Abstract
The rapid activation and feedback regulation of many G protein signaling cascades raises the possibility that the critical signaling proteins may be tightly coupled. Previous studies show that the PDZ domain containing protein INAD, which functions in Drosophila vision, coordinates a signaling complex by binding directly to the light-sensitive ion channel, TRP, and to phospholipase C (PLC). The INAD signaling complex also includes rhodopsin, protein kinase C (PKC), and calmodulin, though it is not known whether these proteins bind to INAD. In the current work, we show that rhodopsin, calmodulin, and PKC associate with the signaling complex by direct binding to INAD. We also found that a second ion channel, TRPL, bound to INAD. Thus, most of the proteins involved directly in phototransduction appear to bind to INAD. Furthermore, we found that INAD formed homopolymers and the homomultimerization occurred through two PDZ domains. Thus, we propose that the INAD supramolecular complex is a higher order signaling web consisting of an extended network of INAD molecules through which a G protein–coupled cascade is tethered.
Keywords: rhodopsin, TRPL, calmodulin, PKC, PDZ
Receptor-mediated signal transduction, the process by which extracellular signals are transduced across the plasma membrane, is a widespread phenomenon critical for a plethora of cellular events. The proteins in signaling cascades are probably not randomly distributed, but spatiotemporarily organized in such a way to achieve efficiency and specificity. Whether signaling components are physically associated to form a signaling complex or are merely in close proximity to facilitate random collisions is not well understood. Recently, we and others provided evidence that proteins involved in Drosophila phototransduction function as a supramolecular signaling complex (Huber et al., 1996a ; Shieh and Zhu, 1996; Chevesich et al., 1997; Tsunoda et al., 1997). Inactivation no afterpotential D (INAD)1 (Shieh and Niemeyer, 1995), a protein with five tandem protein interaction modules, PDZ domains, appears to be the coordinator (for reviews see Montell, 1997, 1998; Pawson and Scott, 1997). However, all of the components that comprise this signaling complex are not known. One intriguing possibility is that INAD may function as a scaffold for most, if not all, of the proteins in this signaling pathway. However, the mechanism by which a single molecule, INAD, could nucleate an array of proteins is unclear.
PDZ domains are ∼90 amino acid modules, identified initially in PSD-95, DLG, and ZO-1 which mediate protein– protein interactions by binding to the COOH-terminal ends of their targets (Kim et al., 1995, 1996; Kornau et al., 1995; Muller et al., 1996; Hata et al., 1997; Kornau et al., 1997; Tejedor et al., 1997). The crystal structure of two PDZ domains reveals a cradle of β barrels with a conserved hydrophobic pocket and a buried arginine, suggesting a common mechanism for PDZ–target interactions (Cabral et al., 1996; Doyle et al., 1996).
Previous studies have shown that INAD binds directly to TRP (Shieh and Zhu, 1996) and the phospholipase C (PLC) (Chevesich et al., 1997) encoded by the norpA locus (Bloomquist et al., 1988). In addition, protein kinase C (PKC) (Huber et al., 1996a ,b; Tsunoda et al., 1997), calmodulin, and rhodopsin (Chevesich et al., 1997) have been shown to associate with the INAD complex. Whether these latter proteins are linked directly to INAD has not been addressed. In the current paper, we show that rhodopsin, PKC, calmodulin, and TRPL bind directly to INAD. Thus, most of the proteins critical in phototransduction appear to be coupled directly to INAD. In addition, INAD homomultimerized leading to the formation of INAD polymers. Both homomeric and target binding occurred simultaneously through the same PDZ domains indicating that the two interactions were mediated by distinct regions. This observation provided a mechanism by which a multitude of targets can be coupled to the complex through the same PDZ domains. We propose that the INAD signaling complex is composed of an array of INAD molecules to which most, if not all, of the proteins involved in phototransduction are tethered.
Materials and Methods
DNA Constructs for Expression in 293T Cells
The plasmids transfected in 293T cells were all constructed using the pcDNA3 vector (Invitrogen, Carlsbad, CA). The translation initiation codon in each construct was modified to optimize translation initiation (Kozak, 1984). Many of the constructs contained either an NH2-terminal MYC or FLAG epitope tag as indicated. The peptide sequences of the MYC and FLAG tags were MEQKLISEEDL and MDYKDDDDK, respectively. pPKC-F, pINAD, pINAD-M, and pINAD-F consisted of the full-length sequences. pTRPL is the full-length clone reported previously (Xu et al., 1997). pPLC-M consisted of the COOH-terminal 123 residues of PLC fused at the NH2 terminus to a MYC tag and the maltose binding protein. The residues in constructs containing different INAD fragments are indicated in Fig. 3 A.
Figure 3.

INAD interacted with opsin, TRPL, and PKC via either PDZ3L or PDZ4. (A) Schematic indicating the PDZ domains to which the opsin, TRPL, PKC, and PLC interacted in a coimmunoprecipitation assay after expressing the proteins in 293T cells. Those INAD fragments that coimmunoprecipitated with the opsin, TRPL, PKC, or a PLC fragment encoding the last 123 amino acids were indicated with a + whereas those that did not were indicated with a −. N/D, experiments that were not done. (B) Representative results from A indicating that TRPL coimmunoprecipitated with PDZ3L and PDZ4. A plasmid encoding the COOH-terminal end of TRPL (residues 676–1,124) was cotransfected into 293T cells with a second construct encoding INAD or portions of INAD (panel A, left) fused to a MYC tag. TRPL was immunoprecipitated from cell lysates with anti-TRPL antibodies and a Western blot of the immune complexes and total cell lysates (Input) were probed with anti-MYC antibodies. (C) Representative results summarized in A indicating that PKC bound to either PDZ3L or PDZ4. A plasmid encoding FLAG-tagged PKC (pPKC-F) was cotransfected into 293T cells along with a second construct encoding full-length or fragments of INAD fused to MYC tags. Immunoprecipitations were performed with anti-MYC antibodies and Western blots of the immune complexes and total cell lysates were probed with anti-FLAG antibodies. (D) TRPC3 and Shaker B did not interact with PDZ3-4 of INAD in 293T cells. pPDZ3.4-M (PDZ3-4 of INAD with a Myc tag) was coexpressed with either pTRPC3-F or pShB in 293T cells. Cell lysates were immunoprecipitated with anti-MYC or anti-FLAG antibodies and the Western blots were probed with anti-FLAG, anti-MYC, or anti-ShB1 antibodies. (E) Representative results showing that PLC binds to PDZ1 and not other domains such as PDZ3 and PDZ4. A plasmid encoding MYC-tagged PLC (pPLC-M) was cotransfected into 293T cells along with a second construct the various INAD forms (indicated in panel A) fused to MYC tags. Immunoprecipitations were performed with anti-MYC antibodies and Western blots of the immune complexes and total cell lysates were probed with anti-FLAG antibodies. (F) Schematic of INAD–PDZ domains and calmodulin-binding domain. The arbitrary boundaries of the five PDZ domains are indicated in amino acids.
Cell Culture and Coimmunoprecipitations
The coimmunoprecipitation of TRPL and INAD from fly heads was conducted as described (Xu et al., 1997). Fly heads (20 mg) were homogenized in 0.4 ml ice-cold SMART buffer (0.2% dodecyl-β-maltoside, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 500 mM NaCl, 5 mM EDTA, 5 mM EGTA, 2 mg/ml aprotinin, 10 mg/ml leupeptin, 0.1 mM PMSF, 10 mM NaPPi, 50 mM NaF, pH 7.3) and centrifuged at 16,000 g for 15 min to remove debris. Anti-TRPL antibodies (or nonimmune serum) and 50 μl of protein A–agarose beads supernatant was subsequently added to the supernatant and then the mixture was then rotated at 4°C for 2 h. After five washes with SMART buffer, the immunoprecipitates were eluted with SDS sample buffer, fractionated by SDS-PAGE, probed with rabbit anti-INAD antibodies and then detected by enhanced chemiluminescence (Amersham Corp., Arlington Heights, IL). The coimmunoprecipitation of rhodopsin and INAD was performed as described (Chevesich et al., 1997) using 1% CHAPS instead of 0.2% dodecyl-β-maltoside in the homogenizing buffer. The immunocomplexes were fractionated by SDS-PAGE, probed with anti-INAD antibodies, incubated with 125I-conjugated protein A, and then processed for autoradiography.
293T cells were grown in DME/FCS at 37°C, 5.5% CO2. Lipofectamine (GIBCO BRL, Gaithersburg, MD) was used in the transfections according to the manufacturer's instructions. 36–48 h after transfection, the cells from single 100-mm dishes were lysed with 1 ml of cold IPB buffer (1% Triton X-100 and protease inhibitor cocktail [Boehringer Mannheim Biochemicals, Indianapolis, IN]) in PBS and centrifuged to remove cellular debris. The subsequent immunoprecipitation protocol was similar to that described (Chevesich et al., 1997; Xu et al., 1997) using 0.5 ml of supernatant, the appropriate antibodies, and 50 μl of protein A–agarose beads or protein G–Sepharose. The mixture was then rotated for 2 h at 4°C. The proteins in the immunocomplexes were washed three times with IPB buffer with 500 ml, solubilized in SDS sample buffer, fractionated by SDS-PAGE, transferred to polyvinylene difluoride membrane, probed with the appropriate antibodies, and then detected by the enhanced chemiluminescence method (Amersham Corp.).
Calmodulin Overlay Assay
GST–INAD fusion proteins used in the overlay assay were constructed using pGEX vectors (Pharmacia Biotech., Inc., Piscataway, NJ). The fusion proteins were produced in Epicurian coli BL21(DE3)pLysS cells (Stratagene, La Jolla, CA) as described by the manufacturer. The calmodulin overlay assay was performed using biotinylated calmodulin (Life Technologies, Inc., Gaithersburg, MD) in the presence of Ca2+ as described by the manufacturer.
Glutathione Column-binding Experiments
pGST–INAD was constructed by subcloning full-length INAD into pGEX5.1 (Pharmacia Biotech., Inc.). 100 ml of bacteria culture (BL-21) was induced with IPTG and lysed by sonication after addition of 10 ml TBST (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5% Triton X-100, 1 mM PMSF, 1 mM EDTA, 1 mM DTT). After removing the debris by centrifugation, the GST and GST fusion proteins were purified using glutathione beads (Pharmacia Biotech., Inc.). 0.5 μg of purified GST–INAD or 1 μg GST (negative control) was immobilized on 20 μl of glutathione-agarose beads (Pharmacia Biotech., Inc.). [35S]methionine probes were made by coupled transcription/translation using the TNT kit (Promega Corp., Madison, WI). The in vitro translation of the opsin was performed in the presence of microsomes. Equal amounts of 35S probe were incubated in a final volume of 200 μl (TBST buffer) at 4°C for 1–2 h with the glutathione beads bound to GST–INAD or GST control. The mixture was washed three times in TBST containing 500 mM NaCl and the bound proteins were eluted with SDS sample buffer. The elutes were fractionated by SDS-PAGE and the 35S-labeled proteins were detected using a PhosphorImager (model BAS-1500; Fugix Inc., Kanagawa, Japan).
Sucrose Gradient Ultracentrifugation
INAD fragments (PDZ1-2 and PDZ3-4) were translated in vitro with [35S]methionine (25–50 μl each, TNT system; Promega Inc.) and immediately incubated on ice for 5 h and subsequently loaded on a 10 ml 5–20% linear sucrose gradient in TBS (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM DTT, and protease inhibitors) and centrifuged at 130,000 g for 18 h at 18°C with a rotor (model SW 41; Beckman Instrs., Fullerton, CA). Fractions (0.3 ml) were collected and 40 μl of each was resolved by SDS-PAGE and the 35S-labeled proteins were detected using a PhosphorImager. The intensity of individual protein bands was quantified with the PhosphorImager and plotted as a function of fraction numbers. PSL/mm2 are the units assigned arbitrarily by the PhosphorImager software. Protein size markers were catalase (250 kD), β-amylase (200 kD), BSA (66 kD), and carbonic anhydrase (29 kD).
Results
Direct Binding of Rhodopsin, TRPL, PKC, and Calmodulin to INAD
Currently, there are no examples of seven transmembrane domain receptors that interact directly with a PDZ containing protein; although, metabotropic glutamate receptors have been reported to bind to Homer, a protein with a small region which may be distantly related to PDZ domains (Brakeman et al., 1997). A suggestion that the major rhodopsin encoded by the ninaE locus (O'Tousa et al., 1985; Zuker et al., 1985) may interact with INAD is that it coimmunoprecipitated with the TRP channel from wild-type but not InaDP215 mutant fly heads (Chevesich et al., 1997). To test whether the rhodopsin interacts with INAD in vivo, we performed a coimmunoprecipitation experiment using extracts from Drosophila heads. INAD coimmunoprecipitated with rhodopsin but not in a control reaction carried out with nonimmune serum (Fig. 1 A). In addition, we coexpressed the proteins in vitro using a mammalian tissue culture system, 293T cells, and found that the ninaE opsin and INAD coimmunoprecipitated (Fig. 1 B). In a control experiment, we found that INAD did not immunoprecipitate nonspecifically with the NINAE antibodies since INAD was not detected on the Western blot after performing immunoprecipitations using extracts from 293T cells expressing INAD, but not the opsin (Fig. 1 B). Furthermore, the coimmunoprecipitation of the opsin and INAD did not appear to be due to nonspecific interaction of membrane proteins with INAD since two other membrane proteins did not coimmunoprecipitate with INAD in 293T cells. These included a human store-operated channel, TRPC3 (Fig. 1 I) (Wes et al., 1995; Zhu et al., 1996; Xu et al., 1997), and the Shaker K+ channel (Fig. 1 J) (for review see Jan and Jan, 1990), which is expressed in Drosophila photoreceptor cells (Hardie, 1991). Evidence that the association between the opsin and INAD was direct was that in vitro–translated opsin bound to GST–INAD immobilized on a glutathione–Sepharose column but not to the GST control (Fig. 1 C).
Figure 1.
INAD directly interacted with rhodopsin, TRPL, PKC, and calmodulin. (A) Rhodopsin and INAD coimmunoprecipitated from fly heads. Fly head extracts were used for immunoprecipitations with anti-rhodopsin (Rh) antibodies or nonimmune serum (NIS). A Western blot of the immune complexes was probed with rabbit anti-INAD antibodies followed by 125I- labeled protein A. Horseradish peroxidase-conjugated secondary antibodies were used in subsequent experiments (B–H). (B) INAD and opsin coimmunoprecipitated from 293T cells. pINAD (encoding full-length INAD) was transfected into 293T cells or cotransfected with pRh1 (encoding full length opsin [Op]). The cell lysates were immunoprecipitated with rabbit anti-rhodopsin antibodies or NIS and the Western blot containing the immunoprecipitates and total cell lysates (Input) was probed with anti-INAD antibodies. The volume of lysates loaded in the input lanes was 20% of that used in the corresponding lanes containing the immunoprecipitates. The same ratio was followed in subsequent experiments. (C) Opsin bound to a GST–INAD fusion protein. In vitro–translated opsin proteins labeled with [35S]methionine (Op) were incubated with a GST–INAD fusion protein or GST alone bound to glutathione–Sepharose beads. SDS sample buffer was added to the beads, the eluates were fractionated by SDS-PAGE, and then the dried gel was exposed using a PhosphorImager. (D) TRPL and INAD coimmunoprecipitated from fly heads. Immunoprecipitations were performed using fly head extracts and anti-TRPL antibodies or NIS. The immune complexes were fractionated by SDS-PAGE and a Western blot was probed with anti-INAD antibodies. (E) TRPL and INAD coimmunoprecipitated from 293T cells. INAD was expressed in 293T cells or coexpressed with TRPL and immunoprecipitations were performed with anti-TRPL antibodies. The immune complexes and total cell lysates (Input) were fractionated by SDS-PAGE and the Western blot was probed with anti-INAD antibodies. (F) TRPL bound to GST– INAD. In vitro–translated COOH-terminal TRPL labeled with 35S (C-TRPL; residues 676–1,124) was incubated with a GST–INAD fusion protein immobilized on glutathione–Sepharose beads. The bound proteins were eluted with SDS sample buffer, the eluates were fractionated by SDS-PAGE, and then the dried gel was exposed using a PhosphorImager. (G) PKC and INAD coimmunoprecipitated from 293T cells. pPKC-F (PKC with a FLAG epitope tag) and pINAD-M (INAD with a MYC tag) were cotransfected into 293T cells. Immunoprecipitations were performed with anti-MYC antibodies, and a Western blot of the immunocomplexes was probed with anti-FLAG antibodies. (H) PKC bound to GST– INAD. In vitro–translated 35S-PKC was incubated with a GST– INAD fusion protein immobilized on glutathione–Sepharose beads. The bound proteins were eluted with SDS sample buffer, fractionated by SDS-PAGE, and then exposed using a PhosphorImager. (I) TRPC3 and INAD did not coimmunoprecipitate from 293T cells. pTRPC3-F (TRPC3 with a FLAG tag) and pINAD were cotransfected in 293T cells, immunoprecipitations were carried out with anti-FLAG or anti-INAD antibodies, and Western blots were probed with either anti-INAD or anti-FLAG antibodies. To demonstrate that TRPC3 and INAD were immunoprecipitated by the primary antibodies, the same Western blots were reprobed with anti-FLAG or INAD antibodies (results are shown at bottom). (J) Shaker B and INAD did not coimmunoprecipitate from 293T cells. pShB (full-length ShB1 cDNA under the cytomegalovirus promoter; gift of M. Li, Johns Hopkins University School of Medicine, Baltimore, MD) and pINAD were coexpressed in 293T cells. Immunoprecipitations were carried out with anti-INAD antibodies and the Western blot was probed with anti-ShB antibodies (gift of M. Li). Bottom panel, the same blot reprobed with anti-INAD antibodies. (K) CaMKII did not coimmunoprecipitate with INAD from 293T cells. pCKII-M (CaMKII with a Myc tag) and pINAD were cotransfected in 293T cells, immunoprecipitations were performed with anti-INAD antibodies, and then the Western blot was probed with anti-Myc antibodies. Bottom panel, the same blot reprobed with anti-INAD antibodies.
In addition to TRP, another cation influx channel subunit, TRPL (Phillips et al., 1992), functions in phototransduction (Niemeyer et al., 1996) by forming a heteromultimeric channel with TRP (Gillo et al., 1996; Xu et al., 1997). To investigate whether TRPL was an INAD-interacting protein, we first carried out an in vivo coimmunoprecipitation experiment and found that INAD associated with TRPL in fly photoreceptor cells (Fig. 1 D). Since TRPL heteromultimerizes with TRP (Gillo et al., 1996; Xu et al., 1997), it was possible that TRPL associated with INAD through TRP. Therefore, we tested whether TRPL and INAD coimmunoprecipitated after coexpressing the two proteins in 293T cells. INAD was detected after immunoprecipitating cell extracts with TRPL antibodies but not with nonimmune serum (Fig. 1 E). Furthermore, INAD was not detected after immunoprecipitating with TRPL antibodies using extracts expressing only INAD. The interactions of TRP (Shieh and Zhu, 1996) and TRPL with INAD appeared to be specific since a highly related member of the TRP family, human TRPC3, did not coimmunoprecipitate with INAD (Fig. 1 I). Evidence that TRPL and INAD directly interacted was that 35S-labeled TRPL bound to INAD–GST fusion proteins immobilized on a column (Fig. 1 F).
PKC, encoded by the inaC locus (Smith et al., 1991), is another molecule known to function in the signaling cascade. It is required for adaptation and for terminating the photoresponse (Smith et al., 1991; Hardie et al., 1993). Recently, it has been shown that PKC is part of the INAD-signaling complex (Huber et al., 1996a ; Tsunoda et al., 1997). These previous experiments were performed using fly head extracts; therefore, it was not addressed whether PKC binds directly to INAD. Moreover, it has been suggested that PKC may be linked to the signaling complex through PLC, rather than through INAD (Huber et al., 1996a ). To address whether PKC was also an INAD binding protein, we conducted two assays: a coimmunoprecipitation experiment using the 293T cell expression system (Fig. 1 G) and in vitro binding using a GST–INAD fusion protein (Fig. 1 H). We found that PKC interacted with INAD in both of these assays, indicating PKC directly bound to INAD. The coimmunoprecipitation between PKC and INAD in 293T cells appeared to be specific since another cytoplasmic protein kinase which is present in Drosophila photoreceptor cells, calmodulin-dependent protein kinase II (Kahn and Matsumoto, 1997), did not coimmunoprecipitate with INAD (Fig. 1 K).
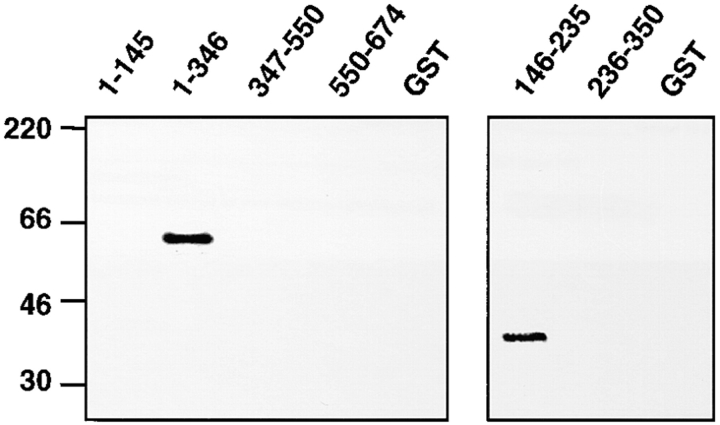
Calmodulin, a Ca2+ regulatory protein which functions in light adaptation and termination of the light response (Porter et al., 1993, 1995; Arnon et al., 1997a ,b; Scott et al., 1997), may also be an INAD-interacting protein since INAD from fly head extracts binds to a calmodulin affinity column in a Ca2+-dependent manner (Chevesich et al., 1997). However, the observation that calmodulin interacts with two other INAD binding proteins, TRPL and TRP (Warr and Kelly, 1996; Chevesich et al., 1997), suggests that the detected interaction might be indirect. To determine whether calmodulin is an INAD-binding protein, we generated a series of GST–INAD fusion proteins and found that calmodulin interacted with INAD fusions in an overlay assay (Fig. 2). Furthermore, calmodulin interacted with the linker region between PDZ1 and PDZ2 (Fig. 2 and Fig. 3 F).
Figure 2.
Calmodulin interacted directly with INAD. Total bacterial extracts expressing GST–INAD fusion proteins (residues included in each construct are indicated above each lane) or GST alone were fractionated by SDS-PAGE and transferred to polyvinylene difluoride. The membranes were then probed with biotin-labeled calmodulin. Left, protein size markers.
Opsin, TRPL, and PKC Bound to INAD through PDZ3 and PDZ4
To map the regions in INAD mediating the interactions with the opsin, TRPL, and PKC, we generated a series of INAD constructs and coexpressed them with each target protein in 293T cells (Fig. 3). We found that either PDZ3 or PDZ4 was sufficient to interact with the opsin, TRPL, and PKC. However, binding of each target to PDZ3 required an extra 28 amino acids COOH-terminal to PDZ3 (PDZ3L). A requirement for additional residues for target binding to a PDZ domain is not unique since a similar COOH-terminal extension is necessary for binding of target peptides to the PDZ domain in neuronal nitric acid synthase (Stricker et al., 1997). The association of the targets with PDZ3L required PDZ3 rather than just the extra 28 COOH-terminal amino acids since truncation of the NH2-terminal portion of PDZ3L-obliterated binding (see below). Binding of the opsin, TRPL, and PKC to PDZ3L and PDZ4 appeared to be specific since none of eight other protein or protein fragments tested bound to PDZ3L or PDZ4. These included TRPC3 (Fig. 3 D), Shaker B (Fig. 3 D), calmodulin (Fig. 2), and the PLC encoded by the norpA locus (Fig. 3 E). Furthermore, consistent with a recent report that PLC bound to a GST–PDZ1 fusion protein (van Huizen et al., 1998), we found that the COOH-terminal 123 residues of PLC expressed in 293T cells coimmunoprecipitated with either PDZ1 or PDZ1-PDZ2 (Fig. 3 E). These data suggest that the lack of interaction between these latter two PDZ domains and either the opsin, TRPL, or PKC was not due improper folding of PDZ1 or PDZ1-PDZ2. Studies in other labs, using bacterial fusion proteins, indicated that PLC bound to either PDZ1 and PDZ5 (van Huizen et al., 1998) or PDZ5 only (Tsunoda et al., 1997). Although the PLC expressed in 293T cells did not bind to PDZ5, we did find that PLC interacted with PDZ5 expressed in E. coli (data not shown). The lack of interaction of PLC with PDZ5 in 293T cells may be due to interference by posttranslational modifications of PDZ5, endogeous proteins that bind to either PLC or PDZ5 precluding the PLC/PDZ5 interaction, or misfolding of PDZ5.
Mutation of the COOH-terminal Residues in PKC Reduces but Does Not Eliminate Binding to INAD
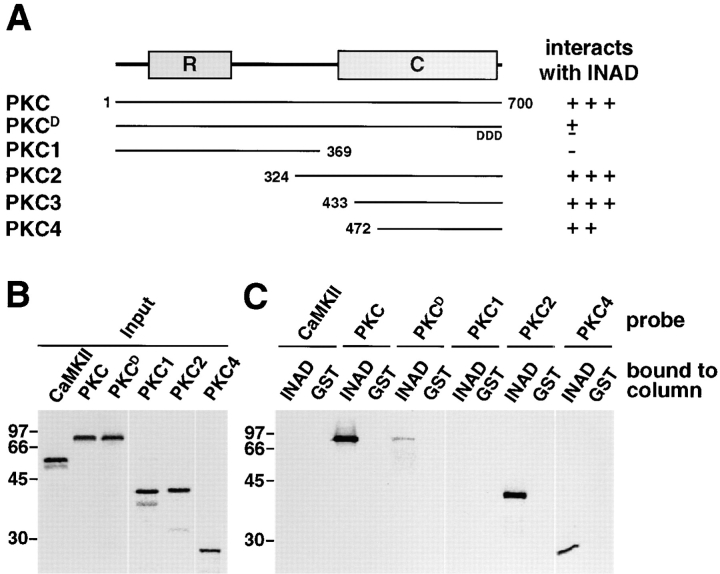
It has been shown that the COOH-terminal three residues of target proteins (often S/TXV) are essential for binding to PDZ domains (for reviews see Saras and Heldin, 1996; Kornau et al., 1997). Mutations in the COOH termini of the Shaker-type K+ channel, inwardly rectifier K+ channel, and NMDA receptor disrupt their interaction with PSD-95 (Kim et al., 1995; Kornau et al., 1995; Cohen et al., 1996). Moreover, the crystal structures of the third PDZ domain in PSD-95 and DLG have been solved, demonstrating that each possesses a hydrophobic pocket and a buried arginine residue that accommodates a COOH-terminal peptide (Cabral et al., 1996; Doyle et al., 1996). To test whether INAD interacted with its targets in a similar way, we changed each of the last three residues in PKC (T-I-I) to aspartic acid (PKCD) and coexpressed the derivative with full-length INAD in 293T cells. Binding to INAD was not abolished as a consequence of the mutation (data not shown). To directly compare whether the binding of PKCD to INAD was reduced relative to wild-type PKC, we performed column-binding assays. Although PKCD still bound to INAD, the interaction was significantly reduced (approximately eightfold). It was possible that the residual binding was due to the presence of a second INAD binding site in PKC since another INAD-binding protein, PLC (Chevesich et al., 1997), has recently been shown to contain two sites (van Huizen et al., 1998). Alternatively, there may be a single binding site in PKC which is close to but not at the extreme COOH terminus. If so, then mutation of the flanking COOH-terminal residues may disrupt but not obliterate binding. To differentiate between these possibilities, we attempted to further map the binding site(s). We found that all of the INAD-binding capacity was contained in the COOH-terminal third of PKC that included most of the catalytic domain (residues 472–700; Fig. 4, A and B). Smaller derivatives of the catalytic domain were all unstable in 293T cells, suggesting that they might have been misfolded. Thus, it was not feasible to further map the INAD binding site(s) in 293T cells or using the column-binding assay.
Figure 4.
COOH-terminal fragment of PKC specifically interacted with INAD. (A) Schematic showing that COOH-terminal fragments of PKC interacted with INAD in column-binding assays. The regulatory domain (R) and catalytic domain (C) of PKC are indicated. PKCD was the full-length PKC with the last three residues substituted with aspartic acids. Polypeptides interacting or not interacting with INAD were indicated with a + or −, respectively. Based on quantification using a PhosphorImager (refer to Materials and Methods), the relative levels of the interactions were arbitrarily assigned from − to +++. The NH2- and COOH-terminal residues included in each protein fragments were indicated. (B) The input probe (with [35S]methionine) used in C are shown. CaMKII and various PKC fragments shown in A were in vitro– translated using a TNT kit (Promega Corp.), separated by SDS-PAGE, and detected after using a PhosphorImager (1-h exposure). (C) A COOH-terminal fragment of PKC interacted with INAD in the column-binding assay. 35S-labeled probes from B were incubated with a GST–INAD or GST-bound glutathione column. After several washes, the beads were eluted with SDS sample buffer, the elutes were fractionated by SDS-PAGE, and then the signals were detected using a PhosphorImager (12-h exposures).
Homomultimerization of INAD through Two PDZ Domains May Lead to Formation of an INAD Network
The finding that PDZ3 and PDZ4 bound multiple targets indicated that a single INAD molecule would not have the capacity to nucleate the entire signaling complex unless INAD functions as a homomultimeric protein. To address this hypothesis, we coexpressed full-length INAD fused with MYC or FLAG epitope tags in 293T cells and found that INAD–FLAG coimmunoprecipitated with INAD– MYC (Fig. 5 A). Further evidence that INAD homomultimerized was obtained by demonstrating that 35S-INAD bound to a GST–INAD fusion immobilized on a glutathione column (Fig. 5 B). Homomultimerization of a vertebrate PDZ domain-containing protein expressed in postsynaptic densities, PSD-95, has recently been reported and the homomeric binding is mediated through NH2-terminal disulfide bonds (Hsueh et al., 1997). In contrast, we found that the INAD homomultimerization occurred through PDZ domains (either PDZ3 or PDZ4, Fig. 5, D and E). Furthermore, INAD PDZ3 and PDZ4 could form either homomeric or heteromeric interactions (Fig. 5 G). The PDZ–PDZ interaction appeared to be specific to PDZ3 and PDZ4 since neither PDZ1, PDZ2, nor PDZ5 bound to any of portion of INAD including PDZ3 or PDZ4. In addition, another protein with multiple PDZ domains, SAP 102 (Muller et al., 1996), did not coimmunoprecipitate with either full-length INAD (Fig. 5 C) or PDZ3–PDZ4 (Fig. 5 F).
Figure 5.
INAD homomultimerized in vitro through PDZ3 and PDZ4. (A) INAD homomultimerized in 293T cells. MYC- and FLAG-tagged INAD (INAD-M and INAD-F) were coexpressed in 293T cells. Immunoprecipitations were performed with anti-MYC antibodies or NIS and the Western blot was probed with anti-FLAG antibodies. (B) INAD displayed homomeric interactions in a column binding assay. In vitro–translated 35S-INAD was coincubated with GST–INAD or GST immobilized on a glutathione column. The 35S-INAD was eluted with SDS sample buffer, fractionated by SDS-PAGE, and then exposed using a PhosphorImager. (C) SAP 102 did not interact with INAD. A plasmid encoding MYC-tagged SAP 102 (gift of R. Huganir, Johns Hopkins University School of Medicine) was cotransfected with pINAD in 293T cells. Cell lysates were immunoprecipitated with anti-INAD antibodies and a Western blot was probed with anti-MYC or anti-INAD antibodies. (D and E) INAD homomultimerized via either PDZ3 or PDZ4. Constructs containing MYC-tagged fragments of INAD were cotransfected in 293T cells with plasmids encoding full-length INAD (pINAD). Cell lysates were immunoprecipitated with anti-MYC antibodies and Western blots were probed with anti-INAD antibodies. Constructs that coimmunoprecipitated with pINAD were indicated with a + whereas those that did not coimmunoprecipitate were indicated with a −. E, representative results summarized in panel D). (F) SAP 102 did not interact with INAD PDZ3-PDZ4 (PDZ3.4-F). SAP 102-M and pPDZ3.4-F (PDZ3-PDZ4 with a FLAG tag) were coexpressed in 293T cells. The coimmunoprecipitation was performed with anti-FLAG antibodies and the Western blot was probed with anti-MYC or anti-FLAG antibodies. (G) PDZ3 and PDZ4 displayed homophilic and heterophilic interactions. MYC- or FLAG-tagged INAD PDZ3 or PDZ4 constructs (PDZ3L-M, PDZ3L-F, PDZ4L-M, and PDZ4L-F) were cotransfected into 293T cells as indicated. Cell lysates were immunoprecipitated with anti-MYC antibodies or nonimmune serum (NIS). The Western blots were probed with anti-FLAG antibodies. (H and I) PDZ3-PDZ4 formed polymers. INAD fragments including either PDZ1 and PDZ2 (PDZ1-2) or PDZ3 and PDZ4 (PDZ3-4) were translated in vitro with [35S]methionine and fractionated by sucrose gradient centrifugation. Both PDZ1-2 and PDZ3-4 were loaded onto the same gradient. Collected fractions were resolved by SDS-PAGE and processed for autoradiography shown in I. The protein bands in panel I were quantified with a PhosphorImager and corresponding readings were plotted versus fraction numbers in H. Fraction numbers as well as those fractions that contained the peak levels of the marker proteins included in the gradient were indicated.
The observation that homomeric interactions occurred through either PDZ3 or PDZ4 raised the possibility that INAD may form a homopolymer rather than just a dimer. To address this possibility, we translated in vitro a segment of INAD including just PDZ3-PDZ4 and fractionated the products by sucrose gradient sedimentation. Although a proportion of PDZ3-4 fractionated near the predicted molecular weight of the dimer (52 kD), a significant amount sedimented as a much larger protein of ∼200 kD (Fig. 5, H and I). PDZ1-PDZ2 loaded onto the same gradient sedimented with a single peak near its predicted monomer molecular weight of ∼39 kD (Fig. 5, H and I). Thus, PDZ1-2 did not homomultimerize or interact with PDZ3-PDZ4. The data that a proportion of PDZ3-PDZ4 sedimented as a protein ≥200 kD suggested that INAD may be capable of forming homopolymers with a subunit composition of ≥8. In contrast to PDZ1-PDZ2 which fractionated with a single peak, four small peaks were detected with PDZ3-PDZ4 which roughly corresponded to the predicted sizes of molecules with 1, 2, 4, and 6 subunits. Since the PDZ1-PDZ2 monomer and marker proteins distributed over many fractions, the PDZ3-PDZ4 peaks may have been small due to a similar broad distribution of the PDZ3-PDZ4 monomer, dimer, and higher order forms.
Homomultimerization of INAD Did Not Prevent PDZ–Target Interactions
The findings that INAD can form homomultimers through PDZ3 and PDZ4 raises the question as to whether homomultimerization precludes INAD–target interaction or vice versa (Fig. 6 A). An indication that PDZ–PDZ and PDZ– target interactions share the same or overlapping interaction interface is that a synthetic peptide corresponding to the COOH-terminal sequence of the NMDA receptor type 2B blocks the PDZ–PDZ interactions between neuronal nitric acid synthase and PSD-95 (Brenman et al., 1996). To investigate whether homomultimerization and PDZ–target interactions can occur simultaneously, we took advantage of the finding that PDZ3 alone was sufficient to promote homotypic interactions, whereas PDZ3L was required for binding to the opsin, TRPL, or PKC. Therefore, we tested whether PDZ3 coimmunoprecipitated with the targets after coexpressing PDZ3 and PDZ3L with either TRPL or PKC. We found that TRPL or PKC coimmunoprecipitated with PDZ3 in the presence but not in the absence of PDZ3L (Fig. 6, B and C). These results indicated that PDZ3 and PDZ3L formed a ternary complex with the target proteins and suggested that the INAD PDZ–PDZ and PDZ–target interactions were mediated via different interfaces. Consistent with this latter proposal, an NH2-terminal truncation that removed the first and second putative β barrel from PDZ3L (Fig. 6 D, PDZ3LΔN) disrupted interaction with PKC; however, homomeric binding still occurred. Furthermore, as described above, the COOH-terminal extension in PDZ3L was required for binding to PKC but not for the PDZ–PDZ association. The extra COOH-terminal residues in PDZ3L were not sufficient for binding to PKC since PDZ3LΔN did not bind PKC (Fig. 6 D).
Figure 6.
PDZ–PDZ interactions did not preclude PDZ–target interactions. (A) Models for the PDZ–PDZ and PDZ–target interaction: PDZ–PDZ interaction precludes binding of targets, PDZ–target binding prevents PDZ-PDZ interaction, PDZ–PDZ and PDZ–target binding can occur simultaneously. (B) TRPL, PDZ3, and PDZ3L formed a ternary complex. COOH-terminal TRPL, MYC-tagged PDZ3 (PDZ3-M) and FLAG-tagged PDZ3 long-form (PDZ3L-F) were expressed in duplicate or triplicate in 293T cells as indicated. Cell lysates were immunoprecipitated with anti-TRPL or anti-MYC antibodies and the Western blots were probed with either anti-MYC (left two panels) or anti-FLAG antibodies (right two panels). (C) PKC, PDZ3, and PDZ3L formed a ternary complex. FLAG-tagged PKC (PKC-F), MYC-tagged PDZ3 (PDZ3-M), MYC-tagged PDZ3 long-form (PDZ3L-M) and FLAG-tagged PDZ3 long-form (PDZ3L-F) were doubly- or triply-expressed in 293T cells as indicated. Cell lysates were immunoprecipitated with anti-MYC antibodies or nonimmune serum and the Western blots were probed with anti-FLAG antibodies. The third panel in B and the fourth panel in C are duplicates. (D) Homomultimerization and target binding were mediated through overlapping but different regions in PDZ3. Plasmids encoding various portions of PDZ3 and linker regions (MYC-tagged) were coexpressed with pPKC-F or pINAD in 293T cells. Coimmunoprecipitations were carried out using anti-MYC antibodies. +, PDZ3 fragments that interacted with PKC or INAD in the assay.
Discussion
INAD Links Most, If Not All, of the Components of the Phototransduction Cascade
The view that signaling through G protein–coupled cascades occurs via random stochastic collisions between membrane receptors and effector molecules has been widely held for many years (Tolkowsky and Levitzki, 1978). However, alternative proposals suggesting that signaling cascades are comprised of components that are physically coupled have been presented but have received less attention (Rodbell, 1992). The major conclusion from the current work is that Drosophila vision is mediated by a massive supramolecular complex and that assembly of such a complex is facilitated by homomultimerization of the scaffold protein INAD. Previous studies have shown that at least two proteins required in Drosophila vision, TRP and the PLC encoded by the norpA locus, bind directly to INAD, a protein with five PDZ domains (Shieh and Zhu, 1996; Chevesich et al., 1997). In addition, PKC (Huber et al., 1996a ), rhodopsin, and calmodulin (Chevesich et al., 1997), have been implicated as INAD-binding proteins; however, direct binding has not been demonstrated. Here, we show that at least five additional proteins bind INAD. These include four target proteins: rhodopsin, TRPL, PKC, and calmodulin. In addition, we found that INAD formed homophilic interactions. Thus, INAD associated directly with a minimum of seven proteins including a receptor (rhodopsin), an effector (PLC), regulators (PKC and calmodulin), and ion channels (TRP and TRPL). These findings raise the possibility that nearly all of the proteins that function in Drosophila phototransduction are linked to INAD.
In a previous study, it was reported that INAD did not bind to TRPL or rhodopsin (Tsunoda et al., 1997); thus, INAD was not considered to have the capacity to coordinate some of the key proteins critical in phototransduction. Specifically, these workers reported that neither TRPL nor rhodopsin coimmunoprecipitate with INAD from fly heads. Nevertheless, we found that INAD not only coimmunoprecipitated with both of these proteins from fly heads but interacted in several in vitro assays. Consistent with these results, we had previously shown that rhodopsin and TRP coimmunoprecipitate from wild-type but not InaDP215 heads (Chevesich et al., 1997). Although these earlier results did not demonstrate that rhodopsin and INAD interact directly, they did show that the association of TRP and rhodopsin in vivo depends on the presence of wild-type INAD. The negative coimmunoprecipitation results in the previous study (Tsunoda et al., 1997) was presumably due to differences in the procedures that were used. For example, the primary antibodies used in our coimmunoprecipitations were directed against the target proteins, whereas in the former analysis, anti-INAD antibodies were used instead. To provide evidence that INAD associated directly with rhodopsin, PKC, and TRPL, we performed column-binding studies. The results of these analyses were further supported by coimmunoprecipitation experiments after expressing INAD and the target proteins in tissue culture cells.
INAD Targets Bind to Multiple PDZ Modules
It has previously been suggested that each of the five PDZ domains in INAD is a distinct binding module that interacts with a different target protein (Tsunoda et al., 1997). However, the concept that there exists a simple one-to-one correspondence between PDZ domains and target proteins appears to be an oversimplification. Other than calmodulin, which bound to the linker region between PDZ1 and PDZ2, each of the INAD targets appears to interact with more than one PDZ domain. We found that the opsin, TRPL, and PKC each associated with PDZ3 and PDZ4. TRP may also bind to PDZ4, in addition to PDZ3, since it coimmunoprecipitated with a fragment that included both PDZ4 and PDZ5 (data not shown). The interaction of these targets with PDZ3 and PDZ4 appeared to be specific since eight other proteins tested in the in vitro assays did not bind full-length INAD or any fragment of it. These included human TRPC3 as well as proteins normally expressed in Drosophila photoreceptor cells, such as Shaker and CaM kinase II. TRPC3 did not bind any portion of INAD despite having a similar overall level of identity with TRP as TRPL. An additional protein that binds multiple domains in INAD is PLC. It has recently been demonstrated that PLC binds to both PDZ1 and PDZ5 (van Huizen et al., 1998). Although we found that PLC bound only to PDZ1 in 293T cells, PLC did bind PDZ5 in an overlay assay (data not shown). Thus, it appears that target binding to INAD typically occurs via more than one PDZ domain and that multiple assays should be used before concluding that a particular PDZ– target interaction does not occur.
Homomultimerization of INAD May Increase the Capacity of the Complex to Simultaneously Link Multiple Targets
Binding of multiple targets to the same PDZ domains should in principle preclude formation of a single complex comprised of all the potential targets. Therefore, a conundrum concerns the mechanism by which INAD could coordinate such a supramolecular signaling complex. A potential resolution to this problem is the finding that INAD is capable of forming homomultimers. Evidence that this was the case was provided by column-binding experiments, coimmunoprecipitation assays, and by examining the distribution of PDZ3-PDZ4 on sucrose gradients. Consistent with the proposal that INAD may form homomultimers in vivo, the original missense allele, InaDP215 (Pak, 1979) exhibits a partially dominant phenotype. Dominant phenotypes often require protein–protein interactions and result from poisoning of a wild-type protein with an altered derivative, although other interpretations cannot be excluded.
The mechanism underlying the INAD homomultimerization differs from that in PSD-95 since INAD uses PDZ– PDZ binding whereas PSD-95 dimer formation is mediated through disulfide bonds NH2-terminal to the PDZ domains (Hsueh et al., 1997). Since the homomultimerization occurred through the same PDZ domains, PDZ3 and PDZ4, that bind to several targets, the homophilic interactions could potentially prevent target binding. However, we found that the homomeric interaction did not preclude target binding, thereby providing a mechanism whereby several proteins could be coupled to the supramolecular signaling complex via the same PDZ domains. The simultaneous homophilic and target binding indicated that the two types of interactions occurred through different interfaces. Further support for this conclusion is that homophilic association required less than a complete PDZ domain.
In contrast to the effects on homomultimerization, deletion of the NH2-terminal region of PDZ3 disrupted binding to PKC. Thus, the residues in PDZ3 that may bond with the COOH terminus of target proteins were required. Consistent with this observation, mutation of the COOH-terminal three residues of PKC greatly reduced, although did not eliminate, interaction with INAD. The residual binding to INAD may reflect a second binding site which is internal since PLC has been shown to bind to INAD via two sites (van Huizen et al., 1998). However, we were unable to map this site due to instability and possible misfolding of smaller derivatives of the COOH-terminal PKC catalytic domain.
Possible Functions of the Signalplex in Drosophila Vision
A potential function of the supramolecular complex may be to facilitate Ca2+-dependent feedback regulation. The activities of several proteins required in phototransduction, such as PLC, are inhibited by a rise in Ca2+ concentration. Close association of the Ca2+ influx channels to other signaling proteins may serve to circumvent the considerable Ca2+ buffering capacity in the photoreceptor cells. Indeed, the InaDP215 missense mutation causes a defect in termination of the photoresponse (Shieh and Niemeyer, 1995). The association of calmodulin with INAD could serve as one of the calcium sensors that operates in the negative feedback regulation. Null InaD flies show a dramatic defect in the amplitude of the photoresponse indicating that a second function of the INAD complex may be in activation (Tsunoda et al., 1997). Drosophila phototransduction is activated within ∼20 milliseconds (Ranganathan et al., 1991) and the rapid opening of the TRP and TRPL cation influx channels may be directly mediated by a second messenger produced in the signaling complex.
All of the defects in InaD flies might be due to physical detachment of the proteins from the signaling complex. Interestingly, some INAD–interacting proteins (TRP, PLC, and PKC) if detached from the complex, in InaD mutants, are no longer spatially restricted to the microvillar portion of the photoreceptor cells, the rhabdomeres (Chevesich et al., 1997; Tsunoda et al., 1997) and are unstable (Tsunoda et al., 1997). However, other INAD binding proteins such as rhodopsin are still in the rhabdomeres and are stably expressed in InaD flies (Tsunoda et al., 1997).
We propose that those proteins that require interaction with INAD for normal spatial localization may be constitutively bound to INAD wheras other proteins, such as rhodopsin and TRPL, which do not depend on INAD for rhabdomere localization or stability, may interact dynamically with INAD. Since TRP and TRPL form heteromultimers (Xu et al., 1997) and TRP requires INAD for rhabdomere-specific localization, we suggest that TRP/ TRPL heteromultimers interact dynamically rather than constitutively with INAD. Since TRP is much more abundant than TRPL (Gillo et al., 1996; Xu et al., 1997), only a small proportion of TRP is complexed with TRPL and expected to remain in the rhabdomeres in InaD mutants. In fact, a proportion of TRP remains in the rhabdomeres of InaD photoreceptors (Chevesich et al., 1997). Thus, the view that all INAD binding proteins interact stably and require the interaction for proper localization (Tsunoda et al., 1997) does not appear to be correct. Since rhodopsin is the most abundant protein in the rhabdomeres and is therefore present in excess over INAD, only a fraction of rhodopsin could associate with INAD at any given time. It is intriguing to speculate such a fraction might consist of photoactivated rhodopsin.
The INAD supramolecular complex may not be a particle, as suggested by the term transducisome (Tsunoda et al., 1997), consisting of a single INAD monomer to which a maximum of five target proteins bind. Instead, the visual cascade appears to be mediated through a more complicated higher order signaling web or complex (signalplex) consisting of an extended network of INAD homomultimers to which more than five targets bind. An indication that INAD forms homomultimers in vivo, is that the original InaD missense allele is partially dominant, yet the null allele is recessive. Most of these targets appear to bind to more than one PDZ module and several targets appear to associate with INAD via the same PDZ domains (Fig. 7). Thus, the nature of the INAD signalplex appears to be more complicated than previously envisioned. The full extent and complexity of the INAD array remains to be determined.
Figure 7.
A schematic model of the phototransduction signalplex. INAD homomultimerizes through PDZ3 and PDZ4. Rhodopsin, PKC, and TRPL bind to either PDZ3 or PDZ4. Calmodulin binds to the linker region between PDZ3 and PDZ4. Binding of TRP to PDZ3 (Shieh and Zhu, 1996) and PLC to PDZ1 and PDZ5 is based on work described in separate reports (Tsunoda et al., 1997; van Huizen et al., 1998). The individual PDZ domains (1–5) in each INAD monomer are indicated by the ovals.
Acknowledgments
We thank H.-S. Li for helpful discussions, J. Chevesich for performing Fig. 1 A, M. Li for helpful discussions and contributing a norpA subclone, R. Huganir for the clone of SAP 102, J. Yu (all five from Johns Hopkins University School of Medicine, Baltimore, MD), for technical assistance, D. Smith (University of Texas Southwestern Medical Center, Dallas, TX) and C. Zuker (University of California San Diego, La Jolla, CA) for the inaC cDNA, S. Subramaniam (Johns Hopkins University School of Medicine) for rhodopsin antibodies, J. Saras (Ludwig Institute for Cancer Research, Uppsala, Sweden) for assistance in identifying the positions of the five PDZ domains in INAD, and D. Montell (Johns Hopkins University School of Medicine) for critically reading the manuscript.
This work was supported by a grant from the National Eye Institute (EY08117) to C. Montell.
Abbreviations used in this paper
- INAD
inactivation no afterpotential D, PLC, phospholipase C
- PKC
protein kinase C
Footnotes
Address all correspondence to Craig Montell, Department of Biological Chemistry and Department of Neuroscience, The Johns Hopkins University School of Medicine, Baltimore, MD 21205. Tel.: (410) 955-1199. Fax: (410) 614-9573. E-mail: cmontell@bs.jhmi.edu
References
- Arnon A, Cook B, Gillo B, Montell C, Selinger Z, Minke B. Calmodulin regulation of light adaptation and store-operated dark current in Drosophilaphotoreceptors. Proc Natl Acad Sci USA. 1997a;94:5894–5899. doi: 10.1073/pnas.94.11.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon A, Cook B, Montell C, Selinger Z, Minke B. Calmodulin regulation of calcium stores in phototransduction of Drosophila. . Science. 1997b;275:1119–1121. doi: 10.1126/science.275.5303.1119. [DOI] [PubMed] [Google Scholar]
- Bloomquist BT, Shortridge RD, Schneuwly S, Perdew M, Montell C, Steller H, Rubin G, Pak WL. Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell. 1988;54:723–733. doi: 10.1016/s0092-8674(88)80017-5. [DOI] [PubMed] [Google Scholar]
- Brakeman PR, Lanahan AA, Obrien R, Roche K, Barners CA, Huganir RL, Worley PF. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu Z, Huang F, Xia H, Peters MF, et al. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and α1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- Cabral JH, Petosa C, Sutcliffe MJ, Raza S, Byron O, Poy F, Marfatia SM, Chishti AH, Liddington RC. Crystal structure of a PDZ domain. Nature. 1996;382:649–652. doi: 10.1038/382649a0. [DOI] [PubMed] [Google Scholar]
- Chevesich J, Kreuz AJ, Montell C. Requirement for the PDZ domain protein, INAD, for localization of the TRP store-operated channel to a signaling complex. Neuron. 1997;18:95–105. doi: 10.1016/s0896-6273(01)80049-0. [DOI] [PubMed] [Google Scholar]
- Cohen NA, Brenman JE, Snyder SH, Bredt DS. Binding of the inward rectifier K channel Kir 2.3 to PSD-95 is required by protein kinase A phosphorylation. Neuron. 1996;17:759–767. doi: 10.1016/s0896-6273(00)80207-x. [DOI] [PubMed] [Google Scholar]
- Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- Gillo B, Chorna I, Cohen H, Cook B, Manistersky I, Chorev M, Arnon A, Pollock JA, Selinger Z, Minke B. Coexpression of Drosophila TRP and TRP-like proteins in Xenopus oocytes reconstitutes capacitative CA2+entry. Proc Natl Acad Sci USA. 1996;93:14146–14151. doi: 10.1073/pnas.93.24.14146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie RC. Voltage-sensitive potassium channels in Drosophilaphotoreceptors. J Neurosci. 1991;11:3079–3095. doi: 10.1523/JNEUROSCI.11-10-03079.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie RC, Peretz A, Suss-Toby E, Rom-Glass A, Bishop SA, Selinger Z, Minke B. Protein kinase C is required for light adaptation in Drosophilaphotoreceptors. Nature. 1993;363:634–637. doi: 10.1038/363634a0. [DOI] [PubMed] [Google Scholar]
- Hata MIY, Takeuchi M, Ichtchenko K, Toyoda A, Hirao K, Takai Y, Rosahi TW, Sudhof TC. Binding of neuroligins to PSD-95. Science. 1997;277:1511–1515. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- Hsueh Y-P, Kim E, Sheng M. Disulfide-linked head-to-tail multimerization in the mechanism of ion channel clustering by PSD-95. Neuron. 1997;18:803–814. doi: 10.1016/s0896-6273(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Huber A, Sander P, Gobert A, Bähner M, Hermann R, Paulsen R. The transient receptor potential protein (Trp), a putative store-operated Ca2+channel essential for phosphoinositide-mediated photoreception, forms a signaling complex with NorpA, InaC and InaD. EMBO (Eur Mol Biol Organ) J. 1996a;15:7036–7045. [PMC free article] [PubMed] [Google Scholar]
- Huber A, Sander P, Paulsen R. Phosphorylation of the InaDgene product, a photoreceptor membrane protein required for recovery of visual excitation. J Biol Chem. 1996b;271:11710–11717. doi: 10.1074/jbc.271.20.11710. [DOI] [PubMed] [Google Scholar]
- Jan, L.Y., and Y.N. Jan. 1990. How might the diversity of potassium channels be generated? TINS (Trends Neurosci.). 13:415–419. [DOI] [PubMed]
- Kahn ES, Matsumoto H. Calcium/calmodulin-dependent kinase II phosphorylates Drosophila visual arrestin. J Neurochem. 1997;68:169–175. doi: 10.1046/j.1471-4159.1997.68010169.x. [DOI] [PubMed] [Google Scholar]
- Kim E, Cho K-O, Rothschild A, Sheng M. Heteromultimerization and NMDA receptor-clustering activity of chapsyn-110, a member of the PSD-95 family of proteins. Neuron. 1996;17:103–113. doi: 10.1016/s0896-6273(00)80284-6. [DOI] [PubMed] [Google Scholar]
- Kim E, Niethammmer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K+channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- Kornau H-C, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- Kornau H-C, Seeburg PH, Kennedy MB. Interaction of ion channels and receptors with PDZ domain proteins. Curr Opin Neurobiol. 1997;7:368–373. doi: 10.1016/s0959-4388(97)80064-5. [DOI] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translation start site in eukaryotic mRNAs. Nucl Acids Res. 1984;12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C. New light on TRP and TRPL. Mol Pharmacol. 1997;52:755–763. doi: 10.1124/mol.52.5.755. [DOI] [PubMed] [Google Scholar]
- Montell C. TRP trapped in fly signaling web. Curr Opin Neurobiol. 1998;8:389–397. doi: 10.1016/s0959-4388(98)80066-4. [DOI] [PubMed] [Google Scholar]
- Muller BM, Kistner U, Kindler S, Chung WJ, Kuhlendahl S, Fenster SD, Lau L-F, Veh RW, Huganir RL, Gundelfinger ED, Garner CC. SAP 102, a novel postsynaptic protein that interacts with NMDA receptor complexes in vivo. Neuron. 1996;17:255–265. doi: 10.1016/s0896-6273(00)80157-9. [DOI] [PubMed] [Google Scholar]
- Niemeyer BA, Suzuki E, Scott K, Jalink K, Zuker CS. The Drosophila light-activated conductance is composed of the two channels TRP and TRPL. Cell. 1996;85:651–659. doi: 10.1016/s0092-8674(00)81232-5. [DOI] [PubMed] [Google Scholar]
- O'Tousa JE, Baehr W, Martin RL, Hirsh J, Pak WL, Applebury ML. The Drosophila ninaEgene encodes an opsin. Cell. 1985;40:839–850. doi: 10.1016/0092-8674(85)90343-5. [DOI] [PubMed] [Google Scholar]
- Pak, W.L. 1979. Study of photoreceptor function using Drosophila mutants. In Genetic Approaches to the Nervous System. X.O. Breakfield, editor. Elsevier Science, New York. 67–99.
- Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor protein. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- Phillips AM, Bull A, Kelly LE. Identification of a Drosophila gene encoding a calmodulin-binding protein with homology to the trpphototransduction gene. Neuron. 1992;8:631–642. doi: 10.1016/0896-6273(92)90085-r. [DOI] [PubMed] [Google Scholar]
- Porter JA, Minke B, Montell C. Calmodulin binding to Drosophila NinaC required for termination of phototransduction. EMBO (Eur Mol Biol Organ) J. 1995;18:4450–4459. doi: 10.1002/j.1460-2075.1995.tb00124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JA, Yu M, Doberstein SK, Pollard TS, Montell C. Dependence of calmodulin localization in the retina on the ninaCunconventional myosin. Science. 1993;262:1038–1042. doi: 10.1126/science.8235618. [DOI] [PubMed] [Google Scholar]
- Ranganathan R, Harris GL, Stevens CF, Zuker CS. A Drosophilamutant defective in extracellular calcium-dependent photoreceptor deactivation and desensitization. Nature. 1991;354:230–232. doi: 10.1038/354230a0. [DOI] [PubMed] [Google Scholar]
- Rodbell M. The role of GTP-binding proteins in signal transduction: from the sublimely simple to the conceptually complex. Current Topics in Cellular Regulation. 1992;32:1–47. doi: 10.1016/b978-0-12-152832-4.50003-3. [DOI] [PubMed] [Google Scholar]
- Saras J, Heldin C-H. PDZ domains bind carboxy-terminal sequences of target proteins. TIBS (Trends Biosci) 1996;21:455–458. doi: 10.1016/s0968-0004(96)30044-3. [DOI] [PubMed] [Google Scholar]
- Scott K, Sun Y, Beckingham K, Zuker CS. Calmodulin regulation of Drosophila light-activated channels and receptor function mediates termination of the light response in vivo. Cell. 1997;91:375–383. doi: 10.1016/s0092-8674(00)80421-3. [DOI] [PubMed] [Google Scholar]
- Shieh B-H, Niemeyer B. A novel protein encoded by the InaDgene regulates recovery of visual transduction in Drosophila. Neuron. 1995;14:201–210. doi: 10.1016/0896-6273(95)90255-4. [DOI] [PubMed] [Google Scholar]
- Shieh B-H, Zhu M-Y. Regulation of the TRP Ca2+channel by INAD in Drosophila photoreceptors. Neuron. 1996;16:991–998. doi: 10.1016/s0896-6273(00)80122-1. [DOI] [PubMed] [Google Scholar]
- Smith DP, Ranganathan R, Hardy RW, Marx J, Tsuchida T, Zuker CS. Photoreceptor deactivation and retinal degeneration mediated by a photoreceptor-specific protein kinase C. Science. 1991;254:1478–1484. doi: 10.1126/science.1962207. [DOI] [PubMed] [Google Scholar]
- Stricker NL, Christopherson KS, Yi BA, Schatz PJ, Raab RW, Dawes G, Bassett DEJ, Bredt DS, Li M. PDZ domain of neuronal nitric oxide synthase recognizes novel C-terminal peptide sequences. Nat Biotechnol. 1997;15:336–342. doi: 10.1038/nbt0497-336. [DOI] [PubMed] [Google Scholar]
- Tejedor FJ, Bokhari A, Rogero O, Gorczyca M, Zhang J, Kim E, Sheng M, Budnik V. Essential role for dlg in synaptic clustering of Shaker K+ channels in vivo. . J Neurosci. 1997;17:152–159. doi: 10.1523/JNEUROSCI.17-01-00152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkowsky AM, Levitzki A. Mode of coupling between the β-adrenergic receptor and adenylate cyclase in turkey erythrocytes. Biochemistry. 1978;17:3795–3810. doi: 10.1021/bi00611a020. [DOI] [PubMed] [Google Scholar]
- Tsunoda S, Sierralta J, Sun Y, Bodner R, Suzuki E, Becker A, Socolich M, Zuker CS. A multivalent PDZ-domain protein assembles signalling complexes in a G-protein-coupled cascade. Nature. 1997;388:243–249. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- van Huizen R, Miller K, Chen D-M, Li Y, Lai Z-C, Raab RW, Stark WS, Shortridge RD, Li M. Two distantly positioned PDZ domains mediate multivalent INAD-phospholipase C interactions essential for G protein-coupled signaling. EMBO (Eur Mol Biol Organ) J. 1998;17:2285–2297. doi: 10.1093/emboj/17.8.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr CG, Kelly LE. Identification and characterization of two distinct calmodulin-binding sites in the Trpl ion-channel protein of Drosophila melanogaster. . Biochem J. 1996;314:497–503. doi: 10.1042/bj3140497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wes PD, Chevesich J, Jeromin A, Rosenberg C, Stetten G, Montell C. TRPC1, a human homolog of a Drosophilastore-operated channel. Proc Natl Acad Sci USA. 1995;92:9652–9656. doi: 10.1073/pnas.92.21.9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X-ZS, Li H-S, Guggino WB, Montell C. Coassembly of TRP and TRPL produces a distinct store-operated conductance. Cell. 1997;89:1155–1164. doi: 10.1016/s0092-8674(00)80302-5. [DOI] [PubMed] [Google Scholar]
- Zhu X, Jiang M, Peyton M, Boulay G, Hurst R, Stefani E, Birnbaumer L. trp, a novel mammalian gene family essential for agonist-activated capacitative Ca2+entry. Cell. 1996;85:661–671. doi: 10.1016/s0092-8674(00)81233-7. [DOI] [PubMed] [Google Scholar]
- Zuker CS, Cowman AF, Rubin GM. Isolation and structure of a rhodopsin gene from D. melanogaster. Cell. 1985;40:851–858. doi: 10.1016/0092-8674(85)90344-7. [DOI] [PubMed] [Google Scholar]