Abstract
Cell proliferation is strictly controlled during differentiation. In T cell development, the cell cycle is normally arrested at the CD4+CD8+ stage, but the mechanism underlying such differentiation-specific exit from the cell cycle has been unclear. Fbxw7 (also known as Fbw7, Sel-10, hCdc4, or hAgo), an F-box protein subunit of an SCF-type ubiquitin ligase complex, induces the degradation of positive regulators of the cell cycle, such as c-Myc, c-Jun, cyclin E, and Notch. FBXW7 is often mutated in a subset of human cancers. We have now achieved conditional inactivation of Fbxw7 in the T cell lineage of mice and found that the cell cycle is not arrested at the CD4+CD8+ stage in the homozygous mutant animals. The mutant mice manifested thymic hyperplasia as a result of c-Myc accumulation and eventually developed thymic lymphoma. In contrast, mature T cells of the mutant mice failed to proliferate in response to mitogenic stimulation and underwent apoptosis in association with accumulation of c-Myc and p53. These latter abnormalities were corrected by deletion of p53. Our results suggest that Fbxw7 regulates the cell cycle in a differentiation-dependent manner, with its loss resulting in c-Myc accumulation that leads to hyperproliferation in immature T cells but to p53-dependent cell-cycle arrest and apoptosis in mature T cells.
Control of cell proliferation is fundamental to the development of multicellular organisms. Regulation of total cell mass during development is dependent on termination of cell proliferation at the appropriate time as well as on the initial stimulation of such proliferation (1). In most vertebrate cell lineages, for example, precursor cells divide a limited number of times before they stop and undergo terminal differentiation into specialized postmitotic cells. However, the mechanisms responsible for spatiotemporal regulation of the cell cycle during development have remained largely unclear. In particular, the mechanisms that control the exit of cells from the cell cycle during differentiation of the various cell lineages are not known.
The basic helix-loop-helix transcription factor c-Myc is a key regulator of exit from and reentry into the cell cycle. The observations that ectopic expression of c-Myc alone is sufficient to promote the reentry of quiescent cells into the cell cycle in the absence of serum mitogens (2) and that the level of endogenous c-Myc expression is high at the G0-G1 transition, low in proliferating cells, and undetectable in most resting cells (3) are consistent with the hypothesis that c-Myc controls the decision of whether to divide or not in mammalian cells. Overexpression of c-Myc is also implicated in the development of diverse human tumors.
Regulation of the abundance of c-Myc is achieved at several levels, one of which is control of c-Myc stability mediated by posttranslational modification. We and others have shown that the F-box protein Fbxw7 (also known as Fbw7, Sel-10, hCdc4, or hAgo), the substrate-recognition subunit of an SCF-type ubiquitin ligase complex, interacts with and mediates the ubiquitylation of c-Myc in a manner dependent on its phosphorylation on Thr58 (4, 5). Mutation of this residue is common in many human malignancies and results in marked stabilization of c-Myc (6, 7). Accumulation of c-Myc is also apparent in mouse Fbxw7−/− cells (4), as well as in lymphomas that arise in Fbxw7+/− mice (8). These data suggest that Fbxw7 is a negative regulator of c-Myc abundance and may thereby induce exit of cells from the cell cycle (9).
To investigate the role of Fbxw7-mediated ubiquitylation in control of the cell cycle during development in a physiologically relevant manner, we and others have generated mice that are deficient in Fbxw7. However, the Fbxw7−/− embryos die in utero at embryonic day 10.5, manifesting marked abnormalities in vascular development as a result of dysregulation of Notch signaling (10, 11). This early embryonic mortality has thus impeded study of the role of Fbxw7 in cell-cycle control. To overcome this obstacle, we have generated mice with Fbxw7 conditionally ablated only in the T cell lineage, in which cell proliferation and differentiation have been extensively characterized.
T cell progenitors are produced in the bone marrow, undergo maturation in the thymus, and finally populate the peripheral lymphoid organs. In the thymus, most immature T cells initially express neither CD4 nor CD8 and are therefore referred to as double-negative (DN) cells (12). Maturation of DN cells to cells that express both CD4 and CD8 (double-positive [DP] cells) requires a sequence of events that are closely associated with rearrangement of the TCR. DP cells differentiate into single-positive (SP) T cells that express either CD4 or CD8. SP T cells exit the thymus and are routed to peripheral lymphoid organs, such as the spleen and lymph nodes, where they participate in a variety of immune responses. Early DN cells remain in G0 phase of the cell cycle. Extensive proliferation normally occurs within the CD25−CD44− (stage IV) population of DN cells, resulting in a several hundredfold increase in cell number. The cell cycle is rapidly restrained, however, when T cells become DP, and SP cells in the peripheral lymphoid organs remain in G0 phase for long periods until they encounter cognate antigen bound at the surface of an antigen-presenting cell. Such activation by antigen induces extensive proliferation of peripheral SP T cells, most of which are subsequently destined to die.
We now show that the loss of Fbxw7 in immature T cells of mice results in their failure to exit the cell cycle at the DP stage, leading to thymic hyperplasia and the subsequent development of lymphoma. Among known targets of Fbxw7, only c-Myc and Notch accumulated in the Fbxw7-deficient thymocytes, and Notch did not appear to contribute to the hyperproliferation phenotype. In contrast to immature T cells, the accumulation of c-Myc apparent in Fbxw7-null mature T cells induced expression of p53, which in turn led to cell-cycle arrest and apoptosis. We therefore conclude that Fbxw7 is required for the regulation of c-Myc abundance and for cell-cycle exit in immature T cells but for the activation of proliferation in mature T cells. We propose that Fbxw7 is a key regulator that determines the fate of cells to divide or not to divide by targeting c-Myc for degradation.
RESULTS
Conditional inactivation of Fbxw7 in the T cell lineage
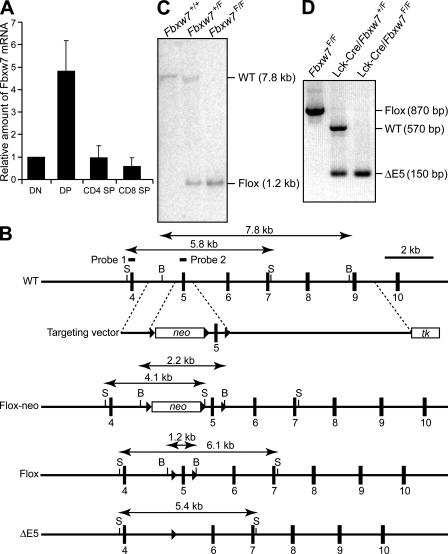
To investigate the mechanisms underlying cell-cycle exit during differentiation, we took advantage of several aspects of T cell development, including the existence of many well-characterized differentiation markers, the coincidence of cell-cycle exit and reentry with clearly defined developmental stages, and the fact that, given the nonessential nature of T cells for survival, mice are not killed by genetic manipulation of these cells. The increased abundance of Fbxw7 messenger RNA (mRNA) at the DP stage of T cell development (Fig. 1 A) led us to hypothesize that Fbxw7 might contribute to the cell-cycle exit that normally occurs at this stage.
Figure 1.
Generation of mice with T cell–specific deficiency of Fbxw7. (A) RT and real-time PCR analysis of the relative abundance of Fbxw7 mRNA in DN, DP, CD4 SP, and CD8 SP mouse thymocytes isolated by flow cytometry. Data are means ± SD of values from three independent experiments. (B) Schematic representations of the wild-type mouse Fbxw7 allele, the targeting vector, the Fbxw7 allele with a loxP-neo cassette (Flox-neo), the floxed Fbxw7 allele (Flox), and the floxed Fbxw7 allele after removal of exon 5 by Cre recombinase (ΔE5). The expected sizes of bands in a Southern blot analysis with probe 1 (for StuI [S] fragments) or probe 2 (for BamHI [B] fragments) are indicated. Exons are shown as numbered closed boxes, and loxP sites are shown as closed triangles. (C) Southern blot analysis with probe 2 of BamHI-digested DNA from the tail of mice of the indicated genotypes. The wild-type and floxed alleles give rise to hybridizing fragments of 7.8 and 1.2 kb, respectively. (D) PCR analysis of genomic DNA from the thymus of Fbxw7F/F, Lck-Cre/Fbxw7+/F, and Lck-Cre/Fbxw7F/F mice with the primers Floxed 1 and Floxed 2 (see Materials and methods). The positions of amplified fragments corresponding to wild-type, floxed, and ΔE5 alleles are indicated.
We established mouse embryonic stem (ES) cells that harbor a “floxed” Fbxw7 allele in which exon 5 (which encodes the F-box domain) is flanked by loxP sites. This allele was generated by homologous recombination and transient transfection of the cells with a vector for Cre recombinase to remove the introduced neo cassette (Fig. 1 B). Mice harboring the floxed allele were produced by microinjection of the mutant ES cells into blastocysts and breeding of the resultant chimeric animals with mice of the C57BL/6 strain (Fig. 1 C). Mice homozygous for the floxed Fbxw7 allele (Fbxw7F/F) had no apparent defects, indicating that the allele is fully functional. Mice homozygous for germline deletion of exon 5 of Fbxw7 (Fbxw7ΔE5/ΔE5) died in utero (of 61 live-born offspring of Fbxw7+/ΔE5 intercrosses, 18 were Fbxw7+/+, 43 were Fbxw7+/ΔE5, and 0 were Fbxw7ΔE5/ΔE5) and were indistinguishable from the Fbxw7−/− mice that we previously described (10), indicating that deletion of exon 5 abolishes the function of Fbxw7. To ablate Fbxw7 only in the T cell lineage, we crossed Fbxw7F/F mice with mice harboring a Cre transgene under the control of the promoter for the Lck or CD4 gene (Lck-Cre or CD4-Cre). We confirmed that almost all floxed alleles were inactivated by Cre in thymocytes of the resulting offspring (Fig. 1 D and Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20062299/DC1).
The cell cycle is not arrested in Fbxw7-deficient DP cells
The thymus of Lck-Cre/Fbxw7F/F mice was larger than that of control (Lck-Cre/Fbxw7+/F) mice (Fig. 2 A). Flow cytometric analysis also revealed that both the percentage (Fig. 2 B) and absolute number (Fig. 2 C) of DP thymocytes were increased in Lck-Cre/Fbxw7F/F mice compared with littermate controls, whereas the absolute numbers of DN, CD4 SP, and CD8 SP thymocytes did not differ between the two genotypes. The number of DP thymocytes was also increased in CD4-Cre/Fbxw7F/F mice, albeit to a lesser extent than in Lck-Cre/Fbxw7F/F mice (see Fig. 3 B). It is possible that the onset of cell-cycle arrest induced by Fbxw7 occurs slightly earlier than that of expression of CD4.
Figure 2.
Failure of cell-cycle arrest in DP thymocytes of Lck-Cre/Fbxw7F/Fmice. (A) Gross appearance of the thymus of Lck-Cre/Fbxw7+/F and Lck-Cre/Fbxw7F/F mice at 8 wk of age. Bar, 5 mm. (B) Representative flow cytometric analysis of surface expression of CD4 and CD8 on thymocytes from Lck-Cre/Fbxw7+/F or Lck-Cre/Fbxw7F/F mice at 8 wk of age. The percentages of DN, DP, and SP populations are indicated. (C) Absolute cell numbers for total thymocytes and thymocyte subsets determined as in B. Data are means ± SD of values from 13 Fbxw7F/F (control) and 19 Lck-Cre/Fbxw7F/F mice. **, P < 0.01 using the Student's t test. (D) BrdU incorporation into thymocytes in vivo. The proportion of each thymocyte subset in S phase was determined by measurement of incorporation of BrdU after its intraperitoneal injection in 8-wk-old Lck-Cre/Fbxw7F/F or Fbxw7F/F (control) mice. Data are means ± SD of values from four animals of each genotype. **, P < 0.01 using the Student's t test. (E) Increased expression of Notch1, Notch3, and c-Myc in total thymocytes from Lck-Cre/Fbxw7F/F mice. Total thymocytes of 8-wk-old Lck-Cre/Fbxw7+/F or Lck-Cre/Fbxw7F/F mice were subjected to immunoblot analysis with antibodies to the indicated proteins. (F) Accumulation of Notch1, Notch3, and c-Myc specifically in DP thymocytes of Lck-Cre/Fbxw7F/F mice. Thymocyte subsets of Fbxw7F/F (control) or Lck-Cre/Fbxw7F/F mice were analyzed as in E.
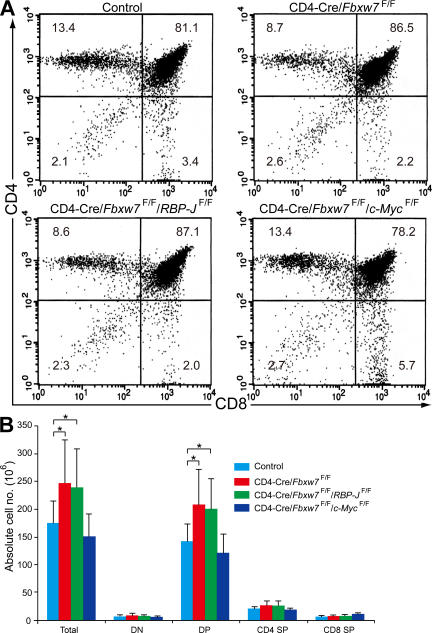
Figure 3.
c-Myc is responsible for the overproliferation phenotype of Fbxw7-deficient DP thymocytes. (A) Representative flow cytometric analysis of surface expression of CD4 and CD8 on thymocytes from Fbxw7F/F/RBP-JF/F (control), CD4-Cre/Fbxw7F/F, CD4-Cre/Fbxw7F/F/RBP-JF/F, or CD4-Cre/Fbxw7F/F/c-MycF/F mice at 12 wk of age. The respective percentages are indicated. (B) Absolute cell numbers for total thymocytes and thymocyte subsets determined as in A. Data are means ± SD of values from 7 Fbxw7F/F/RBP-JF/F (control), 11 CD4-Cre/Fbxw7F/F, 6 CD4-Cre/Fbxw7F/F/RBP-JF/F, and 5 CD4-Cre/Fbxw7F/F/c-MycF/F mice. *, P < 0.05 using the Student's t test.
The size of the thymic cell population is largely dependent on the extents of apoptosis and proliferation during differentiation. Susceptibility to spontaneous apoptosis or to that induced by antibodies to anti-CD3 or dexamethasone was determined in vitro by propidium iodide staining and was found to be largely unaffected in thymocytes from Lck-Cre/Fbxw7F/F mice (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20062299/DC1). We therefore measured the incorporation of BrdU into thymocytes 3 h after its intraperitoneal administration. The extent of BrdU incorporation into total thymocytes of Lck-Cre/Fbxw7F/F mice was about twice that into those of control mice (Fig. 2 D). Although incorporation of BrdU into DN thymocytes did not differ between Lck-Cre/Fbxw7F/F and control mice, incorporation of BrdU into DP thymocytes was markedly greater for Lck-Cre/Fbxw7F/F mice. The incorporation of BrdU into CD4 SP cells did not differ substantially between the two genotypes. The incorporation of BrdU into CD8 SP cells was much greater than that into CD4 SP cells for both genotypes and was highly variable among individuals (unpublished data), probably because this fraction contains both immature SP cells and mature CD8 SP cells. These data indicated that Fbxw7 is indispensable for exit from the cell cycle at the DP stage. However, the cell cycle appeared to be arrested at the SP stage even in the absence of Fbxw7, suggesting that the requirement for Fbxw7 in cell-cycle arrest changes between the DP and SP stages, probably as a result of positive selection (see Discussion).
c-Myc (not Notch) is responsible for overproliferation of Fbxw7-null DP cells
With the use of immunoblot analysis, we next examined the expression of substrates of Fbxw7, including cyclin E, c-Myc, Notch1, Notch3, and c-Jun, in total thymocytes (Fig. 2 E) as well as in thymocyte subsets (Fig. 2 F) of Lck-Cre/Fbxw7F/F or control mice. The amounts of Notch1, Notch3, and c-Myc were increased in total thymocytes as well as in the DP subset of Lck-Cre/Fbxw7F/F mice. The half-lives of Notch1, Notch3, and c-Myc in the DP subset of Lck-Cre/Fbxw7F/F mice were also increased compared with those in control mice (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20062299/DC1). The levels of cyclin E and phosphorylated c-Jun were not affected bydeletion of Fbxw7, suggesting that the abnormal accumulation of Notch or c-Myc (or of other unknown targets of Fbxw7) may be responsible for promotion of the cell cycle in Fbxw7-deficient DP cells.
To determine whether accumulation of Notch or c-Myc is responsible for the failure of cell-cycle arrest in Fbxw7-deficient DP thymocytes, we examined the effect of loss of Fbxw7 in the absence either of RBP-J, an essential mediator of Notch signaling, or of c-Myc. The early inactivation of RBP-J mediated by Lck-Cre resulted in developmental arrest of T cells at the DN stage, whereas its later inactivation mediated by CD4-Cre did not affect the absolute number or the production rate of CD4+ or CD8+ mature T cells (13). Loss of c-Myc also results in developmental arrest of the T cell lineage at the DN stage (14). We therefore generated CD4-Cre/Fbxw7F/F/RBP-JF/F mice and CD4-Cre/Fbxw7F/F/c-MycF/F mice to ablate the RBP-J and c-Myc genes at the DP stage. The percentage and absolute number of DP thymocytes in CD4-Cre/Fbxw7F/F/RBP-JF/F mice were increased as in CD4-Cre/Fbxw7F/F mice, whereas those in CD4-Cre/Fbxw7F/F/c-MycF/F mice were similar to those in controls (Fig. 3). These genetic data thus suggested that the hyperproliferation phenotype of Fbxw7-deficient DP thymocytes is dependent on c-Myc but not on Notch. We thus concluded that the accumulation of c-Myc in Fbxw7-deficient DP thymocytes is responsible for the failure of cell-cycle exit and the consequent hyperproliferation phenotype.
Loss of Fbxw7 in T cells predisposes to thymic lymphoma
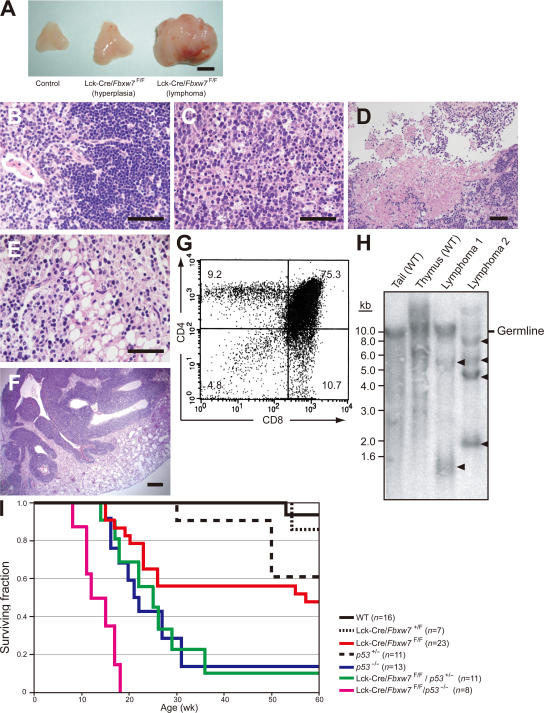
Lck-Cre/Fbxw7F/F mice developed aggressive lymphomas characterized by massive thymic enlargement (Fig. 4 A). Histopathologic examination of the tumors revealed that the architecture of the thymus was completely effaced and that they contained a uniform population of immature lymphoid cells (Fig. 4, B and C), with some areas of necrosis (Fig. 4 D). Fat and lung tissue adjacent to the thymus manifested infiltration by tumor cells (Fig. 4, E and F). Flow cytometric analysis revealed that the lymphomas typically showed an immature T cell immunophenotype characterized by expression of both CD4 and CD8 (Fig. 4 G).
Figure 4.
Lck-Cre/Fbxw7F/Fmice develop T cell lymphoma. (A) Thymi from Fbxw7F/F (control; left) and Lck-Cre/Fbxw7F/F (middle and right) littermates at 14 wk of age. Those from the Lck-Cre/Fbxw7F/F mice were hyperplastic (middle) or tumorous (right). Bar, 5 mm. (B and C) Hematoxylin-eosin staining of sections of the thymus from an Fbxw7F/F mouse (B) or from an Lck-Cre/Fbxw7F/F mouse that developed lymphoma (C). The normal thymic structure was destroyed and replaced with large, atypical lymphoma cells in the latter animal. Bars, 50 μm. (D) Necrotic lesions in the thymus of an Lck-Cre/Fbxw7F/F mouse with lymphoma. Bar, 50 μm. (E and F) Infiltration of lymphoma cells into the fat (E) and lung (F) of Lck-Cre/Fbxw7F/F mice. Bars: (E) 50 μm; (F) 500 μm. (G) Surface expression of CD4 and CD8 on thymic lymphoma cells from an Lck-Cre/Fbxw7F/F mouse. (H) Southern blot analysis of genomic DNA from the tail and thymus of wild-type mice and from two thymic lymphomas of Lck-Cre/Fbxw7F/F mice. The DNA was digested with BamHI and probed with a 2.1-kb EcoRI fragment of the TCR-Jβ2 locus. The positions of germline and rearranged (arrowheads) fragments are indicated. (I) Kaplan-Meier plot of the overall survival of mice of the indicated genotypes.
To examine whether the lymphomas were monoclonal in origin, we subjected DNA from two specimens to Southern blot analysis with a genomic fragment of the TCRβ locus as the probe. A single nonrearranged (germline) fragment and several prominent rearranged fragments were detected in the lymphoma DNA, whereas the wild-type thymic DNA showed a smear pattern caused by random rearrangement as well as one germline band (Fig. 4 H). These data indicated that the lymphomas in Lck-Cre/Fbxw7F/F mice arise by clonal expansion. Transplantation of 5 × 106 lymphoma cells into the peritoneal cavity of nude mice resulted in the development of marked ascites, peritoneal lymphadenopathy, extraperitoneal invasion of tumor cells, and hepatosplenomegaly after 3 wk (Fig. S4 A, available at http://www.jem.org/cgi/content/full/jem.20062299/DC1). Flow cytometry revealed that both the original tumor cells and the tumor cells in the recipient nude mice expressed CD4 and CD8, although both types of cells exhibited a slight reduction in the level of CD8 expression (Fig. S4 B). Southern blot analysis confirmed that the clonality of tumor cells in the recipient animals was almost identical to that of the original thymoma (Fig. S4 C). These results thus indicated that the lymphomas of the mutant mice are capable of lymphomatogenesis in nude mice.
These lymphomas were fatal. Specifically, 12 out of 23 Lck-Cre/Fbxw7F/F mice developed tumors, and these animals either died or became terminally ill (and were killed) between 14 and 60 wk of age (Fig. 4 I). There were no marked differences in gross appearance, surface expression of CD4 or CD8, or histopathologic characteristics between early-onset (<20 wk of age) and late-onset (>20 wk of age) tumors (Fig. S5, available at http://www.jem.org/cgi/content/full/jem.20062299/DC1). We bred Lck-Cre/Fbxw7F/F mice with p53−/− mice to generate Lck-Cre/Fbxw7F/F/p53−/− mice. The double-mutant mice developed thymic lymphomas at a markedly increased frequency and with a reduced latency compared with both parental groups (Fig. 4 I). However, there were no substantial differences in gross appearance or in surface expression of CD4 or CD8 between tumors arising in Lck-Cre/Fbxw7F/F mice and those in Lck-Cre/Fbxw7F/F/p53−/− mice (Fig. S6). All tumors in mice of both genotypes showed a DP phenotype; no SP lymphomas were observed.
Fbxw7-deficient mature T cells fail to proliferate in response to antigenic stimulation
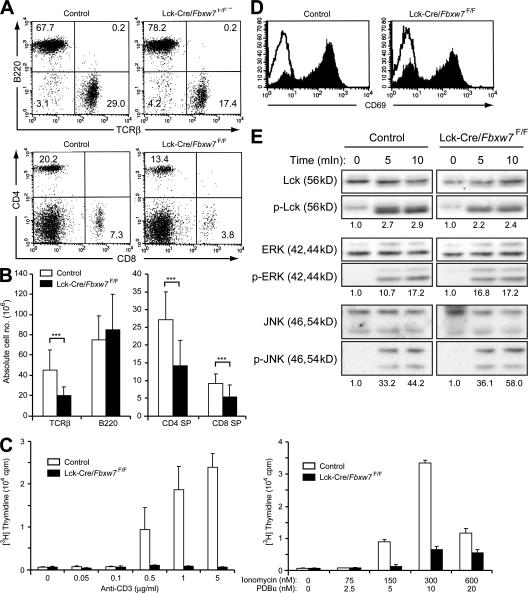
Expansion of the DP population did not result in an increase in the numbers of SP thymocytes in Fbxw7-deficient mice, suggesting that the DP thymocytes had lost the ability to undergo positive selection or that the proliferation or survival of SP cells was impaired. The percentages and numbers of CD4+ or CD8+ T cells were markedly reduced, whereas the number of B cells was unaffected, in the peripheral lymphoid organs, such as the spleen and lymph nodes, of Lck-Cre/Fbxw7F/F mice (Fig. 5, A and B; unpublished data). To investigate the mechanism underlying this deficiency of mature T cells, we measured the incorporation of [3H]thymidine into splenic T cells in response to mitogenic stimulation. Fbxw7-deficient T cells did not respond to stimulation with either anti-CD3 or the combination of phorbol 12,13-dibutyrate and ionomycin, whereas these agents stimulated the proliferation of control T cells in a concentration-dependent manner (Fig. 5 C). To exclude the possibility that TCR signaling was impaired in the Fbxw7-deficient splenic T cells, we first examined the induction of the early activation marker CD69 in response to stimulation with anti-CD3. The induction of CD69 expression was similar in Fbxw7-null and control T cells (Fig. 5 D). Furthermore, phosphorylation of Lck, extracellular signal-regulated kinase (ERK), and c-Jun N-terminal kinase (JNK) in response to TCR activation was not altered in Fbxw7-null T cells (Fig. 5 E). These results thus excluded the possibility of a signaling defect in Fbxw7-null T cells. Fbxw7 thus appeared to be indispensable for the proliferation of mature T cells in response to mitogenic stimulation, a response that is necessary for maintenance of the T cell population in peripheral lymphoid organs.
Figure 5.
Proliferative defect of Fbxw7-deficient mature T cells. (A) Representative flow cytometric analysis of surface expression of TCRβ and B220 (top) or of CD4 and CD8 (bottom) on spleen cells from Fbxw7F/F (control) or Lck-Cre/Fbxw7F/F mice at 8 wk of age. The respective percentages are indicated. (B) Absolute cell numbers of splenocyte subsets determined as in A. Data are means ± SD of values from 8 Fbxw7F/F (control) and 14 Lck-Cre/Fbxw7F/F mice. ***, P < 0.005 using the Student's t test. (C) Splenic T cells from Lck-Cre/Fbxw7+/F (control) or Lck-Cre/Fbxw7F/F mice were stimulated for 48 h with the indicated concentrations of anti-CD3ε (left) or ionomycin and phorbol 12,13-dibutyrate (PDBu; right), after which the incorporation of [3H]thymidine was measured. Data are means ± SD of values from three independent experiments. (D) Splenic T cells from Fbxw7F/F (control) or Lck-Cre/Fbxw7F/F mice were left unstimulated (open trace) or stimulated with 5 μg/ml anti-CD3ε for 7 h (closed trace) and were then subjected to flow cytometric analysis for detection of the early activation antigen CD69. (E) Immunoblot analysis of phospho-Lck and Lck (loading control), phospho-ERK and ERK (loading control), and phospho-JNK and JNK (loading control) in splenic T cells from Fbxw7F/F (control) or Lck-Cre/Fbxw7F/F mice at 0, 5, and 10 min after stimulation with 5 μg/ml anti-CD3ε. The intensity of the bands corresponding to the phosphorylated proteins was normalized by that of total proteins. The value at time = 0 is defined as 1.
Loss of Fbxw7 in mature T cells induces p53 expression and cell-cycle arrest
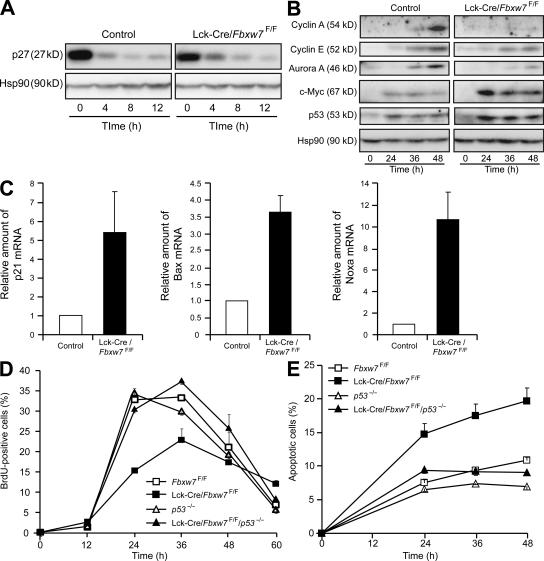
To explore the mechanism underlying the failure of cell-cycle progression in mature T cells of Fbxw7-null mice, we initially examined the down-regulation of the cyclin-dependent kinase inhibitor p27 at the G0-G1 transition. We found that it occurred normally in the Fbxw7-deficient cells (Fig. 6 A), suggesting that the G0-G1 transition was not impaired by the lack of Fbxw7. However, the subsequent expression of cyclin A, cyclin E, and Aurora A in these cells was markedly inhibited or delayed, which was suggestive of a defect at the G1-S transition (Fig. 6 B). Abnormal accumulation of c-Myc was apparent in the activated mature T cells of Lck-Cre/Fbxw7F/F mice, as it was in the thymocytes of these animals. We also found that the abundance of p53 was increased in these cells compared with that in control cells (Fig. 6 B). Indeed, the amounts of mRNAs derived from target genes of p53, including those for p21, Bax, and Noxa, were also increased in the activated T cells of Lck-Cre/Fbxw7F/F mice (Fig. 6 C).
Figure 6.
Abnormal accumulation of c-Myc and induction of p53 in stimulated splenic T cells from Lck-Cre/Fbxw7F/F mice. (A) Immunoblot analysis of p27 and Hsp90 (loading control) in splenic T cells from Fbxw7F/F (control) or Lck-Cre/Fbxw7F/F mice at 0, 4, 8, and 12 h after stimulation with 5 μg/ml anti-CD3ε. (B) Immunoblot analysis of cyclin A, cyclin E, Aurora A, c-Myc, p53, and Hsp90 in splenic T cells from Fbxw7F/F (control) or Lck-Cre/Fbxw7F/F mice at 0, 24, 36, and 48 h after stimulation with 5 μg/ml anti-CD3ε. (C) RT and real-time PCR analysis of p53-dependent gene expression in splenic T cells from Fbxw7F/F (control) or Lck-Cre/Fbxw7F/F mice after stimulation for 24 h with 5 μg/ml anti-CD3ε. Normalized data for p21, Bax, and Noxa mRNAs are expressed relative to the corresponding values for cells from control mice and are means ± SD of values from three independent experiments. (D) Splenic T cells from mice of the indicated genotypes were stimulated with 5 μg/ml anti-CD3ε for the indicated times and exposed to BrdU during the final 1 h of incubation. They were then stained with anti-BrdU, and the percentage of BrdU-positive cells was determined by flow cytometry. Data are means ± SD of values from three independent experiments. (E) Splenic T cells stimulated as in D were stained with propidium iodide, and the percentage of sub-G1 (apoptotic) cells was determined by flow cytometry. Data are means ± SD of values from three independent experiments.
We therefore examined G1-S progression in T cells from Lck-Cre/Fbxw7F/F mice after antigenic stimulation. Flow cytometric analysis revealed that progression into S phase was inhibited and that the proportion of apoptotic cells was increased (Fig. 6, D and E). Both the G1-S arrest and increased level of apoptosis in Fbxw7-deficient T cells were reversed by deletion of p53 (i.e., in cells from Lck-Cre/Fbxw7F/F/p53−/− mice). These data suggested that abnormal activation of p53, likely caused by c-Myc accumulation, is responsible for the growth inhibition and induction of apoptosis in mature T cells of Lck-Cre/Fbxw7F/F mice. Fbxw7 thus appears to be required for maintenance of c-Myc at an appropriate concentration and for progression of the cell cycle in response to antigenic stimulation in mature T cells.
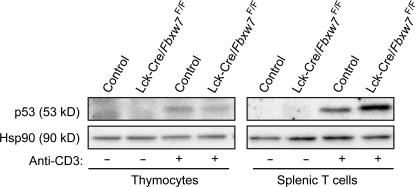
Finally, we compared the extent of p53 induction in response to anti-CD3 stimulation between control and Fbxw7-deficient thymocytes and splenic T cells by immunoblot analysis. Stimulation with anti-CD3 resulted in similar small increases in the abundance of p53 in the mutant and control thymocytes (Fig. 7). In contrast, the up-regulation of p53 induced by such stimulation was more pronounced in splenic T cells than in thymocytes and was markedly greater in splenic T cells from the mutant mice than in those from control animals. These differences in the induction of p53 between thymocytes and splenic T cells from the mutant and control mice appear to be reflected in the differences in the proliferative characteristics of these cells.
Figure 7.
Exaggerated induction of p53 in splenic T cells from Lck-Cre/Fbxw7F/F mice. Thymocytes and splenic T cells isolated from Fbxw7F/F (control) or Lck-Cre/Fbxw7F/F mice were incubated in the absence or presence of 5 μg/ml anti-CD3ε for 24 h, after which cell lysates were subjected to immunoblot analysis with antibodies to p53 and to Hsp90 (loading control).
DISCUSSION
How an organism regulates cell proliferation during development is a central issue in developmental biology. In most vertebrate cell lineages, precursor cells divide a limited number of times before becoming quiescent and undergoing terminal differentiation into specialized postmitotic cells (1). Characterization of the mechanisms by which cell-cycle regulators are controlled during this process is important not only for our understanding of development but also because loss of such control is thought to be a major cause of cancer (9). In most tissues, however, how cells exit the cell cycle during differentiation has remained largely unknown. Our observations with conditionally Fbxw7-deficient mice now indicate that Fbxw7 is required for cell-cycle arrest during T cell differentiation.
We have shown that the loss of Fbxw7 during T cell development results in excessive accumulation of c-Myc, and that this effect (or, albeit less likely, the accumulation of other unidentified substrates of Fbxw7) in turn results in opposite phenotypes in a manner dependent on developmental stage, such as the failure of cell-cycle arrest, leading to tumorigenesis, in immature T cells versus the failure of cell-cycle progression and induction of apoptosis in mature T cells. This apparent discrepancy can be explained by the difference in the effect of c-Myc accumulation on the expression of p53, a mediator of the checkpoint response to such accumulation. Thus, although the abundance of p53 is not increased in immature T cells of Lck-Cre/Fbxw7F/F mice, in spite of the accumulation of c-Myc, the up-regulation of p53 expression in response to mitogenic stimulation is exaggerated in mature T cells of such mice, likely as a result of excessive c-Myc accumulation.
c-Myc plays a central role in normal cell-cycle progression, especially during the transition from G0 to S phase (15). c-Myc is the product of an early response gene, it promotes the entry of cells into G1 phase in response to mitogenic stimulation (16, 17), and its expression is maintained throughout the cell cycle (18, 19). These observations prompted us to hypothesize that c-Myc normally prevents cells from exiting the cell cycle, and that Fbxw7-dependent degradation of c-Myc is required for such exit. During T cell development, the expression of Fbxw7 increases in association with cell-cycle arrest at the DP stage. Furthermore, DP thymocytes of Lck-Cre/Fbxw7F/F mice are defective in cell-cycle arrest and manifest hyperplasia and an increased susceptibility to tumorigenesis. These observations are thus consistent with our hypothesis.
The increase in the number of DP thymocytes in Lck-Cre/Fbxw7F/F mice does not result in an increase in the number of SP cells. There are at least two possible explanations for this observation. First, Fbxw7 may be necessary for efficient positive selection of thymocytes during the transition from the DP to the SP stage. However, positive selection is thought not to require cell-cycle progression. Furthermore, our analysis of TCR signaling revealed no impairment in Fbxw7-deficient mice, suggesting that this explanation is unlikely. Second, an increase in the level of apoptosis in SP cells of the mutant mice might counteract and mask an increase in their generation from DP cells. This scenario is consistent with the decrease in the percentage as well as in the number of splenic T cells in Fbxw7-deficient mice. Such increased apoptosis in SP cells would likely be attributable to the induction of p53 in response to c-Myc accumulation, as was apparent in splenic T cells of the mutant mice.
In addition to its role in the cell cycle, c-Myc contributes to the regulation of apoptosis, as has been revealed by studies of various cell types under a wide variety of physiological conditions, with both under- and overexpression of c-Myc having been found to result in cell death (20). c-Myc induces apoptosis through at least two distinct pathways (21). First, it induces the expression of p19ARF, an inhibitor of the ubiquitin ligase MDM2, resulting in the stabilization of p53 (22). Ablation of either p19ARF or p53 allows c-Myc to immortalize primary cells and greatly facilitates c-Myc–induced lymphomagenesis (23). However, the p19ARF gene does not appear to be a direct target of c-Myc, and the mechanism through which c-Myc activates p19ARF expression is unclear. The p19ARF gene is regulated directly by E2F transcription factors that function downstream of c-Myc. Whether c-Myc induces the expression of p19ARF by increasing the activity of E2F1 is controversial and may be dependent on cell type (24, 25). Second, c-Myc promotes the release of cytochrome c from mitochondria in a p19ARF- and p53-independent manner (26), and this effect is mediated in part by the BH3-only protein Bim, whose expression is induced by c-Myc (27); again, the induction of Bim expression by c-Myc is probably not a direct effect. The roles of c-Myc in both the cell cycle and apoptosis are thus dependent, at least in part, on p53. The opposite phenotypes of immature and mature Fbxw7-deficient T cells are therefore likely attributable to c-Myc accumulation and differential induction of p53.
The factors that determine whether a cell divides or dies warrant further investigation. We have shown that the response to Fbxw7 loss is substantially changed between the DP and SP stages of T cell development. The expression of many sets of genes associated with cell proliferation, survival, migration, or maturation, as well as with allelic exclusion or lineage commitment, changes markedly during positive selection of T cells (28, 29). These changes in gene expression are controlled by transcription factors responsive to specific signals in immature thymocytes. Although recent studies have implicated a small number of transcription factors in the regulation of gene expression during positive selection (28), the mechanism by which the accumulation of c-Myc induces the expression of p53 in Fbxw7-deficient T cells remains to be determined.
Given that Fbxw7 is responsible for the degradation of c-Myc and other oncoproteins, it is thought to function as a tumor suppressor. Indeed, mutations in FBXW7 have been detected in certain human malignancies (30–36). In addition, Fbxw7+/− mice manifest an increased susceptibility to radiation-induced tumorigenesis, although most of the induced tumors retain and express the wild-type allele, indicating that Fbxw7 is a haploinsufficient tumor suppressor gene (8). We have now shown that Lck-Cre/Fbxw7F/F mice manifest spontaneous tumorigenesis in the thymus at earlier ages than do Lck-Cre/Fbxw7+/F mice. This tumor development was further promoted by the deletion of p53, suggesting that p53 inhibits tumorigenesis induced by the loss of Fbxw7, although we did not detect induction of p53 expression in Fbxw7-null thymocytes. The low level of p53 expression in these cells is thus likely not sufficient to impede the promotion of the cell cycle induced by loss of Fbxw7 but may partially antagonize the uncontrolled proliferation that leads to tumor formation.
How does the loss of Fbxw7 function result in cancer development? Dysregulation of cyclin E is a potential contributor to tumorigenesis in Fbxw7-deficient cells, given that increased levels of cyclin E have been associated with a variety of malignancies and that constitutive expression of cyclin E results in genomic instability (37). Furthermore, RNA interference–mediated depletion of cyclin E in cancer cell lines in which Fbxw7 is also disrupted resulted in a marked decrease in the extent of chromosomal instability (36). However, expression of cyclin E is not always increased in cancer cells in which Fbxw7 is mutated (unpublished data) (8, 31). In addition, expression of cyclin E is unaffected in Fbxw7−/− embryos (10), although it is increased in the placenta of these embryos (11). Furthermore, although the frequency of radiation-induced lymphomas is increased in Fbxw7+/− mice, the abundance of cyclin E is not increased in these tumors (8). In the present study, the abundance of cyclin E was not affected in Fbxw7-deficient T cells. Collectively, these various observations suggest that accumulation of cyclin E in response to a loss of Fbxw7 is not a major cause of carcinogenesis. Whereas acute inactivation of Fbxw7 by RNA interference results in an increase in the level of cyclin E in many cell lines (unpublished data) (4, 31, 36, 38), such an acute effect of loss of Fbxw7 may be compensated for during development. It therefore seems likely that Fbxw7 contributes to cyclin E proteolysis in a context-dependent manner.
Another important substrate of Fbxw7 whose dysregulation might be responsible for cancer development is c-Myc. Deregulated expression of Myc family genes is a common feature of a wide variety of malignancies, including T cell lymphomas. Indeed, T cells appear to be especially sensitive to Myc-dependent tumorigenesis, given that T cell lymphoma is the predominant tumor type in transgenic mice that overexpress c-Myc in hematopoietic progenitor cells (39, 40). Previous transgenic animal studies have revealed frequent inactivating mutations of p53 in Myc-induced lymphomas (21, 23). Likewise, Myc-dependent T cell lymphomagenesis is accelerated by events that abrogate Myc-induced apoptosis, such as disruption of p53 pathway function (41, 42). The synergy between Fbxw7- and p53-null genotypes in the development of mouse thymic lymphomas detected in the present study is reminiscent of the observation that mice harboring a CD2-Myc transgene and a homozygous p53-null mutation (p53−/−/CD2-myc) develop thymic lymphomas with a greatly increased frequency and reduced latency compared with both parental mice (41). Although we did not detect an increase in p53 abundance in Fbxw7-null thymocytes, p53 may still play a role in antagonizing tumorigenesis in Fbxw7-deficient mice.
In contrast to T cells with an excess of c-Myc, those that lack c-Myc exhibit a pronounced proliferative defect (43, 44) and fail to develop beyond the DN stage (14). This inability of c-Myc–deficient T cells to differentiate into DP thymocytes may reflect an important role for c-Myc in proliferation mediated by pre-TCR signaling (45). Furthermore, the expression of c-Myc is down-regulated, possibly by a dedicated pathway, in naive quiescent peripheral T cells (46) and is subsequently up-regulated during the proliferative response to antigen-mediated TCR activation (47). Our data now demonstrate a physiological role for a ubiquitin ligase in cell-cycle exit during differentiation, providing the basis for a new paradigm in the relation between cell proliferation and differentiation at the molecular level.
MATERIALS AND METHODS
Construction of a targeting vector and generation of mice with a floxed Fbxw7 allele.
Cloned genomic DNA corresponding to the Fbxw7 locus was previously isolated from a 129/Sv mouse genomic library (Stratagene). The 5′ and 3′ regions of homology in the targeting vector composed a 1.2-kb fragment of the fourth intron generated by PCR with the appropriate primers and an 8-kb DraI-XhoI fragment spanning the fourth and ninth introns, respectively. The neomycin resistance gene (neo) flanked by loxP sites was isolated from the plasmid pL2-Neo(2) (a gift from D.R. Littman, New York University, New York, NY) (48) and inserted into the DraI site upstream of the fifth exon of Fbxw7. A loxP site was also inserted into the HpaI site downstream of the fifth exon. The PGK-tk-poly(A) cassette (a gift from D.R. Littman) was ligated at the 3′ end of the targeting construct. The maintenance, transfection, and selection of ES cells were performed as previously described (49, 50). The recombination event was confirmed by Southern blot analysis with a probe (probe 1) located outside of the 5′ homology region (Fig. 1 B and Fig. S1). ES clones that had undergone homologous recombination were transfected with 30 μg pMC-Cre (a gift from D.R. Littman) to excise the loxP-neo cassette. Colonies were screened for acquired sensitivity to G418, and loss of the cassette was confirmed by Southern blot analysis with probe 2 (Fig. 1 B). ES clones were injected into C57BL/6 mouse blastocysts to generate chimeric mice. Germline transmission of the floxed Fbxw7 allele was achieved by crossing chimeras with C57BL/6 mice and was confirmed by Southern blot analysis of tail DNA with probe 2. Heterozygous offspring were intercrossed to produce homozygous mutant animals. All mouse experiments were approved by the animal ethics committee of Kyushu University.
Generation of conditional knockout mice.
Mice homozygous for the floxed Fbxw7 allele (Fbxw7F/F) were crossed with Lck-Cre (51) or CD4-Cre (a gift from C.B. Wilson, University of Washington, Seattle, WA) (52) transgenic mice. Deletion of exon 5 of the floxed Fbxw7 allele was detected by PCR with the primers Floxed 1 (5′-CCTATAGGGAATTATGTTATTT-3′) and Floxed 2 (5′-CTCACAGCCAAGTTATTCTGTT-3′). Fbxw7F/F mice were also crossed with RBP-JF/F mice (a gift from T. Honjo, Kyoto University, Kyoto, Japan) (13), c-MycF/F mice (17), or p53−/− mice (Taconic). All animal experiments were performed with littermates of a mixed genetic background.
Flow cytometry and antibodies.
The following monoclonal antibodies were used: FITC-conjugated anti-CD3ε (145-2C11), anti-CD4 (RM4-5), anti-CD8α (53-6.7), anti-TCRβ (H57-597), anti-B220 (RA3-6B2), and anti-CD69 (H1.2F3); PE-conjugated anti-CD8α (53-6.7); PE- and Cy5-conjugated anti-CD3ε (145-2C11), anti-CD4 (RM4-5), anti-CD8α (53-6.7), and anti-B220 (RA3-6B2); and allophycocyanin-conjugated anti-CD4 (RM4-5; all were obtained from BD Biosciences). For determination of BrdU incorporation in vivo, mice were administered two 1-mg intraperitoneal injections of BrdU with a 2-h interval. The thymus was removed 1 h after the second injection, and BrdU incorporation was evaluated with a BrdU flow kit (Becton Dickinson). All analyses were performed with FACSCalibur or FACSAria instruments (Becton Dickinson).
RT and real-time PCR analysis.
Total RNA was extracted from thymocytes or stimulated splenic T cells by the guanidinium thiocyanate–phenol-chloroform method, purified, and subjected (1 μg) to complementary DNA (cDNA) synthesis with random hexanucleotide primers (ReverTra Ace α; Toyobo). The cDNA was added to a quantitative PCR mixture that contained 1× SYBR green PCR Master Mix (Applied Biosystems) and 200 nM of gene-specific primers. Assays were performed in triplicate with a sequence detector (ABI Prism 7700; Applied Biosystems). The PCR protocol included incubation at 60°C for 30 s and 95°C for 5 s, followed by 40 cycles. The sequences of PCR primers were as follows: 5′-GGACCCGAGAAGACCTCCTT-3′ (sense) and 5′-GCACATCACTCAGAATTTCAATGG-3′ (antisense) for acidic ribosomal phosphoprotein P0 (ARBP; a gift from Y. Ishikawa, Tohoku University, Sendai, Japan); 5′-GCCTGGAGAAACCTGCCAAGTATG-3′ (sense) and 5′-GAGTGGGAGTTGCTGTTGAAGTCG-3′ (antisense) for GAPDH; 5′-TGCAAAGTCTCAGATTATACC-3′ (sense) and 5′-ACTTCTCTGGTCCGCTCCAGC-3′ (antisense) for Fbxw7; 5′-TGTCTGAGCGGCCTGAAGATTC-3′ (sense) and 5′-GCAGAAGACCAATCTGCGCTTG-3′ (antisense) for p21; 5′-TTTCATCCAGGATCGAGCAGGG-3′ (sense) and 5′-GTCCAGTTCATCTCCAATTCGCC-3′ (antisense) for Bax; and 5′-ACTCAGGAAGATCGGAGACAAAGTG-3′ (sense) and 5′-ACACTCGTCCTTCAAGTCTGCTGG-3′ (antisense) for Noxa. Primers for p21, Bax, and Noxa were gifts from M. Nishiyama (Kyushu University, Fukuoka, Japan). Reactions for ARBP or GAPDH mRNAs were performed concurrently on the same plate as those for the test mRNAs, and results were normalized by the corresponding amounts of ARBP or GAPDH mRNAs.
Immunoblot analysis.
Total protein extracts were prepared from thymocytes or splenic T cells of 8-wk-old mice with radioimmunoprecipitation buffer. The 30-μg protein extracts were subjected to immunoblot analysis, as previously described (53), with anti-Notch3 (M-20), anti–c-Myc (N-262), anti–cyclin E (M-20), anti–cyclin A (H-432), or anti-p53 (Pab240; all obtained from Santa Cruz Biotechnology, Inc.); anti–cleaved Notch1 (Val1744), anti–Ser63-phosphorylated c-Jun II, anti-JNK, anti–phospho-JNK (Thr183, Tyr185), anti-ERK (p44/42 mitogen-activated protein kinase), or anti–phospho-ERK (Thr202, Tyr204; all obtained from Cell Signaling Technology); anti–Aurora A, anti–phospho-Lck (Tyr505), or anti-p27 (all obtained from BD Biosciences); or anti-Lck (3A5; Sigma-Aldrich). As a control, each membrane was stripped and probed with anti-Hsp70 or anti-Hsp90 (BD Biosciences).
Confirmation of clonal expansion by Southern blot analysis.
The genomic DNA prepared from normal thymocytes or thymic lymphoma cells was digested with BamHI and probed with a 2.1-kb EcoRI fragment of the TCR-Jβ2 locus (a gift from T. Sato, Tokai University, Isehara, Japan).
T cell proliferation assay.
Splenic T cells were purified to ∼90% homogeneity from 8-wk-old mice with the use of a T cell enrich column (R&D Systems). 105 purified T cells were stimulated in triplicate with plate-bound anti-CD3ε (145-2C11) at various concentrations in 96-well plates containing RPMI 1640 medium supplemented with 10% FBS. The cells were stimulated for 48 h and exposed to 0.5 μCi [3H]thymidine per well for the final 6 h of incubation, after which the incorporation of [3H]thymidine was measured with a scintillation counter (1205 Betaplate; Wallac). In other experiments, the cells were incubated with 10 μM BrdU for the last 1 h and stained with FITC-labeled anti-BrdU (BD Biosciences).
Transplantation experiments.
5 × 106 lymphoma cells were collected, resuspended in PBS, and injected intraperitoneally into nude mice in a total volume of 0.5 ml. Mice were evaluated for clinical symptoms of malignancy (tachypnea, palpable lymphadenopathy, or ascites) and killed when an obvious tumor burden was detected. All animal experiments were approved by the appropriate institutional review committee.
Statistical analysis.
Data are presented as means ± SD and were analyzed by the Student's t test. P < 0.05 was considered statistically significant.
Online supplemental material.
Fig. S1 depicts the generation of mice with T cell–specific Fbxw7 deficiency. Fig. S2 shows that the loss of Fbxw7 does not affect spontaneous, anti-CD3–induced, or glucocorticoid-induced cell death. In Fig. S3, degradation of Notch1, Notch3, and c-Myc is delayed in DP thymocytes of Lck-Cre/Fbxw7F/F mice. Fig. S4 depicts the tumorigenicity of lymphoma cells from Lck-Cre/Fbxw7F/F mice in nude mice. In Fig. S5, there is no difference in phenotype between early- and late-onset tumors of Lck-Cre/Fbxw7F/F mice. Fig. S6 shows that Lck-Cre/Fbxw7F/F/p53−/− mice develop thymic lymphomas derived from DP thymocytes. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20062299/DC1.
Supplemental Material
Acknowledgments
We thank D.R. Littman for pL2-Neo(2), MC1-tk, and pMC-Cre plasmids; C.B. Wilson for CD4-Cre transgenic mice; T. Honjo for RBP-JF/F mice; I. Taniuchi for advice on vector construction; T. Sato for the probe for the TCR-Jβ2 genomic region; M. Nishiyama for p21, Bax, and Noxa primers for real-time PCR; Y. Ishikawa for ARBP primers for real-time PCR; Y. Yamada, K. Oyamada, and N. Nishimura for technical assistance; E. Susaki, Y. Kotake, M. Yada, and members of our laboratories for comments on the manuscript; and A. Ohta and M. Kimura for help in preparing the manuscript.
The authors have no conflicting financial interests.
Abbreviations used: ARBP, acidic ribosomal phosphoprotein P0; DN, double negative; DP, double positive; ERK, extracellular signal-regulated kinase; ES, embryonic stem; JNK, c-Jun N-terminal kinase; mRNA, messenger RNA; SP, single positive.
References
- 1.Conlon, I., and M. Raff. 1999. Size control in animal development. Cell. 96:235–244. [DOI] [PubMed] [Google Scholar]
- 2.Eilers, M., S. Schirm, and J.M. Bishop. 1991. The MYC protein activates transcription of the alpha-prothymosin gene. EMBO J. 10:133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwood, E.M., L. Kretzner, and R.N. Eisenman. 1992. Myc and Max function as a nucleoprotein complex. Curr. Opin. Genet. Dev. 2:227–235. [DOI] [PubMed] [Google Scholar]
- 4.Yada, M., S. Hatakeyama, T. Kamura, M. Nishiyama, R. Tsunematsu, H. Imaki, N. Ishida, F. Okumura, K. Nakayama, and K.I. Nakayama. 2004. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 23:2116–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welcker, M., A. Orian, J. Jin, J.A. Grim, J.W. Harper, R.N. Eisenman, and B.E. Clurman. 2004. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. USA. 101:9085–9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahram, F., N. von der Lehr, C. Cetinkaya, and L.G. Larsson. 2000. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood. 95:2104–2110. [PubMed] [Google Scholar]
- 7.Grandori, C., S.M. Cowley, L.P. James, and R.N. Eisenman. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16:653–699. [DOI] [PubMed] [Google Scholar]
- 8.Mao, J.H., J. Perez-Losada, D. Wu, R. Delrosario, R. Tsunematsu, K.I. Nakayama, K. Brown, S. Bryson, and A. Balmain. 2004. Fbxw7/Cdc4 is a p53-dependentw, haploinsufficient tumour suppressor gene. Nature. 432:775–779. [DOI] [PubMed] [Google Scholar]
- 9.Nakayama, K.I., and K. Nakayama. 2006. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer. 6:369–381. [DOI] [PubMed] [Google Scholar]
- 10.Tsunematsu, R., K. Nakayama, Y. Oike, M. Nishiyama, N. Ishida, S. Hatakeyama, Y. Bessho, R. Kageyama, T. Suda, and K.I. Nakayama. 2004. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J. Biol. Chem. 279:9417–9423. [DOI] [PubMed] [Google Scholar]
- 11.Tetzlaff, M.T., W. Yu, M. Li, P. Zhang, M. Finegold, K. Mahon, J.W. Harper, R.J. Schwartz, and S.J. Elledge. 2004. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc. Natl. Acad. Sci. USA. 101:3338–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellmeier, W., S. Sawada, and D.R. Littman. 1999. The regulation of CD4 and CD8 coreceptor gene expression during T cell development. Annu. Rev. Immunol. 17:523–554. [DOI] [PubMed] [Google Scholar]
- 13.Tanigaki, K., M. Tsuji, N. Yamamoto, H. Han, J. Tsukada, H. Inoue, M. Kubo, and T. Honjo. 2004. Regulation of alphabeta/gammadelta T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 20:611–622. [DOI] [PubMed] [Google Scholar]
- 14.Douglas, N.C., H. Jacobs, A.L. Bothwell, and A.C. Hayday. 2001. Defining the specific physiological requirements for c-Myc in T cell development. Nat. Immunol. 2:307–315. [DOI] [PubMed] [Google Scholar]
- 15.Spencer, C.A., and M. Groudine. 1991. Control of c-myc regulation in normal and neoplastic cells. Adv. Cancer Res. 56:1–48. [DOI] [PubMed] [Google Scholar]
- 16.Heikkila, R., G. Schwab, E. Wickstrom, S.L. Loke, D.H. Pluznik, R. Watt, and L.M. Neckers. 1987. A c-myc antisense oligodeoxynucleotide inhibits entry into S phase but not progress from G0 to G1. Nature. 328:445–449. [DOI] [PubMed] [Google Scholar]
- 17.de Alboran, I.M., R.C. O'Hagan, F. Gartner, B. Malynn, L. Davidson, R. Rickert, K. Rajewsky, R.A. DePinho, and F.W. Alt. 2001. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 14:45–55. [DOI] [PubMed] [Google Scholar]
- 18.Hann, S.R., C.B. Thompson, and R.N. Eisenman. 1985. c-myc oncogene protein synthesis is independent of the cell cycle in human and avian cells. Nature. 314:366–369. [DOI] [PubMed] [Google Scholar]
- 19.Mateyak, M.K., A.J. Obaya, S. Adachi, and J.M. Sedivy. 1997. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 8:1039–1048. [PubMed] [Google Scholar]
- 20.Pelengaris, S., and M. Khan. 2003. The many faces of c-MYC. Arch. Biochem. Biophys. 416:129–136. [DOI] [PubMed] [Google Scholar]
- 21.Dang, C.V., K.A. O'Donnell, and T. Juopperi. 2005. The great MYC escape in tumorigenesis. Cancer Cell. 8:177–178. [DOI] [PubMed] [Google Scholar]
- 22.Zindy, F., C.M. Eischen, D.H. Randle, T. Kamijo, J.L. Cleveland, C.J. Sherr, and M.F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eischen, C.M., J.D. Weber, M.F. Roussel, C.J. Sherr, and J.L. Cleveland. 1999. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 13:2658–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baudino, T.A., K.H. Maclean, J. Brennan, E. Parganas, C. Yang, A. Aslanian, J.A. Lees, C.J. Sherr, M.F. Roussel, and J.L. Cleveland. 2003. Myc-mediated proliferation and lymphomagenesis, but not apoptosis, are compromised by E2f1 loss. Mol. Cell. 11:905–914. [DOI] [PubMed] [Google Scholar]
- 25.Leone, G., R. Sears, E. Huang, R. Rempel, F. Nuckolls, C.H. Park, P. Giangrande, L. Wu, H.I. Saavedra, S.J. Field, et al. 2001. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol. Cell. 8:105–113. [DOI] [PubMed] [Google Scholar]
- 26.Juin, P., A.O. Hueber, T. Littlewood, and G. Evan. 1999. c-Myc-induced sensitization to apoptosis is mediated through cytochrome c release. Genes Dev. 13:1367–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egle, A., A.W. Harris, P. Bouillet, and S. Cory. 2004. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc. Natl. Acad. Sci. USA. 101:6164–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kersh, G.J. 2004. Transcriptional control of thymocyte positive selection. Immunol. Res. 29:125–138. [DOI] [PubMed] [Google Scholar]
- 29.Laky, K., and B.J. Fowlkes. 2005. Receptor signals and nuclear events in CD4 and CD8 T cell lineage commitment. Curr. Opin. Immunol. 17:116–121. [DOI] [PubMed] [Google Scholar]
- 30.Kwak, E.L., K.H. Moberg, D.C. Wahrer, J.E. Quinn, P.M. Gilmore, C.A. Graham, I.K. Hariharan, D.P. Harkin, D.A. Haber, and D.W. Bell. 2005. Infrequent mutations of Archipelago (hAGO, hCDC4, Fbw7) in primary ovarian cancer. Gynecol. Oncol. 98:124–128. [DOI] [PubMed] [Google Scholar]
- 31.Ekholm-Reed, S., C.H. Spruck, O. Sangfelt, F. van Drogen, E. Mueller-Holzner, M. Widschwendter, A. Zetterberg, and S.I. Reed. 2004. Mutation of hCDC4 leads to cell cycle deregulation of cyclin E in cancer. Cancer Res. 64:795–800. [DOI] [PubMed] [Google Scholar]
- 32.Strohmaier, H., C.H. Spruck, P. Kaiser, K.A. Won, O. Sangfelt, and S.I. Reed. 2001. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 413:316–322. [DOI] [PubMed] [Google Scholar]
- 33.Cassia, R., G. Moreno-Bueno, S. Rodriguez-Perales, D. Hardisson, J.C. Cigudosa, and J. Palacios. 2003. Cyclin E gene (CCNE) amplification and hCDC4 mutations in endometrial carcinoma. J. Pathol. 201:589–595. [DOI] [PubMed] [Google Scholar]
- 34.Spruck, C.H., H. Strohmaier, O. Sangfelt, H.M. Muller, M. Hubalek, E. Muller-Holzner, C. Marth, M. Widschwendter, and S.I. Reed. 2002. hCDC4 gene mutations in endometrial cancer. Cancer Res. 62:4535–4539. [PubMed] [Google Scholar]
- 35.Moberg, K.H., D.W. Bell, D.C. Wahrer, D.A. Haber, and I.K. Hariharan. 2001. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature. 413:311–316. [DOI] [PubMed] [Google Scholar]
- 36.Rajagopalan, H., P.V. Jallepalli, C. Rago, V.E. Velculescu, K.W. Kinzler, B. Vogelstein, and C. Lengauer. 2004. Inactivation of hCDC4 can cause chromosomal instability. Nature. 428:77–81. [DOI] [PubMed] [Google Scholar]
- 37.Spruck, C.H., K.A. Won, and S.I. Reed. 1999. Deregulated cyclin E induces chromosome instability. Nature. 401:297–300. [DOI] [PubMed] [Google Scholar]
- 38.Koepp, D.M., L.K. Schaefer, X. Ye, K. Keyomarsi, C. Chu, J.W. Harper, and S.J. Elledge. 2001. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 294:173–177. [DOI] [PubMed] [Google Scholar]
- 39.Felsher, D.W., and J.M. Bishop. 1999. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell. 4:199–207. [DOI] [PubMed] [Google Scholar]
- 40.Smith, D.P., M.L. Bath, A.W. Harris, and S. Cory. 2005. T-cell lymphomas mask slower developing B-lymphoid and myeloid tumours in transgenic mice with broad haemopoietic expression of MYC. Oncogene. 24:3544–3553. [DOI] [PubMed] [Google Scholar]
- 41.Blyth, K., A. Terry, M. O'Hara, E.W. Baxter, M. Campbell, M. Stewart, L.A. Donehower, D.E. Onions, J.C. Neil, and E.R. Cameron. 1995. Synergy between a human c-myc transgene and p53 null genotype in murine thymic lymphomas: contrasting effects of homozygous and heterozygous p53 loss. Oncogene. 10:1717–1723. [PubMed] [Google Scholar]
- 42.Elson, A., C. Deng, J. Campos-Torres, L.A. Donehower, and P. Leder. 1995. The MMTV/c-myc transgene and p53 null alleles collaborate to induce T-cell lymphomas, but not mammary carcinomas in transgenic mice. Oncogene. 11:181–190. [PubMed] [Google Scholar]
- 43.Trumpp, A., Y. Refaeli, T. Oskarsson, S. Gasser, M. Murphy, G.R. Martin, and J.M. Bishop. 2001. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 414:768–773. [DOI] [PubMed] [Google Scholar]
- 44.Wilson, A., M.J. Murphy, T. Oskarsson, K. Kaloulis, M.D. Bettess, G.M. Oser, A.C. Pasche, C. Knabenhans, H.R. Macdonald, and A. Trumpp. 2004. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 18:2747–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, L., V. Camerini, T.P. Bender, and K.S. Ravichandran. 2002. A nonredundant role for the adapter protein Shc in thymic T cell development. Nat. Immunol. 3:749–755. [DOI] [PubMed] [Google Scholar]
- 46.Buckley, A.F., C.T. Kuo, and J.M. Leiden. 2001. Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc–dependent pathway. Nat. Immunol. 2:698–704. [DOI] [PubMed] [Google Scholar]
- 47.Grumont, R., P. Lock, M. Mollinari, F.M. Shannon, A. Moore, and S. Gerondakis. 2004. The mitogen-induced increase in T cell size involves PKC and NFAT activation of Rel/NF-kappaB-dependent c-myc expression. Immunity. 21:19–30. [DOI] [PubMed] [Google Scholar]
- 48.Gu, H., Y.R. Zou, and K. Rajewsky. 1993. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 73:1155–1164. [DOI] [PubMed] [Google Scholar]
- 49.Nakayama, K., N. Ishida, M. Shirane, A. Inomata, T. Inoue, N. Shishido, I. Horii, D.Y. Loh, and K.I. Nakayama. 1996. Mice lacking p27Kip1 display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell. 85:707–720. [DOI] [PubMed] [Google Scholar]
- 50.Nakayama, K., H. Nagahama, Y.A. Minamishima, M. Matsumoto, I. Nakamichi, K. Kitagawa, M. Shirane, R. Tsunematsu, T. Tsukiyama, N. Ishida, et al. 2000. Targeted disruption of Skp2 results in accumulation of cyclin E and p27Kip1, polyploidy and centrosome overduplication. EMBO J. 19:2069–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolfer, A., A. Wilson, M. Nemir, H.R. MacDonald, and F. Radtke. 2002. Inactivation of Notch1 impairs VDJβ rearrangement and allows pre-TCR-independent survival of early αβ lineage thymocytes. Immunity. 16:869–879. [DOI] [PubMed] [Google Scholar]
- 52.Wolfer, A., T. Bakker, A. Wilson, M. Nicolas, V. Ioannidis, D.R. Littman, P.P. Lee, C.B. Wilson, W. Held, H.R. MacDonald, and F. Radtke. 2001. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nat. Immunol. 2:235–241. [DOI] [PubMed] [Google Scholar]
- 53.Kamura, T., T. Hara, M. Matsumoto, N. Ishida, F. Okumura, S. Hatakeyama, M. Yoshida, K. Nakayama, and K.I. Nakayama. 2004. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27Kip1 at G1 phase. Nat. Cell Biol. 6:1229–1235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.