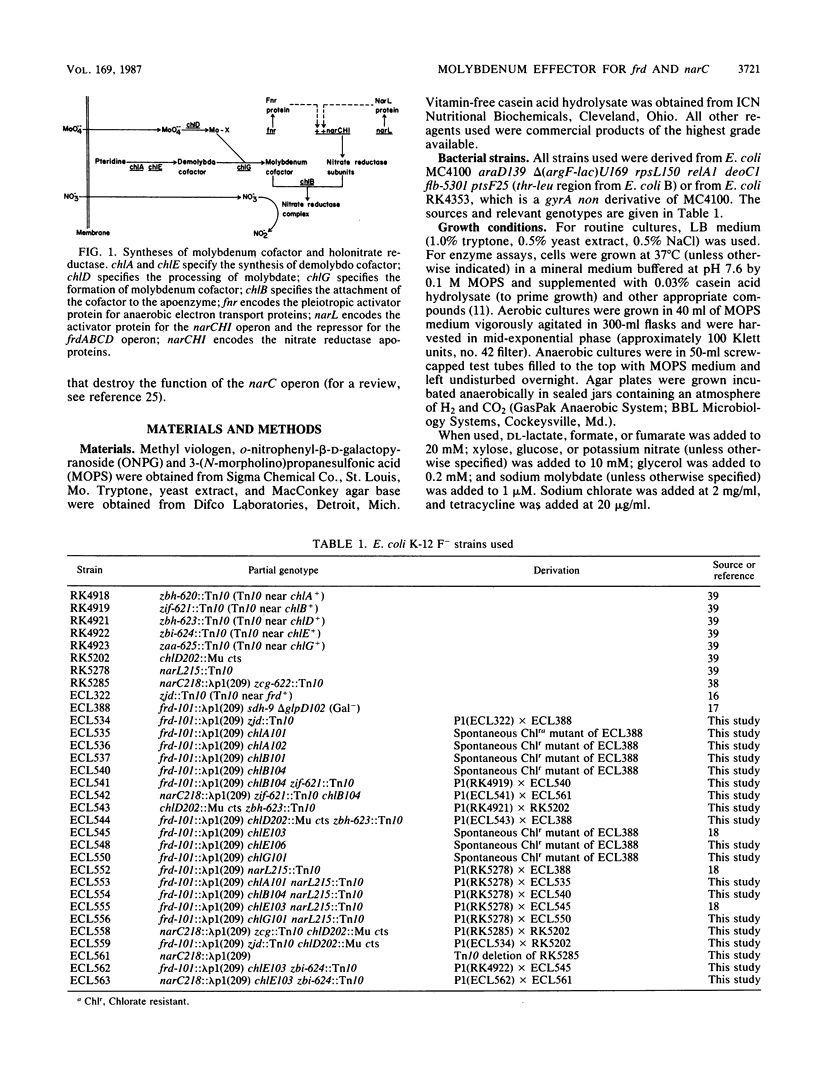
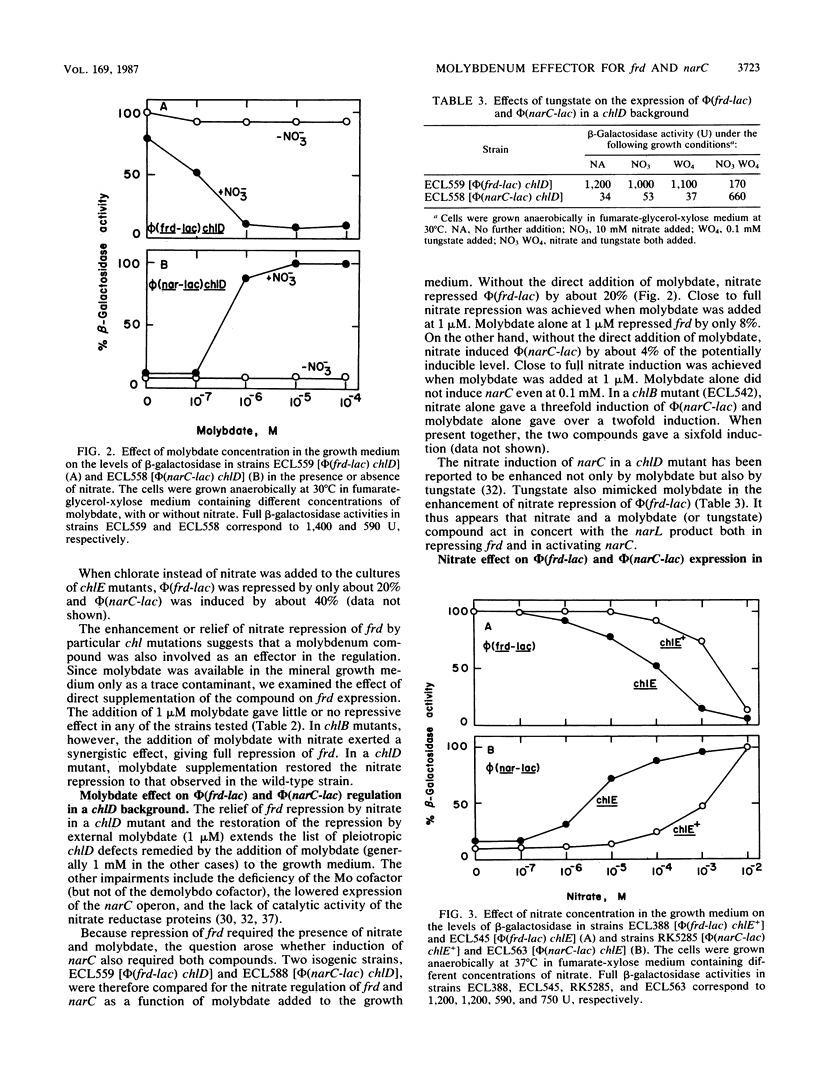
Abstract
In Escherichia coli the presence of nitrate prevents the utilization of fumarate as an anaerobic electron acceptor. The induction of the narC operon encoding the nitrate reductase is coupled to the repression of the frd operon encoding the fumarate reductase. This coupling is mediated by nitrate as an effector and the narL product as the regulatory protein (S. Iuchi and E. C. C. Lin, Proc. Natl. Acad. Sci. USA 84:3901-3905, 1987). The protein-ligand complex appears to control narC positively but frd negatively. In the present study we found that a molybdenum coeffector acted synergistically with nitrate in the regulation of frd and narC. In chlD mutants believed to be impaired in molybdate transport (or processing), full repression of phi(frd-lac) and full induction of phi(narC-lac) by nitrate did not occur unless the growth medium was directly supplemented with molybdate (1 microM). This requirement was not clearly manifested in wild-type cells, apparently because it was met by the trace quantities of molybdate present as a contaminant in the mineral medium. In chlB mutants, which are known to accumulate the Mo cofactor because of its failure to be inserted as a prosthetic group into proteins such as nitrate reductase, nitrate repression of frd and induction of narC were also intensified by molybdate supplementation. In this case a deficiency of the molybdenum coeffector might have resulted from enhanced feedback inhibition of molybdate transport (or processing) by the elevated level of the unutilized Mo cofactor. In addition, mutations in chlE, which are known to block the synthesis of the organic moiety of the Mo cofactor, lowered the threshold concentration of nitrate (< 1 micromole) necessary for frd repression and narC induction. These changes could be explained simply by the higher intracellular nitrate attainable in cells lacking the ability to destroy the effector.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amy N. K. Identification of the molybdenum cofactor in chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1981 Oct;148(1):274–282. doi: 10.1128/jb.148.1.274-282.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett E. L., Jackson C. E., Fukumoto H. T., Chang G. W. Formate dehydrogenase mutants of Salmonella typhimurium: a new medium for their isolation and new mutant classes. Mol Gen Genet. 1979;177(1):95–101. doi: 10.1007/BF00267258. [DOI] [PubMed] [Google Scholar]
- Bonnefoy-Orth V., Lepelletier M., Pascal M. C., Chippaux M. Nitrate reductase and cytochrome bnitrate reductase structural genes as parts of the nitrate reductase operon. Mol Gen Genet. 1981;181(4):535–540. doi: 10.1007/BF00428749. [DOI] [PubMed] [Google Scholar]
- Chippaux M., Giudici D., Abou-Jaoudé A., Casse F., Pascal M. C. Laboratoire de Chimie Bactérienne C.N.R.S., Marsielle, France. Mol Gen Genet. 1978 Apr 6;160(2):225–229. doi: 10.1007/BF00267485. [DOI] [PubMed] [Google Scholar]
- Cole J. A., Wimpenny J. W. Metabolic pathways for nitrate reduction in Escherichia coli. Biochim Biophys Acta. 1968 Jul 16;162(1):39–48. doi: 10.1016/0005-2728(68)90212-0. [DOI] [PubMed] [Google Scholar]
- Cole S. T., Grundström T., Jaurin B., Robinson J. J., Weiner J. H. Location and nucleotide sequence of frdB, the gene coding for the iron-sulphur protein subunit of the fumarate reductase of Escherichia coli. Eur J Biochem. 1982 Aug;126(1):211–216. doi: 10.1111/j.1432-1033.1982.tb06768.x. [DOI] [PubMed] [Google Scholar]
- Cole S. T. Nucleotide sequence coding for the flavoprotein subunit of the fumarate reductase of Escherichia coli. Eur J Biochem. 1982 Mar 1;122(3):479–484. doi: 10.1111/j.1432-1033.1982.tb06462.x. [DOI] [PubMed] [Google Scholar]
- Edwards E. S., Rondeau S. S., DeMoss J. A. chlC (nar) operon of Escherichia coli includes structural genes for alpha and beta subunits of nitrate reductase. J Bacteriol. 1983 Mar;153(3):1513–1520. doi: 10.1128/jb.153.3.1513-1520.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forage R. G., Lin E. C. DHA system mediating aerobic and anaerobic dissimilation of glycerol in Klebsiella pneumoniae NCIB 418. J Bacteriol. 1982 Aug;151(2):591–599. doi: 10.1128/jb.151.2.591-599.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundström T., Jaurin B. Overlap between ampC and frd operons on the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1111–1115. doi: 10.1073/pnas.79.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HIRSCH C. A., RASMINSKY M., DAVIS B. D., LIN E. C. A FUMARATE REDUCTASE IN ESCHERICHIA COLI DISTINCT FROM SUCCINATE DEHYDROGENASE. J Biol Chem. 1963 Nov;238:3770–3774. [PubMed] [Google Scholar]
- Imperial J., Ugalde R. A., Shah V. K., Brill W. J. Mol- mutants of Klebsiella pneumoniae requiring high levels of molybdate for nitrogenase activity. J Bacteriol. 1985 Sep;163(3):1285–1287. doi: 10.1128/jb.163.3.1285-1287.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingledew W. J., Poole R. K. The respiratory chains of Escherichia coli. Microbiol Rev. 1984 Sep;48(3):222–271. doi: 10.1128/mr.48.3.222-271.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Kuritzkes D. R., Lin E. C. Escherichia coli mutant with altered respiratory control of the frd operon. J Bacteriol. 1985 Mar;161(3):1023–1028. doi: 10.1128/jb.161.3.1023-1028.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Kuritzkes D. R., Lin E. C. Three classes of Escherichia coli mutants selected for aerobic expression of fumarate reductase. J Bacteriol. 1986 Dec;168(3):1415–1421. doi: 10.1128/jb.168.3.1415-1421.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Lin E. C. The narL gene product activates the nitrate reductase operon and represses the fumarate reductase and trimethylamine N-oxide reductase operons in Escherichia coli. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3901–3905. doi: 10.1073/pnas.84.11.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. E., Rajagopalan K. V. In vitro system for molybdopterin biosynthesis. J Bacteriol. 1987 Jan;169(1):110–116. doi: 10.1128/jb.169.1.110-116.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. E., Rajagopalan K. V. Involvement of chlA, E, M, and N loci in Escherichia coli molybdopterin biosynthesis. J Bacteriol. 1987 Jan;169(1):117–125. doi: 10.1128/jb.169.1.117-125.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H. M., Gunsalus R. P. Transcription of the Escherichia coli fumarate reductase genes (frdABCD) and their coordinate regulation by oxygen, nitrate, and fumarate. J Bacteriol. 1985 Dec;164(3):1100–1109. doi: 10.1128/jb.164.3.1100-1109.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuritzkes D. R., Zhang X. Y., Lin E. C. Use of phi(glp-lac) in studies of respiratory regulation of the Escherichia coli anaerobic sn-glycerol-3-phosphate dehydrogenase genes (glpAB). J Bacteriol. 1984 Feb;157(2):591–598. doi: 10.1128/jb.157.2.591-598.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lambden P. R., Guest J. R. Mutants of Escherichia coli K12 unable to use fumarate as an anaerobic electron acceptor. J Gen Microbiol. 1976 Dec;97(2):145–160. doi: 10.1099/00221287-97-2-145. [DOI] [PubMed] [Google Scholar]
- Lemire B. D., Robinson J. J., Weiner J. H. Identification of membrane anchor polypeptides of Escherichia coli fumarate reductase. J Bacteriol. 1982 Dec;152(3):1126–1131. doi: 10.1128/jb.152.3.1126-1131.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor C. H., Schnaitman C. A., Normansell D. E. Purification and properties of nitrate reductase from Escherichia coli K12. J Biol Chem. 1974 Aug 25;249(16):5321–5327. [PubMed] [Google Scholar]
- MacGregor C. H., Schnaitman C. A. Restoration of reduced nicotinamide adenine dinucleotide phosphate-nitrate reductase activity of a Neurospora mutant by extracts of various chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1972 Oct;112(1):388–391. doi: 10.1128/jb.112.1.388-391.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor C. H. Synthesis of nitrate reductase components in chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1975 Mar;121(3):1117–1121. doi: 10.1128/jb.121.3.1117-1121.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. B., Amy N. K. Molybdenum cofactor in chlorate-resistant and nitrate reductase-deficient insertion mutants of Escherichia coli. J Bacteriol. 1983 Aug;155(2):793–801. doi: 10.1128/jb.155.2.793-801.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal M. C., Burini J. F., Ratouchniak J., Chippaux M. Regulation of the nitrate reductase operon: effect of mutations in chlA, B, D and E genes. Mol Gen Genet. 1982;188(1):103–106. doi: 10.1007/BF00333001. [DOI] [PubMed] [Google Scholar]
- Pascal M. C., Chippaux M. Involvement of a gene of the chl E locus in the regulation of the nitrate reductase operon. Mol Gen Genet. 1982;185(2):334–338. doi: 10.1007/BF00330808. [DOI] [PubMed] [Google Scholar]
- Rondeau S. S., Hsu P. Y., DeMoss J. A. Construction in vitro of a cloned nar operon from Escherichia coli. J Bacteriol. 1984 Jul;159(1):159–166. doi: 10.1128/jb.159.1.159-166.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw D. J., Guest J. R. Amplification and product identification of the fnr gene of Escherichia coli. J Gen Microbiol. 1982 Oct;128(10):2221–2228. doi: 10.1099/00221287-128-10-2221. [DOI] [PubMed] [Google Scholar]
- Spencer M. E., Guest J. R. Isolation and properties of fumarate reductase mutants of Escherichia coli. J Bacteriol. 1973 May;114(2):563–570. doi: 10.1128/jb.114.2.563-570.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperl G. T., DeMoss J. A. chlD gene function in molybdate activation of nitrate reductase. J Bacteriol. 1975 Jun;122(3):1230–1238. doi: 10.1128/jb.122.3.1230-1238.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., MacGregor C. H. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J Bacteriol. 1982 Aug;151(2):788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V. Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J Bacteriol. 1982 Sep;151(3):1320–1325. doi: 10.1128/jb.151.3.1320-1325.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugalde R. A., Imperial J., Shah V. K., Brill W. J. Biosynthesis of the iron-molybdenum cofactor and the molybdenum cofactor in Klebsiella pneumoniae: effect of sulfur source. J Bacteriol. 1985 Dec;164(3):1081–1087. doi: 10.1128/jb.164.3.1081-1087.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]


