Abstract
The transforming growth factor-β (TGF-β) superfamily of growth factors and cytokines has been implicated in a variety of physiological and developmental processes within the cardiovascular system. Smad proteins are a recently described family of intracellular signaling proteins that transduce signals in response to TGF-β superfamily ligands. We demonstrate by both a mammalian two-hybrid and a biochemical approach that human Smad2 and Smad4, two essential Smad proteins involved in mediating TGF-β transcriptional responses in endothelial and other cell types, can functionally interact with the transcriptional coactivator CREB binding protein (CBP). This interaction is specific in that it requires ligand (TGF-β) activation and is mediated by the transcriptional activation domains of the Smad proteins. A closely related, but distinct endothelial-expressed Smad protein, Smad7, which does not activate transcription in endothelial cells, does not interact with CBP. Furthermore, Smad2,4–CBP interactions involve the COOH terminus of CBP, a region that interacts with other regulated transcription factors such as certain signal transduction and transcription proteins and nuclear receptors. Smad–CBP interactions are required for Smad-dependent TGF-β-induced transcriptional responses in endothelial cells, as evidenced by inhibition with overexpressed 12S E1A protein and reversal of this inhibition with exogenous CBP. This report demonstrates a functional interaction between Smad proteins and an essential component of the mammalian transcriptional apparatus (CBP) and extends our insight into how Smad proteins may regulate transcriptional responses in many cell types. Thus, functional Smad–coactivator interactions may be an important locus of signal integration in endothelial cells.
Keywords: vascular endothelium/P300/transcription factors/signal transduction
The transforming growth factor β (TGF-β) superfamily of cytokines constitutes a group of proteins whose members mediate many biological effects. Within the vasculature, TGF-β has been demonstrated to play an important role in a variety of pathophysiologic processes, including angiogenesis, vascular remodeling, and atherogenesis (1–3). Both in vitro and in vivo, TGF-β has been demonstrated to regulate extracellular matrix elaboration, cellular migration, apoptosis, and cell cycle traverse in vascular smooth muscle and endothelial cells (EC). Over the last several years, the intracellular signaling mechanisms used by this family of effectors have begun to be elucidated (4–6). In the case of TGF-β, its cellular effects seem to be transduced via at least three types of cell surface receptors (types I, II, III). The active form of TGF-β binds to the type II receptor at the cell surface; this complex subsequently interacts with and phosphorylates (activates) the type I receptor, which then propagates downstream signals within the cell. This is accomplished in part by direct interaction of the activated type 1 receptor with members of a newly described family of intracellular signaling molecules known as the Smad proteins (4–9). In the case of TGF-β1, the activated type 1 receptor transiently interacts with a specific Smad, Smad2, and phosphorylates it on a series of COOH-terminal serines (10, 11). Subsequently, Smad2 associates with another distinct Smad, Smad4, in a heteromeric complex that then translocates to the nucleus and influences gene expression by mechanisms that are not well understood (12–14). Although a variety of other intracellular signaling cascades have been implicated in TGF-β signaling, the importance of the Smad proteins is highlighted by the fact that mutations in both Smad2 and Smad4 have been causally linked to specific human malignancies and that disruption of either of these genes in the mouse results in early embryonic lethality (14–17).
Recently, there has been rapid progress in the identification of additional members of the Smad family of proteins in humans and other species. Based on their structures and known functional roles, the mammalian Smad proteins seem to fall into at least three broad classes (5, 6). The first class, typified by Smads 1, 2, 3, and 5 (and more recently Smad8), seems to be capable of interacting with the activated type 1 receptors corresponding to a particular TGF-β superfamily ligand, undergoes receptor-mediated phosphorylation, and subsequently translocates to the nucleus (9, 10, 18–20). As part of this process, these “signaling” (or receptor-associated) Smads bind to a distinct class of Smad, Smad4, that can synergize with certain signaling Smads and act as a transcriptional activator (12, 13, 21). Thus, Smad4 seems to define a second class of Smad protein that does not interact directly with receptors but is required for signaling. Members of a third class of Smad proteins recently described (Smad6 and -7) are capable of inhibiting TGF-β signaling, and in contrast to the other classes of Smads, they demonstrate inducibility in response to a variety of stimuli such as TGF-β or fluid mechanical (flow) stimuli (22–25). These have been termed “inhibitory Smads.” Smad6 and Smad7 are expressed in endothelial cells and have been demonstrated to modulate both TGF-β and biomechanical (fluid shear stress)-induced gene expression (22) in this cell type.
In this paper, we demonstrate that there is functional heterogeneity among the classes of mammalian Smads in their ability to act as transcriptional activators in endothelial cells and that this correlates with their ability to interact with the conserved nuclear transcriptional coactivator CREB binding protein (CBP). Furthermore, specific ligand-activated Smad–CBP interactions seem essential for Smad-mediated transcriptional effects. CBP (and its homologue P300) have been demonstrated to play essential coactivator roles for a growing number of regulated transcription factors including CREB, nuclear receptors, myogenic helix–loop–helix factors, signal transduction and transcription (STAT) proteins, and members of the Rel (NF-κB) family (26–34). Because these coactivators have been proposed as an important locus of integration for signaling pathways in EC and other cell types (26, 27), our data suggest a novel mechanism whereby signals derived from the TGF-β superfamily of cytokines modulate a variety of cellular effects in endothelial cells.
METHODS
Cell Culture.
Primary bovine aortic endothelial cells (BAEC) were isolated as described (22) and cultured in low-glucose-DMEM supplemented with 10% heat-inactivated bovine calf serum, 2 mM l-glutamine, 250 units/ml penicillin G, and 250 μg/ml streptomycin. These were used at passages 3–12. Cos-7 cells were maintained in the same medium.
Expression Constructs and Transfections.
For transient transfections, cells were seeded at 50–70% confluency and transfected by using Lipofectamine (GIBCO/BRL) for 5 h. The cells were allowed to recover overnight in media containing 0.2% serum. Cells were then incubated in the absence or presence of 5 ng/ml human TGF-β1 (Genzyme). After approximately 18 h of incubation, luciferase, and β-galactosidase activity were measured (Tropix). All results are reported as luciferase activity (RLU) normalized to cotransfected β-galactosidase activity (expressed from cotransfected, constitutive expression constructs, e.g., cytomegalovirus β-galactosidase, phosphoglycerate kinase β-galactosidase) and are representative of at least three independent experiments.
Fusion proteins of full-length or COOH-terminal MH2 domain (see Fig. 1) of the Smads (pMSmads 2, 2C, 4, 4C, 6, and 7C) were created by PCR methods from cDNAs and cloned into either the pM (Gal4) or the pVP16 vectors (CLONTECH). All COOH-terminal MH2 domain Smads (Smads 2C, 4C, and 7C) include the linker region of the protein. The primers, which were constructed with unique, flanking restriction sites for cloning in-frame, are as follows: Smad2, 5′-GGA ATT CAT GTC GTC CAT CTT GCC-3′ and 5′-GGT GAA GCT TTA TGA CAT GCT TG-3′; Smad2C, 5′-GGA ATT CTT GAA TCA AAG TAT GGA CA-3′ and 5′-GGT GAA GCT TTA TGA CAT GCT TG-3′; Smad4, 5′-GGA ATT CAT GGA CAA TAT GTC-3′ and 5′-GGT GAA GCT TTC AGT CTA AAG GTT GTG GG-3′; Smad4C, 5′-GGA ATT CAC CAC CTG GAC TGG-3′ and 5′-GGT GAA GCT TTC AGT CTA AAG GTT GTG GG-3′; Smad7C, 5′-GGA ATT CAA ACC AAC TGC AGA C-3′ and 5′-CGG GAT CCC GCT ACC GGC TGT TGA AG-3′. Smads2*P and 2C*P truncation mutants were created as described above using their respective 5′ primers and the following common 3′ primer: 5′-CCC AAG CTT TAG CAA CGC ACT GAA GGG G-3′. This 3′ primer anneals just upstream of the SSXS motif in the MH2 domain of Smad2, deleting the COOH-terminal phosphorylation sites. All constructs were confirmed by sequencing. ORFs of Smads 6 and 7 were subcloned directly into the BamHI/SalI and EcoRI/HindIII sites, respectively, of the pM and pVP16 from the pGBT vectors described previously (22). All of the Smad6 constructs used in these experiments correspond to the ORF reported previously, which encodes a truncated isoform of human Smad6 (22).
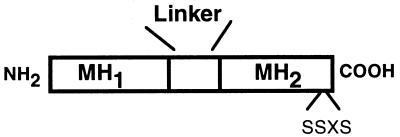
Figure 1.
Schematic of Smad protein structure. A general schematic of the structure of a human Smad protein is shown. Smad proteins consist of two domains (based on sequence conservation among identified Smad proteins) termed MH1 and MH2 (for MAD homology domain) located in their amino- and carboxyl-terminal halves, respectively. These regions are separated by a more variable, proline-rich domain known as the linker region. Certain signaling or receptor-associated Smads, such as Smad1 and Smad2, have a series of serine residues (termed the SSXS motif) present in their carboxyl termini, which are the sites of type 1 receptor-mediated phosphorylation. Human Smad4 and Smad7 do not possess the SSXS sequence.
VP16-CBP fusion protein expression plasmids and the mammalian two-hybrid system have been described previously (26). The reporter plasmid, pFR-Luc (Stratagene), contains five tandem repeats of the GAL4 binding site upstream of the luciferase gene. The p3TP-lux promoter construct, which has been described previously (22), is a chimeric TGF-β responsive promoter derived from sequences of the human PAI-1 and collagenase promoters, and the wild-type human PAI-1 promoter construct (P800) was obtained from D. Loskutoff. 12S E1A, mutE1A (NH2-terminal deletion that does not bind CBP/P300) CBP, and P300 expression constructs, as well as the CBP-VP16 fusion constructs, have been described previously (26). c-Jun-Gal4 and Elk-Gal4 fusion constructs (pMJun, pMElk) consist of the activation domains of c-Jun (amino acids 1–223) and Elk-1 (amino acids 307–427) fused to Gal4 DB.
Immunoprecipitations and Western Blotting.
Antibodies directed against the epitopes used in the immunoprecipitations and Western blots were obtained from commercial suppliers (Boehringer Mannheim, Santa Cruz Biotechnology). The anti-CBP/P300 antisera were obtained from Santa Cruz Biotechnology. Cell lysates were made approximately 24 h after transfection in 1% Triton X-100, 150 mM NaCl, 50 mM Tris⋅HCl, pH 8.0, with protease inhibitors, and immunoprecipitations were performed overnight at 4°C. Proteins were resolved on 10–12% SDS/PAGE denaturing gels, transferred to nitrocellulose by electroblotting and probed with appropriate antisera at 1/1,000 to 1/2,000 as indicated in the figures. Individual proteins were detected with a secondary antibody coupled to peroxidase and visualized with chemiluminescence (ECL).
RESULTS
Smad-Dependent Transcriptional Activation Is Enhanced by CBP.
An increasing number of regulated transcription factors have been shown to interact with the coactivator CBP (35). To investigate whether transcriptional events modulated by TGF-β involve CBP as a coactivator, we tested the ability of overexpressed recombinant CBP to stimulate two TGF-β responsive promoters in cultured endothelial cells. Both the plasminogen activator inhibitor-1 promoter (P800) and the p3TP promoter constructs are responsive to TGF-β, and this response is augmented approximately 2-fold by cotransfection of a plasmid expressing CBP. An expression plasmid for the closely related coactivator P300 gave similar results, although the magnitude of induction was consistently lower than that seen with CBP (data not shown).
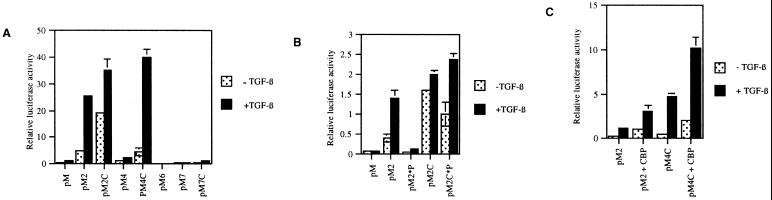
Recently, the conserved family of Smad proteins has been shown to transduce TGF-β superfamily transcriptional signals in species ranging from Drosophila to humans (6, 7, 9). To elucidate whether the Smad proteins involved in mediating TGF-β responses in endothelial cells are using CBP as a coactivator, we used a Gal4-based system to examine the ability of various Smad proteins to enhance transcription in these cells. Fusion proteins between the Gal4 DNA binding domain and either full-length or the COOH-terminal MH2 domains of Smads 2, 4, 6, and 7 were constructed and cotransfected with a luciferase reporter construct containing a minimal promoter coupled to five tandem Gal4 binding sites. As shown in Fig. 2, full-length Smad2 can function as a transcriptional activator in the presence of TGF-β in this system. The relatively low level of luciferase activity seen with the full-length Smad2–Gal4 construct in the absence of TGF-β likely reflects the small amount of endogenous TGF-β present in the endothelial culture system. The full-length Smad4–Gal4 construct reproducibly demonstrates a lower level of transcriptional activity in our hands, which is modestly increased in the presence of added TGF-β. Gal4 constructs containing only the COOH-terminal MH2 domain (including the linker) of these proteins (pM2C, pM4C) generated significantly higher basal and TGF-β stimulated levels of transcriptional activity (Fig. 2). These results are consistent with previous data suggesting that the transcriptional activation domains of these proteins are contained within the MH2 portion of these molecules and that the amino-terminal MH1 domain can mask this activity in the full-length, unstimulated protein (12, 13, 19, 21). In contrast to Smad2 and Smad4, neither Smad6, Smad7, nor the MH2 domain of Smad7 demonstrate any significant ability to up-regulate transcription in this assay. Thus, the various classes of Smad proteins differ in their ability to act as transcriptional activators in endothelial cells. The pathway-restricted Smad (e.g., Smad2), as well as Smad4, possess transcriptional activation domains, whereas the two recently identified “inhibitory Smads” (Smads 6 and 7), which are thought to function in the cytoplasm of the cell (22, 23, 36), demonstrate no transcriptional activation ability.
Figure 2.
Smad2 and Smad4, but not Smad6 or Smad7, can act as transcriptional activators in endothelial cells and are stimulated by CBP. The full-length or carboxyl-terminal domains (C) of Smads 2, 4, 6, and 7 were fused to the DNA binding domain of Gal4 (e.g., pM, Gal4BD alone; pM2, full-length SMad2–Gal4BD fusion; pM2C, C terminus of Smad2–Gal4BD fusion). These constructs were cotransfected into cultured BAEC with a luciferase reporter containing five upstream Gal4 binding sites, and the cells were subsequently treated with TGF-β1 (5 ng/ml) for 18 h, as indicated. (A) The relative transcriptional activity of all of the Gal4 constructs is displayed. Only pM2, pM2C, pM4, and pM4C demonstrate any significant increase in transcriptional activity over the Gal4 plasmid itself. Comparable expression of Gal4 fusion constructs was confirmed by Western blot (data not shown). (B) Mutants harboring deletions of the C-terminal serines that are the sites of type-1 receptor-mediated phosphorylation in Smad2. In the full-length Smad2 protein, the loss of these serines markedly diminishes the response to TGF-β (pM2*P), but this inhibition is reversed by removal of the amino-terminal MH1 domain of Smad2 (pM2C*P). (C) Both the basal and TGF-β stimulated activity of pM2 and pM4C are augmented by coexpression of CBP.
TGF-β receptor-mediated activation has been demonstrated to involve the direct phosphorylation of a series of COOH-terminal serines (SSXS motif) on Smad2. To determine whether the transcriptional activity of the Smad2–Gal4 fusion proteins require receptor-mediated phosphorylation, we constructed two mutants of Smad2 fused to Gal4 that lack these residues, termed pMSmad2*P and pMSmad2C*P, respectively (the latter is the COOH-terminal MH2 domain of Smad2 with the serines deleted). As shown in Fig. 2B, pMSmad2*P is completely unresponsive to TGF-β, but the truncated mutant pMSmad2C*P retains transcriptional activation ability even in the absence of TGF-β stimulation. These results thus confirm that ligand (TGF-β) activation requires the COOH-terminal serines in the full-length Smad2 molecule but that these residues are not required for transcriptional activation in the absence of the inhibitory, NH2-terminal, MH1 domain (pMSmad2C*P). As demonstrated in Fig. 2C, overexpression of CBP significantly enhanced the TGF-β stimulated transcriptional activation activity of both the Smad2 and Smad4C Gal4 constructs. CBP could also stimulate pM2C, pM4, and pM2C*P but had no effect on the Smad6 or Smad7 constructs (data not shown). Taken together, these results suggest that, in this system which recapitulates ligand-activated, Smad-mediated transcription in endothelial cells, CBP can function as a coactivator for Smad2, Smad4-mediated transcription.
Smad2 and Smad4 Specifically Interact with CBP in a Ligand-Dependent Fashion in Vivo.
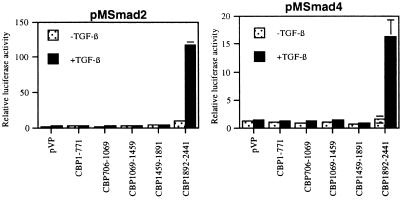
To determine whether Smads 2 and 4 can physically interact with CBP in vivo, we used both two-hybrid and coimmunoprecipitation strategies. In the mammalian two-hybrid assay, an interaction between two proteins is detected by fusing one protein to the Gal4 DNA binding domain, fusing the second protein to the powerful transcriptional activation domain of the VP16 protein, and coexpressing both constructs in a cell type of interest. An interaction between the two test proteins results in activation of the GAL4 luciferase reporter via the VP16 activation domain. Using the Smad–Gal4 fusion proteins described above, we tested the ability of these proteins to interact in endothelial cells with all of the domains of the CBP protein fused to VP16. As shown in the two representative experiments displayed in Fig. 3, both Smad2 and Smad4 interact with only the COOH-terminal 549 amino acids of CBP in a TGF-β dependent manner. All of the Smad2 and Smad4 constructs examined (i.e., pM2, 2C, 2*P, 2C*P, 4, 4C) interacted with CBP, and these interactions were limited to the single COOH-terminal domain of CBP (data not shown). We also tested the ability of Smad1, a distinct human Smad protein closely related to Smad2 but which mediates bone morphogenic protein signaling (19), to interact with CBP. Smad1, or its carboxyl-terminal MH2 domain, also interacted specifically with the COOH-terminal domain of CBP. However, this process required the presence of an activated bone morphogenic protein type-1 receptor and was not observed in the presence of an activated TGF-β receptor alone (data not shown).
Figure 3.
Smad2 and Smad4 can interact specifically with the C terminus of CBP in endothelial cells. Smad2 (pM2) and Smad4 (pM4) were cotransfected into BAEC with plasmids expressing the indicated domains (numbers correspond to amino acids) of the murine CBP protein fused to the potent transcriptional activation domain of VP16. A specific interaction between the Gal4 fusion protein and the VP16 fusion protein results in induction of the Gal4 reporter. The amount of Gal4–Smad vector used was approximately 1/10 used in Fig. 2 to minimize background luciferase activity. The full-length Smad2 and Smad4 proteins selectively interact with the C-terminal domain of CBP (amino acids 1892–2441) in the presence of TGF-β. No reproducible interaction between the other domains of CBP and Smad proteins was observed. All CBP fusion constructs express at comparable levels (data not shown).
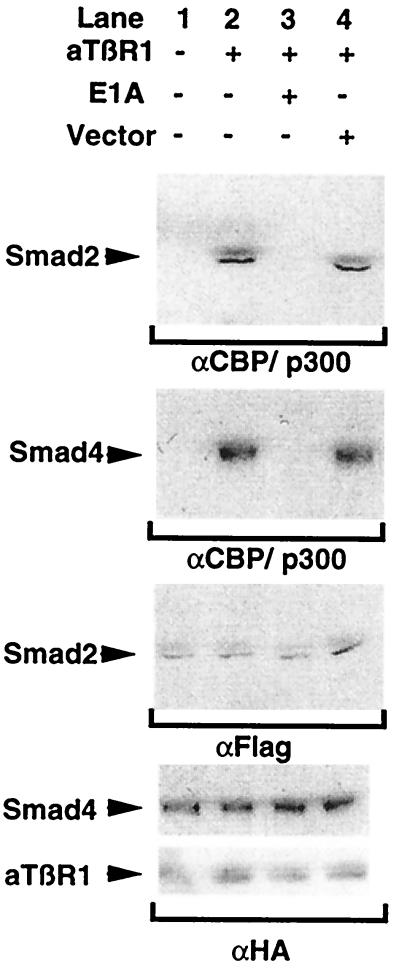
To demonstrate Smad–CBP interactions in cell extracts, we performed a series of coimmunoprecipitations. Preliminary data demonstrated the coimmunoprecipitation of recombinant, full-length CBP (epitope tagged) with tagged Smad2 and Smad4 proteins, but not Smad7, consistent with the mammalian two-hybrid results (data not shown). In the experiment displayed in Fig. 4, we used a polyclonal antisera to CBP/P300 to immunoprecipitate the endogenous molecule and determine whether Smad proteins were associated in endothelial cells. As shown in lane 1, even when Smad2 and Smad4 proteins were overexpressed as epitope-tagged species, there is essentially no detectable interaction with CBP in the absence of TGF-β receptor stimulation. In contrast, if the cells were cotransfected with a constitutively active form of the TGF-β type 1 receptor, significant amounts of both Smad2 and Smad4 proteins coimmunoprecipitate with CBP/P300 (Fig. 4, lane 2). To confirm that these Smad–coactivator interactions were dependent on activation of the Smad pathway, we coexpressed two recently identified inhibitory, endothelial-expressed Smads, Smad6 and Smad7 (22–24). As shown in Fig. 4, lane 3, the expression vector alone (pCIneo) had no effect, whereas both Smad6 and Smad7 completely inhibited the interactions of Smads 2 and 4 with CBP. These results thus confirm that activation of the Smad proteins is required for the CBP interactions seen and that these effects are not solely caused by overexpression of these proteins.
Figure 4.
Smads 2 and 4 can interact with CBP in vivo. Cos-7 cells were transfected with the indicated combinations of epitope-tagged activated TGF-β type-1 receptor, Smad expression construct, or empty expression vector. The cells were then lysed and immunoprecipitated with an anti-CBP/P300 antisera. Coprecipitating Smad proteins were detected by Western blot. The upper two panels demonstrate that significant amounts of both Smad2 and Smad4 protein coimmunoprecipitate with CBP/P300 in the presence of the activated receptor. These interactions are inhibited by the simultaneous expression of either Smad6 or Smad7 but not by cotransfection of an empty expression vector (pCIneo). The bottom panels confirm comparable Smad2, Smad4, and activated receptor expression by immunoprecipitation and Western blotting with antisera against the epitopes fused to these proteins.
Smad-Mediated Transcriptional Activation Requires Smad–CBP Interactions.
The COOH-terminal domain of CBP, which interacts with activated Smads in the mammalian two-hybrid system, is also the site at which this coactivator interacts with a number of other important regulatory proteins (29, 31, 37, 38). One of these, the viral protein E1A, is thought to sequester CBP and thus modulate the transcriptional effects of many effector proteins (34, 39, 40). We used expression of 12S E1A to test whether CBP was required for Smad-mediated transcriptional events. Preliminary experiments demonstrated potent inhibition of the TGF-β responsive promoters P800 and p3TP by 12S E1A (data not shown), so we examined the effect of 12S E1A expression in the Smad–Gal4 system. As shown in Table 1, 12S E1A can almost completely inhibit the stimulated activity of all of the Smad2 and Smad4 constructs in endothelial cells. As a control for nonspecific effects, we also expressed a mutant form of E1A (mutE1A). This molecule harbors an NH2-terminal deletion that renders it unable to interact significantly with CBP or P300. As shown in Table 1, this protein did not inhibit Smad-dependent transcription in this system. As a control for the assay, we also looked at the effects of these E1A proteins on transcription mediated by the activation domains of c-Jun and Elk-1 fused to Gal4 (c-Jun–Gal4, Elk–Gal4), as both of these transcriptional activators have been demonstrated previously to interact functionally with CBP. The results for these constructs were virtually identical to those of the Smad constructs (Table 1), in that they are inhibited by 12S E1A but not by the mutant protein.
Table 1.
12S E1A, but not an amino-terminal truncated mutant of E1A, can inhibit Smad-dependent transcription in BAEC
| Vector | 12S E1A | mut-E1A | |
|---|---|---|---|
| pMSmad2 | 100 | 8 ± 1.2 | 88 ± 11.1 |
| pMSmad2C | 100 | 10 ± 2.1 | 89 ± 12.0 |
| pMSmadC*P | 100 | 3 ± 1.1 | 91 ± 3.4 |
| pMSmad4C | 100 | 3 ± 0.4 | 105 ± 9.3 |
| pMJun | 100 | 7 ± 2.3 | 103 ± 5.1 |
| pMElk | 100 | 2 ± 1.0 | 101 ± 10.0 |
The left side of the table lists the various Smad–Gal4 constructs that were assayed for their ability to activate the Gal4 reporter. The first column represents the level of transcriptional activation seen with maximal stimulation (TGF-β) normalized to 100%. 12S E1A markedly inhibits the transcriptional activity of all of the Smad constructs tested. The mutant form of E1A, which is unable to bind to CBP, does not inhibit the Smad-dependent transcription. Gal4–Jun and Gal4–Elk were also assayed as positive controls for proteins known to require CBP as a coactivator.
E1A can interact with a variety of cellular proteins. To ascertain if the inhibitory effect on Smad-mediated transcription observed was caused by sequestration of limiting amounts of CBP within the cell, we attempted to rescue the E1A-mediated inhibition by titrating in increasing amounts of CBP expression plasmid. As shown in Fig. 5, for both Smad2 and Smad4, as well as the c-Jun activation domain, we could effectively restore transcriptional activation in the presence of 12S E1A by increasing amounts of CBP expression. In fact, all of the Smad constructs inhibited by 12S E1A expression were effectively rescued by coexpression of CBP (data not shown). To determine whether these results correlated with the biochemical association of Smads and CBP, we performed immunoprecipitations in the presence and absence of 12S E1A expression. Fig. 6 is a coimmunoprecipitation experiment demonstrating that in the presence of 12S E1A, the TGF-β stimulated association of Smads 2 and 4 with CBP is absent (compare lanes 3 and 4). Taken together, these results indicate that CBP–Smad interactions seem to be required for Smad-mediated transcriptional activation to occur in endothelial cells.
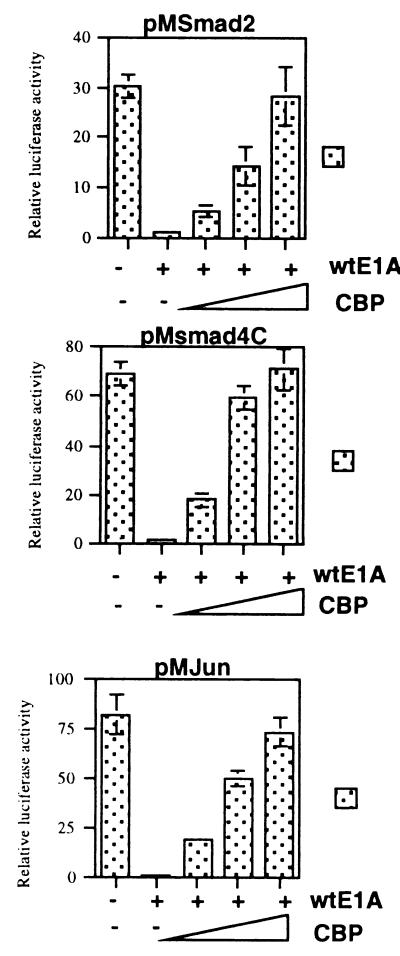
Figure 5.
Overexpression of CBP can rescue 12S E1A-mediated inhibition of Smad transcriptional activation. pM2, pM4C, and pMJun were cotransfected into BAEC with 12S E1A and increasing amounts of CBP. Total DNA was kept constant by the addition of empty vector. In the presence of increasing amounts of CBP expression vector, the inhibition observed in the presence of 12S E1A is reversed.
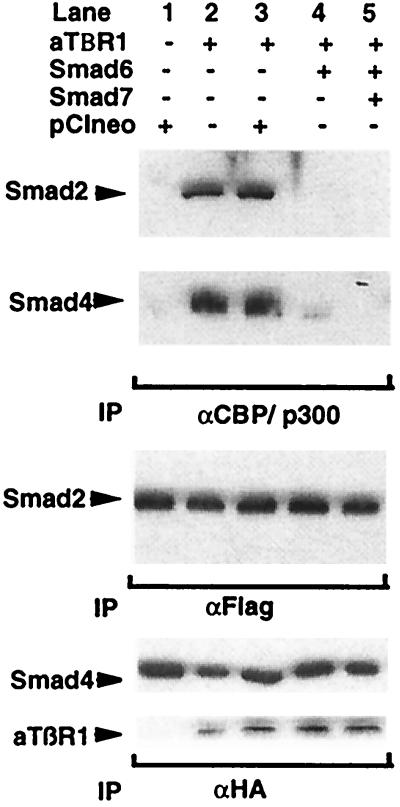
Figure 6.
12S E1A inhibits the association of activated Smad2 and Smad4 with CBP in vivo. Cos-7 cells were transfected with expression constructs expressing epitope-tagged (HA, Flag) versions of Smad2 and Smad4 and the indicated combinations of activated TGF-β type-1 receptor and 12S E1A expression vector. The cells were subsequently lysed and subjected to immunoprecipitation with anti-CBP/p300 antisera, and levels of coimmunoprecipitating Smad proteins were determined by Western blot. The bottom panels confirm comparable Smad2, Smad4, and activated receptor expression.
DISCUSSION
The identification of Smad proteins as critical intracellular mediators of TGF-β superfamily transcriptional responses has been a major advance in understanding how this important family of cytokines and/or growth factors elicit their protean biologic effects (4–6). In this paper, we demonstrate that Smad2 and Smad4, two mediators of TGF-β1 signaling in endothelial and other cells, can act as ligand (TGF-β)-activated transcriptional activators in endothelial cells, whereas two distinct, recently identified endothelial-expressed Smads, Smad6 and Smad7, do not. Furthermore, transcriptional activation by Smad proteins in endothelial cells requires a ligand-induced interaction with the transcriptional coactivator CBP.
The molecular mechanisms by which certain Smad proteins can function as transcriptional activators and/or coactivators are just now being elucidated. Work in the Xenopus system has identified a specific cis-acting activin response element that interacts with a complex consisting of two distinct Xenopus Smad proteins and a forkhead-containing DNA binding protein known as Fast-1 (41). Alone, the individual components of this complex are thought to be unable to activate transcription efficiently, until the activin pathway is activated by receptor–ligand interactions and the active complex is formed. Xenopus Fast-1 can interact with certain mammalian Smad proteins and activate the activin response element when introduced into mammalian cells, suggesting that human homologues of Fast-1 exist (23, 41). In contrast to these Xenopus Smad proteins, a Smad protein from Drosophila has been reported to bind DNA directly, and in vitro, human Smad3 and Smad4, but not Smad2, can interact directly with specific DNA sequences (42, 43). In the studies reported here, we have demonstrated that Smad2 and Smad4, two Smad proteins that can synergize as activators of transcription in response to TGF-β, selectively interact with the COOH-terminal domain of the conserved mammalian transcriptional coactivator CBP. This interaction was demonstrated by both a two-hybrid approach in endothelial cells as well as a biochemical approach using antisera against endogenous CBP. In addition, this interaction seems specific, in that only certain Smad proteins (or truncated forms of these proteins) that contain potential transcriptional activation domains, display any detectable interaction. A closely related endothelial-expressed Smad, Smad7, which does not possess transcriptional activation activity, did not demonstrate any detectable interaction with CBP. Furthermore, both Smad2 and Smad4 interact with CBP in a ligand-dependent manner. In the absence of receptor (TGF-β) stimulation, or in the presence of the inhibitory Smads 6 and 7, no significant interaction with CBP is observed. These results argue strongly that the interactions observed are specifically a result of activation of the TGF-β signaling pathway and are not merely a result of overexpression of recombinant Smad proteins.
The interactions that we have observed between Smads and CBP closely resemble the interactions previously described between nuclear receptors such as the retinoic acid receptor and CBP (31, 38, 44, 45). These nuclear receptors also require ligand activation to interact with CBP and have been demonstrated to bind the same COOH-terminal domain of CBP as the Smads. This domain is adjacent to, but physically distinct from, the C/H3 domain of CBP that interacts with 12S E1A and STAT-1α. It is interesting to note that several members of the nuclear receptor superfamily are capable of mediating both negative and positive transcriptional regulation. It will be interesting to see whether Smad-mediated transcriptional regulation demonstrates similar complexity.
In addition to demonstrating selectivity, Smad–CBP interactions seem to be critical for TGF-β transcriptional responses. In the presence of 12S E1A, a viral molecule that is capable of binding and sequestering limiting amounts of CBP present within the cell, Smad-dependent, TGF-β transcriptional responses were completely inhibited. This response was not seen with a mutant form of E1A that does not interact with CBP, and more important, the 12S E1A-meditated inhibition could be reversed by the addition of exogenous CBP. Furthermore, in the presence of 12S E1A, the biochemical association of Smad2 and Smad4 with endogenous CBP was blocked. These results indicate that 12S E1A can inhibit Smad function by interfering with the assembly of Smad–coactivator complexes and that interactions between CBP and activated Smads are required for Smad-dependent transcriptional responses.
Transcriptional regulation requires at least three general classes of proteins: (i) proteins that recognize specific cis-acting DNA motifs, (ii) proteins that are recruited to promoters by protein–protein interactions and act as coactivators or corepressors, and (iii) proteins that alter the structure of chromatin. CBP, which possesses histone acetyltransferase activity (46), has been demonstrated to interact directly with elements of the basal transcriptional apparatus such as TFIIB and RNA polymerase II, as well as several classes of sequence-specific transcription factors including c-Jun, c-Fos, Sap-1a, Elk-1, STAT-1α, CREB, NF-κB, and members of the nuclear receptor superfamily (26, 27, 29, 31–33, 37, 44, 47). In this capacity, CBP is thought to function as a critical bridge between these sequence-specific factors and the basal transcriptional machinery. Moreover, competition for limiting pools of intracellular CBP is thought to mediate the functional interactions between diverse signaling pathways such as the Ras/AP-1, Jak/STAT, and nuclear receptor-mediated pathways (27, 45, 48). Thus, CBP may function as an important locus of signal integration for diverse signals within the cell.
Our results suggest that Smad proteins such as Smad2 and Smad4 may function in a manner analogous to many of the above- mentioned signal-dependent transcription factors. Namely, either alone or in combination with yet-to-be defined sequence-specific DNA binding molecules (e.g., Fast-1 like factors), these proteins interact with specific cis-acting DNA regulatory elements and modulate transcription by interacting in a specific manner with CBP to facilitate formation of productive transcription initiation complexes. Such a model would predict that there may be complex functional interactions between Smad proteins and other regulated transcription factors that utilize these coactivators. Indeed, it has recently been independently reported that Smad-dependent activation of a TGF-β dependent promoter required a series of TRE or AP-1 sites (49) and that a dominant negative form of Smad3 could inhibit transcriptional responses elicited by diverse stimuli. In addition, we have recently demonstrated that overexpression of Smad proteins can modulate endothelial gene expression in response to fluid mechanical (flow) stimuli (22). These complex interactions between potentially disparate stimuli, the transcription factors they regulate, and Smad proteins likely involve shared interactions with common coactivators such as CBP.
In summary, we have demonstrated that Smad2 and Smad4, two proteins involved in mediating TGF-β transcriptional responses in endothelial and other cell types, can functionally interact with the conserved transcriptional coactivator CBP. Specific Smad–CBP interactions seem to be essential for Smad-mediated transcriptional activation in endothelial cells. This demonstrates a functional interaction between this newly identified class of signaling molecules (Smads) and an essential component of the mammalian transcriptional apparatus (CBP) and extends our insight into how Smad proteins may mediate transcriptional responses in cells. Furthermore, our observations suggest that functional integration at the level of required transcriptional coactivators such as CBP/P300 may play an important role in the many biologic effects characteristic of the TGF-β superfamily of cytokines and growth factors.
Acknowledgments
We thank William Atkinson and Kay Case for expert cell culturing and Keith Anderson and Jeanne Kiely for technical assistance and advice. We are indebted to Katherine Galvin, Jeixing Cai, Yubin Qui, Carlos Gimeno, and Geoffrey Ginsburg at Millennium Pharmaceuticals Inc. for advice and collaborative assistance. J.N.T. is supported by a Howard Hughes Medical Institute Fellowship for Physicians, and J.D.B. is supported by the Sarnoff Endowment for Cardiovascular Science Student Fellowship. This work was supported in part by the grants from the National Heart Lung and Blood Institute to M.A.G. (P50-HL56985, R37-HL-51150) and T.C. (P01-HL-36028) and a sponsored research agreement between the Brigham and Women’s Hospital and Millennium Pharmaceuticals Inc. in collaboration with Eli Lilly Co.
ABBREVIATIONS
- BAEC
bovine aortic endothelial cells
- CBP
CREB binding protein
- STAT
signal transduction and transcription
- TGF-β
transforming growth factor β
References
- 1.Metcalfe J C, Grainger D J. J Hum Hypertens. 1995;9:679. [PubMed] [Google Scholar]
- 2.Grainger D J, Metcalfe J C. Biol Rev Cambr Philos Soc. 1995;70:571–596. doi: 10.1111/j.1469-185x.1995.tb01652.x. [DOI] [PubMed] [Google Scholar]
- 3.Wahl S M. J Exp Med. 1994;180:1587–1590. doi: 10.1084/jem.180.5.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker J C, Harland R M. Curr Opin Gen Dev. 1997;7:467–473. doi: 10.1016/s0959-437x(97)80072-x. [DOI] [PubMed] [Google Scholar]
- 5.Heldin C H, Miyazono K, ten Dijke P. Nature (London) 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 6.Massague J. Cell. 1996;85:947–950. doi: 10.1016/s0092-8674(00)81296-9. [DOI] [PubMed] [Google Scholar]
- 7.Raftery L A, Twombly V, Wharton K, Gelbart W M. Genetics. 1995;139:241–254. doi: 10.1093/genetics/139.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newfeld S J, Chartoff E H, Graff J M, Melton D A, Gelbart W M. Development (Cambridge, UK) 1996;122:2099–2108. doi: 10.1242/dev.122.7.2099. [DOI] [PubMed] [Google Scholar]
- 9.Graff J M, Bansal A, Melton D A. Cell. 1996;85:479–487. doi: 10.1016/s0092-8674(00)81249-0. [DOI] [PubMed] [Google Scholar]
- 10.Eppert K, Scherer S W, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui L C, Bapat B, Gallinger S, Andrulis I L, et al. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 11.Souchelnytskyi S, Tamaki K, Engstrom U, Wernstedt C, ten Dijke P, Heldin C H. J Biol Chem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- 12.Liu F, Pouponnot C, Massague J. Genes Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Nature (London) 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 14.Hahn S A, Schutte M, Hoque A T, Moskaluk C A, da Costa L T, Rozenblum E, Weinstein C L, Fischer A, Yeo C J, Hruban R H, Kern S E. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 15.Takaku K, Oshima M, Miyoshi H, Matsui M, Seldin M F, Taketo M M. Cell. 1998;92:645–656. doi: 10.1016/s0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- 16.Zhou S, Buckhaults P, Zawel L, Bunz F, Riggins G, Le Dai J, Kern S E, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1998;95:2412–2416. doi: 10.1073/pnas.95.5.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waldrip W R, Bikoff E K, Hoodless P A, Wrana J L, Robertson E J. Cell. 1998;92:797–808. doi: 10.1016/s0092-8674(00)81407-5. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Bhushan A, Vale W. Proc Natl Acad Sci USA. 1997;94:12938–12943. doi: 10.1073/pnas.94.24.12938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoodless P A, Haerry T, Abdollah S, Stapleton M, O’Connor M B, Attisano L, Wrana J L. Cell. 1996;85:489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- 20.Kretzschmar M, Liu F, Hata A, Doody J, Massague J. Genes Dev. 1997;11:984–995. doi: 10.1101/gad.11.8.984. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Feng X, We R, Derynck R. Nature (London) 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 22.Topper J N, Cai J, Qiu Y, Anderson K R, Xu Y Y, Deeds J D, Feeley R, Gimeno C J, Woolf E A, Tayber O, et al. Proc Natl Acad Sci USA. 1997;94:9314–9319. doi: 10.1073/pnas.94.17.9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu Y Y, Grinnell B W, Richardson M A, Topper J N, Gimbrone M A, Jr, Wrana J L, et al. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 24.Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Nature (London) 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 25.Nakao A, Afrakhte M, Moren A, Nakayama T, Christian J L, Heuchel R, Itoh S, Kawabata M, Heldin N E, Heldin C H, ten Dijke P. Nature (London) 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 26.Gerritsen M E, Williams A J, Neish A S, Moore S, Shi Y, Collins T. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, et al. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 28.Arany Z, Huang L E, Eckner R, Bhattacharya S, Jiang C, Goldberg M A, Bunn H F, Livingston D M. Proc Natl Acad Sci USA. 1996;93:12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston D M. Nature (London) 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 30.Bannister A J, Oehler T, Wilhelm D, Angel P, Kouzarides T. Oncogene. 1995;11:2509–2514. [PubMed] [Google Scholar]
- 31.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Nature (London) 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 32.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 33.Smith C L, Onate S A, Tsai M J, O’Malley B W. Proc Natl Acad Sci USA. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swope D L, Mueller C L, Chrivia J C. J Biol Chem. 1996;271:28138–28145. doi: 10.1074/jbc.271.45.28138. [DOI] [PubMed] [Google Scholar]
- 35.Janknecht R, Hunter T. Curr Biol. 1996;6:951–954. doi: 10.1016/s0960-9822(02)00636-x. [DOI] [PubMed] [Google Scholar]
- 36.Hata A, Lagna G, Massague J, Hemmati-Brivanlou A. Genes Dev. 1998;12:186–197. doi: 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath C M, Darnell J E., Jr Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 39.Bevilacqua M A, Faniello M C, Quaresima B, Tiano M T, Giuliano P, Feliciello A, Avvedimento V E, Cimino F, Costanzo F. J Biol Chem. 1997;272:20736–20741. doi: 10.1074/jbc.272.33.20736. [DOI] [PubMed] [Google Scholar]
- 40.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 41.Chen X, Rubock M J, Whitman M. Nature (London) 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- 42.Kim J, Johnson K, Chen H J, Carroll S, Laughon A. Nature (London) 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- 43.Zawel L, Dai J L, Buckhaults P, Zhou S, Kinzler K W, Vogelstein B, Kern S E. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 44.Aarnisalo P, Palvimo J J, Janne O A. Proc Natl Acad Sci USA. 1998;95:2122–2127. doi: 10.1073/pnas.95.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T M, Rose D W, Rosenfeld M G, Glass C K. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bannister A J, Kouzarides T. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 47.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. Nature (London) 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 48.Kurokawa R, Kalafus D, Ogliastro M H, Kioussi C, Xu L, Torchia J, Rosenfeld M G, Glass C K. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 49.Yingling J M, Datto M B, Wong C, Frederick J P, Liberati N T, Wang X F. Mol Cell Biol. 1997;17:7019–7028. doi: 10.1128/mcb.17.12.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]