Abstract
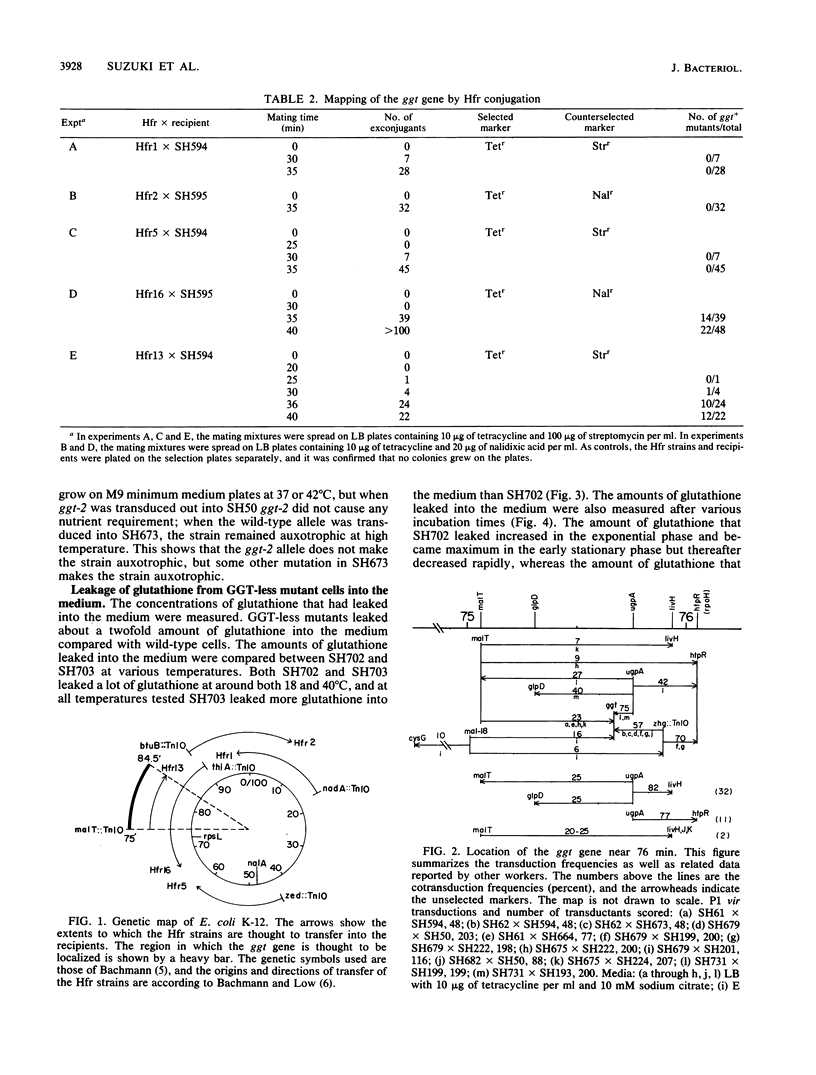
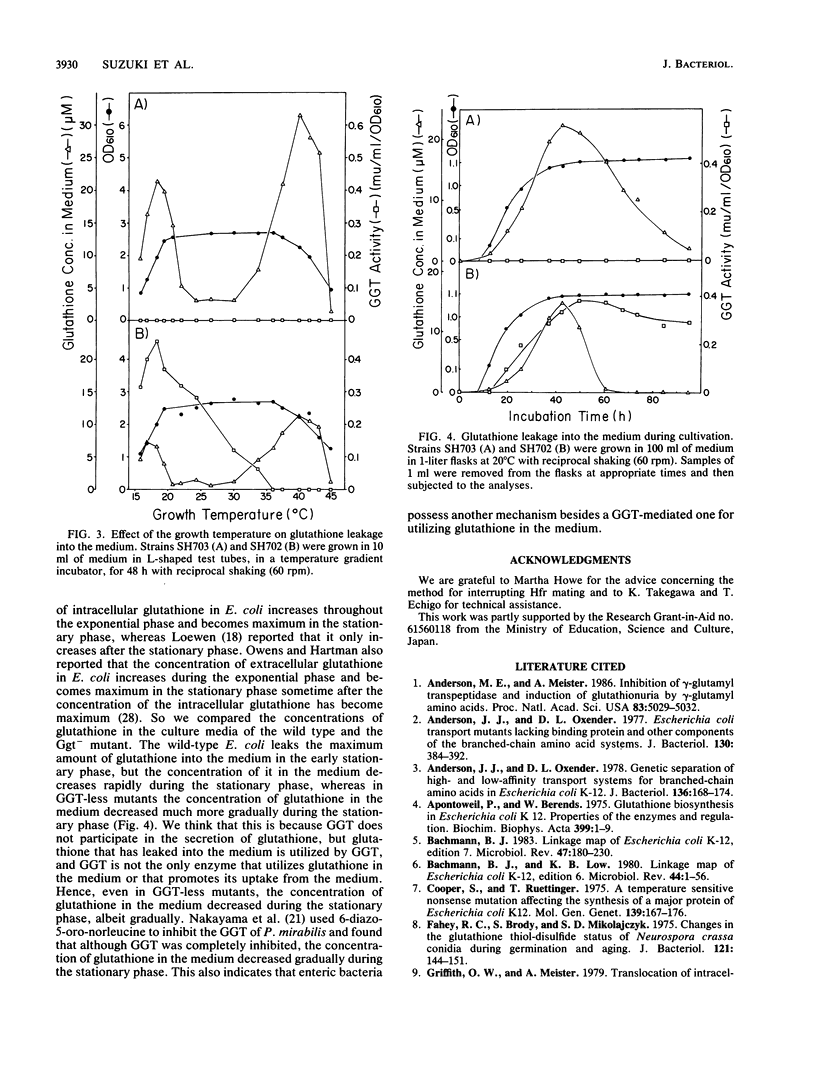
Escherichia coli K-12 mutants lacking gamma-glutamyltranspeptidase (EC 2.3.2.2) were isolated after mutagenesis of cells with ethyl methanesulfonate. They lost the enzyme activity to different extents. The mutations of two mutants that had lost the enzyme activity completely were mapped at 76 min of the E. coli K-12 linkage map. These mutations made the cells neither nutrient requiring nor cold sensitive. The mutants leaked much more glutathione into the medium than the wild type. We propose the symbol ggt for these mutations.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. J., Oxender D. L. Escherichia coli transport mutants lacking binding protein and other components of the branched-chain amino acid transport systems. J Bacteriol. 1977 Apr;130(1):384–392. doi: 10.1128/jb.130.1.384-392.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. J., Oxender D. L. Genetic separation of high- and low-affinity transport systems for branched-chain amino acids in Escherichia coli K-12. J Bacteriol. 1978 Oct;136(1):168–174. doi: 10.1128/jb.136.1.168-174.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M. E., Meister A. Inhibition of gamma-glutamyl transpeptidase and induction of glutathionuria by gamma-glutamyl amino acids. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5029–5032. doi: 10.1073/pnas.83.14.5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apontoweil P., Berends W. Glutathione biosynthesis in Escherichia coli K 12. Properties of the enzymes and regulation. Biochim Biophys Acta. 1975 Jul 14;399(1):1–9. doi: 10.1016/0304-4165(75)90205-6. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980 Mar;44(1):1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S., Ruettinger T. A temperature sensitive nonsense mutation affecting the synthesis of a major protein of Escherichia coli K12. Mol Gen Genet. 1975 Aug 5;139(2):167–176. doi: 10.1007/BF00264696. [DOI] [PubMed] [Google Scholar]
- Fahey R. C., Brody S., Mikolajczyk S. D. Changes in the glutathione thiol-disulfide status of Neurospora crassa conidia during germination and aging. J Bacteriol. 1975 Jan;121(1):144–151. doi: 10.1128/jb.121.1.144-151.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith O. W., Meister A. Translocation of intracellular glutathione to membrane-bound gamma-glutamyl transpeptidase as a discrete step in the gamma-glutamyl cycle: glutathionuria after inhibition of transpeptidase. Proc Natl Acad Sci U S A. 1979 Jan;76(1):268–272. doi: 10.1073/pnas.76.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith O. W., Novogrodsky A., Meister A. Translocation of glutathione from lymphoid cells that have markedly different gamma-glutamyl transpeptidase activities. Proc Natl Acad Sci U S A. 1979 May;76(5):2249–2252. doi: 10.1073/pnas.76.5.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A. D., Zhou Y. N., Gross C., Heilig J., Christie G. E., Calendar R. Mutations in the rpoH (htpR) gene of Escherichia coli K-12 phenotypically suppress a temperature-sensitive mutant defective in the sigma 70 subunit of RNA polymerase. J Bacteriol. 1985 Mar;161(3):939–943. doi: 10.1128/jb.161.3.939-943.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gushima H., Miya T., Murata K., Kimura A. Purification and characterization of glutathione synthetase from Escherichia coli B. J Appl Biochem. 1983 Jun;5(3):210–218. [PubMed] [Google Scholar]
- Guyer M. S., Reed R. R., Steitz J. A., Low K. B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- HAYASHI S., KOCH J. P., LIN E. C. ACTIVE TRANSPORT OF L-ALPHA-GLYCEROPHOSPHATE IN ESCHERICHIA COLI. J Biol Chem. 1964 Sep;239:3098–3105. [PubMed] [Google Scholar]
- Inoue M., Horiuchi S., Morino Y. gamma-Glutamyl transpeptidase in rat ascites tumor cell LY-5. Lack of functional correlation of its catalytic activity with the amino acid transport. Eur J Biochem. 1977 Sep;78(2):609–615. doi: 10.1111/j.1432-1033.1977.tb11774.x. [DOI] [PubMed] [Google Scholar]
- Inoue M., Morino Y. Direct evidence for the role of the membrane potential in glutathione transport by renal brush-border membranes. J Biol Chem. 1985 Jan 10;260(1):326–331. [PubMed] [Google Scholar]
- Loewen P. C. Levels of glutathione in Escherichia coli. Can J Biochem. 1979 Feb;57(2):107–111. doi: 10.1139/o79-013. [DOI] [PubMed] [Google Scholar]
- Milbauer R., Grossowicz N. Gamma-glutamyl transfer reactions in bacteria. J Gen Microbiol. 1965 Nov;41(2):185–194. doi: 10.1099/00221287-41-2-185. [DOI] [PubMed] [Google Scholar]
- Nakayama R., Kumagai H., Tochikura T. Leakage of glutathione from bacterial cells caused by inhibition of gamma-glutamyltranspeptidase. Appl Environ Microbiol. 1984 Apr;47(4):653–657. doi: 10.1128/aem.47.4.653-657.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama R., Kumagai H., Tochikura T. Purification and properties of gamma-glutamyltranspeptidase from Proteus mirabilis. J Bacteriol. 1984 Oct;160(1):341–346. doi: 10.1128/jb.160.1.341-346.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama R., Kumagai H., Tochikura T. gamma-Glutamyltranspeptidase from Proteus mirabilis: localization and activation by phospholipids. J Bacteriol. 1984 Dec;160(3):1031–1036. doi: 10.1128/jb.160.3.1031-1036.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F. C., VanBogelen R. A. Positive regulatory gene for temperature-controlled proteins in Escherichia coli. Biochem Biophys Res Commun. 1981 May 29;100(2):894–900. doi: 10.1016/s0006-291x(81)80257-4. [DOI] [PubMed] [Google Scholar]
- OUCHTERLONY O. Antigen-antibody reactions in gels. IV. Types of reactions in coordinated systems of diffusion. Acta Pathol Microbiol Scand. 1953;32(2):230–240. [PubMed] [Google Scholar]
- Orlowski M., Meister A. The gamma-glutamyl cycle: a possible transport system for amino acids. Proc Natl Acad Sci U S A. 1970 Nov;67(3):1248–1255. doi: 10.1073/pnas.67.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens R. A., Hartman P. E. Export of glutathione by some widely used Salmonella typhimurium and Escherichia coli strains. J Bacteriol. 1986 Oct;168(1):109–114. doi: 10.1128/jb.168.1.109-114.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellefigue F., Butler J. D., Spielberg S. P., Hollenberg M. D., Goodman S. I., Schulman J. D. Normal amino acid uptake by cultured human fibroblasts does not require gamma-glutamyl transpeptidase. Biochem Biophys Res Commun. 1976 Dec 20;73(4):997–1002. doi: 10.1016/0006-291x(76)90221-7. [DOI] [PubMed] [Google Scholar]
- Robins R. J., Davies D. D. The role of glutathione in amino-acid absorption. Lack of correlation between glutathione turnover and amino-acid absorption by the yeast Candida utilis. Biochem J. 1981 Jan 15;194(1):63–70. doi: 10.1042/bj1940063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman J. D., Goodman S. I., Mace J. W., Patrick A. D., Tietze F., Butler E. J. Glutathionuria: inborn error of metabolism due to tissue deficiency of gamma-glutamyl transpeptidase. Biochem Biophys Res Commun. 1975 Jul 8;65(1):68–74. doi: 10.1016/s0006-291x(75)80062-3. [DOI] [PubMed] [Google Scholar]
- Schweizer H., Grussenmeyer T., Boos W. Mapping of two ugp genes coding for the pho regulon-dependent sn-glycerol-3-phosphate transport system of Escherichia coli. J Bacteriol. 1982 Jun;150(3):1164–1171. doi: 10.1128/jb.150.3.1164-1171.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague G. F., Jr, Bell R. M., Cronan J. E., Jr A mutant of Escherichia coli auxotrophic for organic phosphates: evidence for two defects in inorganic phosphate transport. Mol Gen Genet. 1975 Dec 30;143(1):71–77. doi: 10.1007/BF00269422. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Kumagai H., Tochikura T. gamma-Glutamyltranspeptidase from Escherichia coli K-12: formation and localization. J Bacteriol. 1986 Dec;168(3):1332–1335. doi: 10.1128/jb.168.3.1332-1335.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Kumagai H., Tochikura T. gamma-Glutamyltranspeptidase from Escherichia coli K-12: purification and properties. J Bacteriol. 1986 Dec;168(3):1325–1331. doi: 10.1128/jb.168.3.1325-1331.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TALALAY P. S. Glutathione breakdown and transpeptidation reactions in Proteus vulgaris. Nature. 1954 Sep 11;174(4428):516–517. doi: 10.1038/174516b0. [DOI] [PubMed] [Google Scholar]
- Tate S. S., Meister A. gamma-Glutamyl transpeptidase from kidney. Methods Enzymol. 1985;113:400–419. doi: 10.1016/s0076-6879(85)13053-3. [DOI] [PubMed] [Google Scholar]
- Tate S. S., Meister A. gamma-Glutamyl transpeptidase: catalytic, structural and functional aspects. Mol Cell Biochem. 1981 Sep 25;39:357–368. doi: 10.1007/BF00232585. [DOI] [PubMed] [Google Scholar]
- Taylor A. L., Trotter C. D. Revised linkage map of Escherichia coli. Bacteriol Rev. 1967 Dec;31(4):332–353. doi: 10.1128/br.31.4.332-353.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]


