J. Immunol. 161:5880–5885. [PubMed] [Google Scholar]
- 38.Radic, M.Z., J. Mackle, J. Erikson, C. Mol, W.F. Anderson, and M. Weigert. 1993. Residues that mediate DNA binding of autoimmune antibodies. J. Immunol. 150:4966–4977. [PubMed] [Google Scholar]
- 39.Schlissel, M.S., and D. Baltimore. 1989. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 58:1001–1007. [DOI] [PubMed] [Google Scholar]
- 40.Galin, F.S., C.C. Maier, S.R. Zhou, J.N. Whitaker, and J.E. Blalock. 1996. Murine V lambda x and V lambda x-containing antibodies bind human myelin basic protein. J. Clin. Invest. 97:486–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galin, F.S., S.R. Zhou, J.N. Whitaker, and J.E. Blalock. 1996. Preferential association of V lambda x light chains with gamma 2a heavy chains in naturally occurring human myelin basic protein reactive antibodies. J. Neuroimmunol. 70:15–20. [DOI] [PubMed] [Google Scholar]
- 42.Shlomchik, M., M. Mascelli, H. Shan, M.Z. Radic, D. Pisetsky, A. Marshak-Rothstein, and M. Weigert. 1990. Anti-DNA antibodies from autoimmune mice arise by clonal expansion and somatic mutation. J. Exp. Med. 171:265–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jang, Y.J., D. Sanford, H.Y. Chung, S.Y. Baek, and B.D. Stollar. 1998. The structural basis for DNA binding by an anti-DNA autoantibody. Mol. Immunol. 35:1207–1217. [DOI] [PubMed] [Google Scholar]
- 44.Chen, C., Z. Nagy, M.Z. Radic, R.R. Hardy, D. Huszar, S.A. Camper, and M. Weigert. 1995. The site and stage of anti-DNA B-cell deletion. Nature. 373:252–255. [DOI] [PubMed] [Google Scholar]
- 45.Cocca, B.A., S.N. Seal, P. D'Agnillo, Y.M. Mueller, P.D. Katsikis, J. Rauch, M. Weigert, and M.Z. Radic. 2001. Structural basis for autoantibody recognition of phosphatidylserine-beta 2 glycoprotein I and apoptotic cells. Proc. Natl. Acad. Sci. USA. 98:13826–13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radic, M.Z., M.A. Mascelli, J. Erikson, H. Shan, and M. Weigert. 1991. Ig H and L chain contributions to autoimmune specificities. J. Immunol. 146:176–182. [PubMed] [Google Scholar]
- 47.Prak, E.L., M. Trounstine, D. Huszar, and M. Weigert. 1994. Light chain editing in κ-deficient animals: a potential mechanism of B cell tolerance. J. Exp. Med. 180:1805–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Attanavanich, K., and J.F. Kearney. 2004. Marginal zone, but not follicular B cells, are potent activators of naive CD4 T cells. J. Immunol. 172:803–811. [DOI] [PubMed] [Google Scholar]
- 49.Haynes, B.F., J. Fleming, E.W. St Clair, H. Katinger, G. Stiegler, R. Kunert, J. Robinson, R.M. Scearce, K. Plonk, H.F. Staats, et al. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 308:1906–1908. [DOI] [PubMed] [Google Scholar]
- 50.Nemazee, D. 1996. Antigen receptor ‘capacity’ and the sensitivity of self-tolerance. Immunol. Today. 17:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Louzoun, Y., M. Weigert, and G. Bhanot. 2003. Dynamical analysis of a degenerate primary and secondary humoral immune response. Bull. Math. Biol. 65:535–545. [DOI] [PubMed] [Google Scholar]
- 52.Notkins, A.L. 2004. Polyreactivity of antibody molecules. Trends Immunol. 25:174–179. [DOI] [PubMed] [Google Scholar]
- 53.Levine, M.H., A.M. Haberman, D.B. Sant'Angelo, L.G. Hannum, M.P. Cancro, C.A. Janeway Jr., and M.J. Shlomchik. 2000. A B-cell receptor-specific selection step governs immature to mature B cell differentiation. Proc. Natl. Acad. Sci. USA. 97:2743–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prak, E.L., and M. Weigert. 1995. Light chain replacement: a new model for antibody gene rearrangement. J. Exp. Med. 182:541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brard, F., M. Shannon, E.L. Prak, S. Litwin, and M. Weigert. 1999. Somatic mutation and light chain rearrangement generate autoimmunity in anti–single-stranded DNA transgenic MRL/lpr mice. J. Exp. Med. 190:691–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caton, A.J., S.E. Stark, J. Kavaler, L.M. Staudt, D. Schwartz, and W. Gerhard. 1991. Many variable region genes are utilized in the antibody response of BALB/c mice to the influenza virus A/PR/8/34 hemagglutinin. J. Immunol. 147:1675–1686. [PubMed] [Google Scholar]
- 57.Ramsden, D.A., C.J. Paige, and G.E. Wu. 1994. Kappa light chain rearrangement in mouse fetal liver. J. Immunol. 153:1150–1160. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.