Abstract
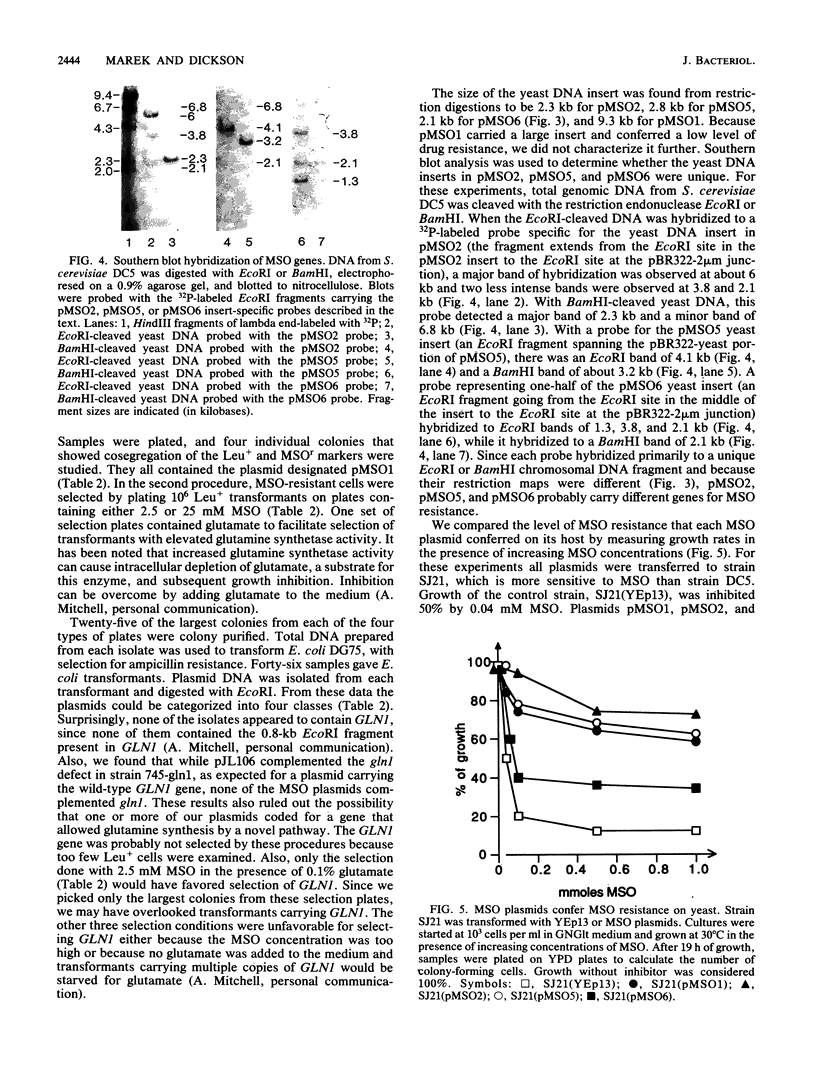
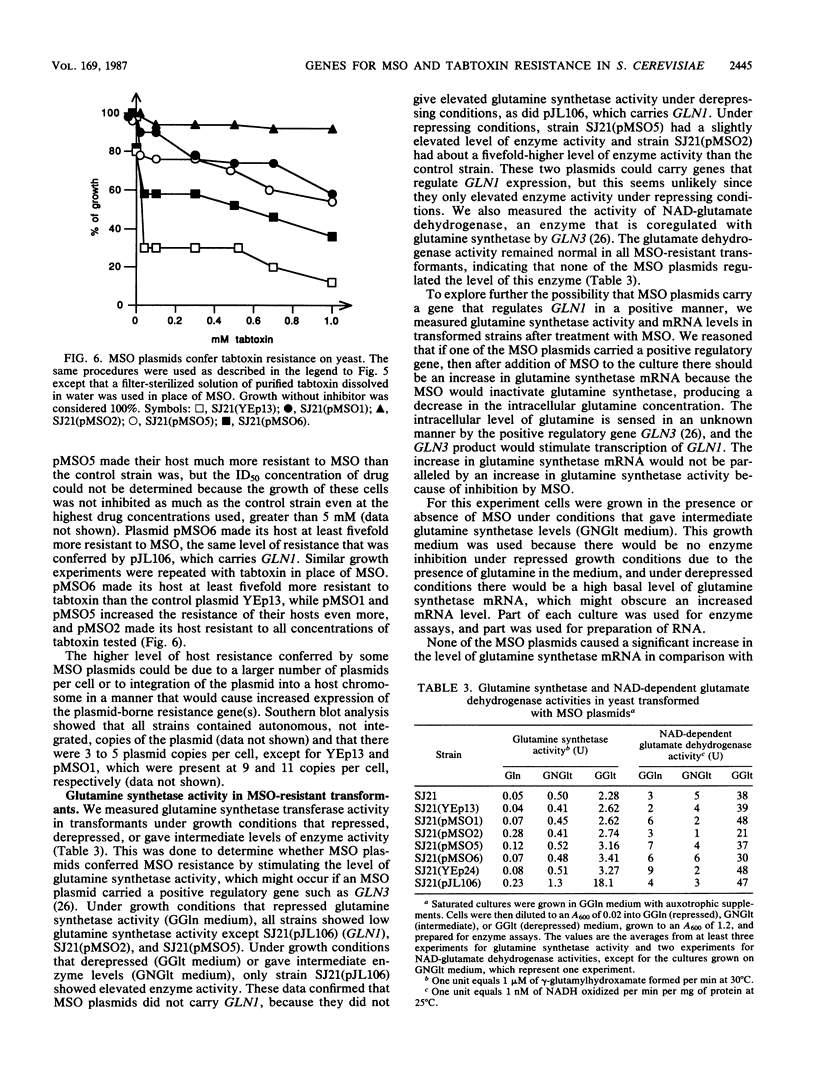
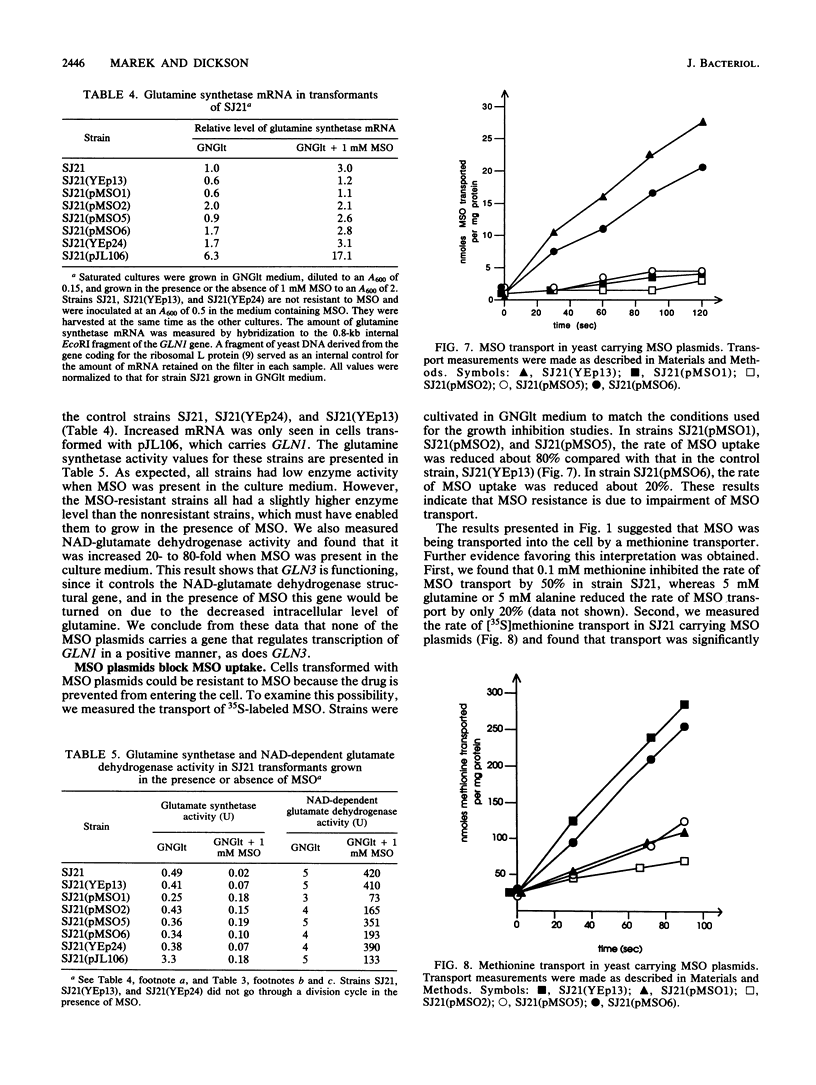
Pseudomonas tabaci produces a toxin, tabtoxin, that causes wildfire disease in tobacco. The primary target of tabtoxin is presumed to be glutamine synthetase. Some effects of tabtoxin in tobacco can be mimicked by methionine sulfoximine (MSO), a compound that is known to inactivate glutamine synthetase. To understand how organisms can be made resistant to tabtoxin and MSO, we used Saccharomyces cerevisiae. We demonstrate that yeast strains carrying the glutamine synthetase gene, GLN1, on a multicopy plasmid overproduced glutamine synthetase and showed increased drug resistance. These and other data indicate that glutamine synthetase is the primary target of tabtoxin and MSO in S. cerevisiae. We also isolated three S. cerevisiae DNA inserts of 2.1, 2.3, and 2.8 kilobases that conferred tabtoxin and MSO resistance when the inserts were present on a multicopy plasmid. These plasmids conferred resistance to MSO by blocking intracellular transport of the drug. Transport appeared to occur by one or more methionine permeases. Resistance to tabtoxin could also occur by blockage of intracellular transport, but the drug was transported by some permease other than a methionine permease. These drug resistance plasmids did not block transport of citrulline, indicating that they did not affect the general amino acid permease.
Full text
PDF



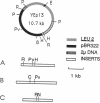
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BENTLEY H. R., McDERMOTT E. E., WHITEHEAD J. K. Action of nitrogen trichloride on certain proteins. II. Synthesis of methionine sulphoximine and other sulphoximines. Proc R Soc Lond B Biol Sci. 1951 Jun;138(891):265–272. doi: 10.1098/rspb.1951.0021. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Carlson P. S. Methionine sulfoximine--resistant mutants of tobacco. Science. 1973 Jun 29;180(4093):1366–1368. doi: 10.1126/science.180.4093.1366. [DOI] [PubMed] [Google Scholar]
- Donn G., Tischer E., Smith J. A., Goodman H. M. Herbicide-resistant alfalfa cells: an example of gene amplification in plants. J Mol Appl Genet. 1984;2(6):621–635. [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Frantz T. A., Peterson D. M., Durbin R. D. Sources of ammonium in oat leaves treated with tabtoxin or methionine sulfoximine. Plant Physiol. 1982 Feb;69(2):345–348. doi: 10.1104/pp.69.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H. M., Warner J. R. Cloning of yeast gene for trichodermin resistance and ribosomal protein L3. Proc Natl Acad Sci U S A. 1981 Jan;78(1):238–242. doi: 10.1073/pnas.78.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gits J. J., Grenson M. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. 3. Evidence for a specific methionine-transporting system. Biochim Biophys Acta. 1967 Jul 3;135(3):507–516. doi: 10.1016/0005-2736(67)90040-5. [DOI] [PubMed] [Google Scholar]
- Grenson M., Hou C., Crabeel M. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. IV. Evidence for a general amino acid permease. J Bacteriol. 1970 Sep;103(3):770–777. doi: 10.1128/jb.103.3.770-777.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. A., Hopper J. E. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6971–6975. doi: 10.1073/pnas.79.22.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Manning J. M., Moore S., Rowe W. B., Meister A. Identification of L-methionine S-sulfoximine as the diastereoisomer of L-methionine SR-sulfoximine that inhibits glutamine synthetase. Biochemistry. 1969 Jun;8(6):2681–2685. doi: 10.1021/bi00834a066. [DOI] [PubMed] [Google Scholar]
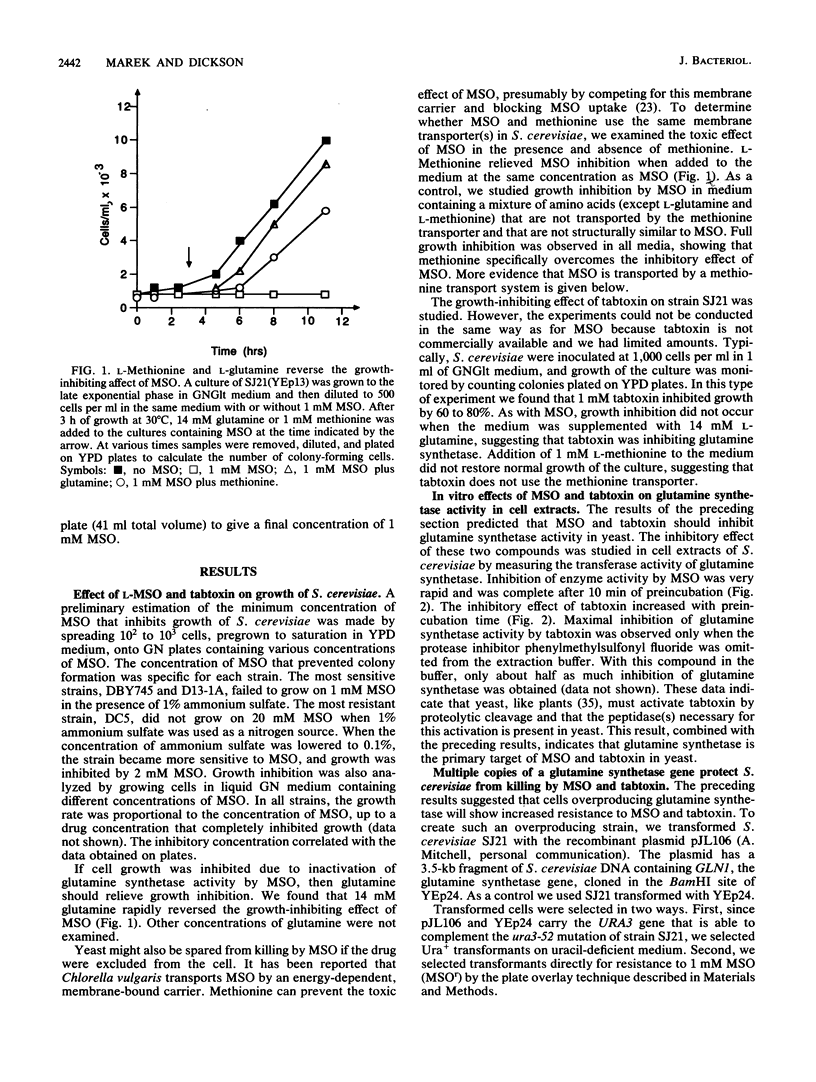
- Meins F., Jr, Abrams M. L. How methionine and glutamine prevent inhibition of growth by methionine sulfoximine. Biochim Biophys Acta. 1972 Apr 14;266(1):307–311. doi: 10.1016/0005-2736(72)90146-0. [DOI] [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B. Purification and properties of glutamine synthetase from Saccharomyces cerevisiae. J Biol Chem. 1983 Jan 10;258(1):119–124. [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B. Regulation of glutamine-repressible gene products by the GLN3 function in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2758–2766. doi: 10.1128/mcb.4.12.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. P. The GLN1 locus of Saccharomyces cerevisiae encodes glutamine synthetase. Genetics. 1985 Oct;111(2):243–258. doi: 10.1093/genetics/111.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
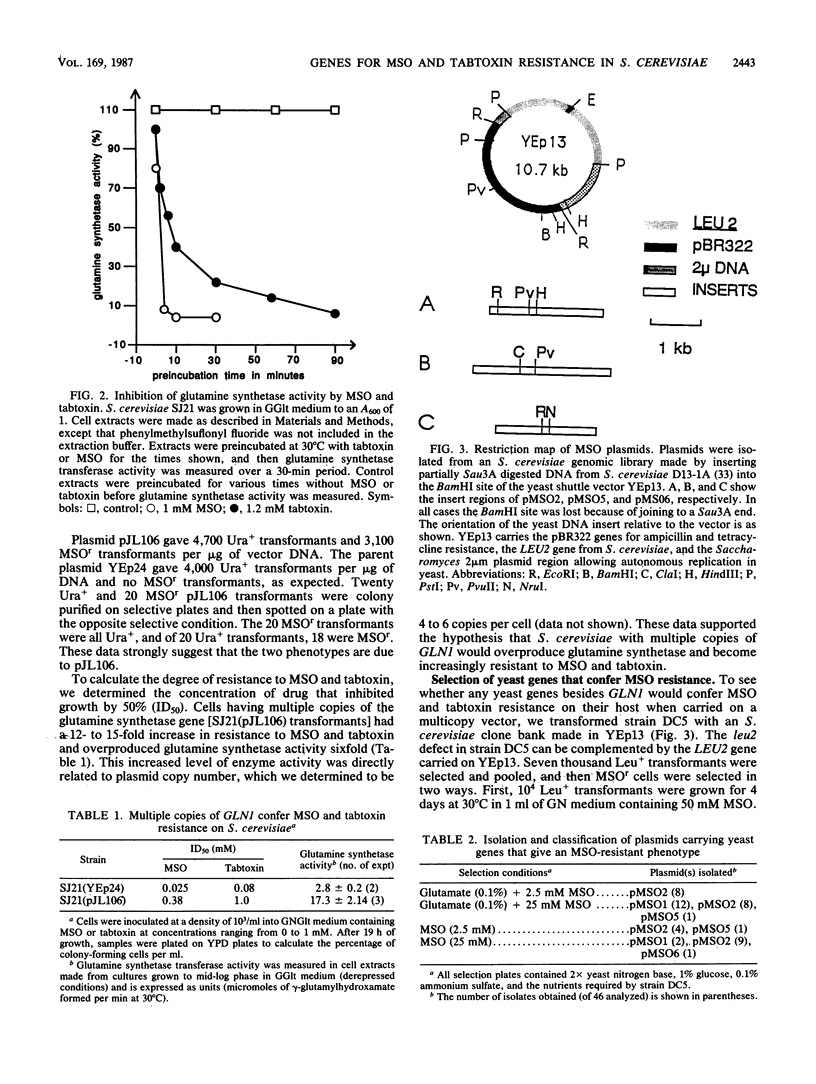
- Rine J., Hansen W., Hardeman E., Davis R. W. Targeted selection of recombinant clones through gene dosage effects. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6750–6754. doi: 10.1073/pnas.80.22.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytka J. Positive selection of general amino acid permease mutants in Saccharomyces cerevisiae. J Bacteriol. 1975 Feb;121(2):562–570. doi: 10.1128/jb.121.2.562-570.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders P. G., Wilson R. H. Amplification and cloning of the Chinese hamster glutamine synthetase gene. EMBO J. 1984 Jan;3(1):65–71. doi: 10.1002/j.1460-2075.1984.tb01762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden S. L., Durbin R. D. Glutamine synthetase inhibition: possible mode of action of wildfire toxin from Pseudomonas tabaci. Nature. 1968 Jul 27;219(5152):379–380. doi: 10.1038/219379a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stewart W. W. Isolation and proof of structure of wildfire toxin. Nature. 1971 Jan 15;229(5281):174–178. doi: 10.1038/229174a0. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A. P., Ringold G. M. Mouse 3T6 cells that overproduce glutamine synthetase. J Biol Chem. 1983 Sep 25;258(18):11260–11266. [PubMed] [Google Scholar]