Abstract
To elucidate the mechanisms underlying chromosomal translocations in diffuse large B cell lymphoma (DLBCL), we investigated the nature and extent of immunoglobulin class switch recombination (CSR) in these tumors. We used Southern blotting to detect legitimate and illegitimate CSR events in tumor samples of the activated B cell–like (ABC), germinal center B cell–like (GCB), and primary mediastinal B cell lymphoma (PMBL) subgroups of DLBCL. The frequency of legitimate CSR was lower in ABC DLBCL than in GCB DLBCL and PMBL. In contrast, ABC DLBCL had a higher frequency of internal deletions within the switch μ (Sμ) region compared with GCB DLBCL and PMBL. ABC DLBCLs also had frequent deletions within Sγ and other illegitimate switch recombinations. Sequence analysis revealed ongoing Sμ deletions within ABC DLBCL tumor clones, which were accompanied by ongoing duplications and activation-induced cytidine deaminase–dependent somatic mutations. Unexpectedly, short fragments derived from multiple chromosomes were interspersed within Sμ in one case. These findings suggest that ABC DLBCLs have abnormalities in the regulation of CSR that could predispose to chromosomal translocations. Accordingly, aberrant switch recombination was responsible for translocations in ABC DLBCLs involving BCL6, MYC, and a novel translocation partner, SPIB.
Diffuse large B cell lymphoma (DLBCL), the most common type of non-Hodgkin's lymphoma, accounts for ∼30% of all lymphoma cases (1). DLBCL can be cured using anthracycline-based chemotherapy regimens in just 40–50% of patients, suggesting that DLBCL represents a heterogeneous disease (2). This concept was supported by several recent gene expression profiling studies distinguishing at least three different molecular DLBCL subgroups (3–7). These three subgroups have been termed germinal center B cell–like (GCB) DLBCL, activated B cell–like (ABC) DLBCL, and primary mediastinal B cell lymphoma (PMBL). The DLBCL subgroups differ not only with respect to their gene expression profiles, but they also have significantly different overall survival rates and use distinct oncogenic pathways (8). By these criteria, the DLBCLs can be viewed as distinct diseases that arise from B lymphocytes at different stages of differentiation.
GCB DLBCL tumors express many genes that are characteristic for normal germinal center B cells, suggesting that this lymphoma subgroup arises from B cells at the germinal center stage of differentiation, probably from the highly proliferative centroblasts (3, 4, 6). Characteristically, GCB DLBCLs express the key transcriptional repressor BCL6 and consequently lack expression of its target genes (9, 10).
In contrast, the gene expression profile of ABC DLBCL suggests that this lymphoma subgroup is derived from B cells that are in the process of differentiating from germinal center B cells to plasma cells (4, 6). This hypothesis was based on the observation that ABC DLBCLs have down-regulated most of the germinal center B cell signature genes, including BCL6. Instead, ABC DLBCLs express a key regulator of the secretory phenotype of plasma cells, XBP-1, as well as many of its downstream target genes (6, 11). Strong support for this hypothesis was provided by the recent demonstration that genomic mutations, deletions, and rearrangements of the gene encoding Blimp-1 are recurrent in ABC DLBCLs but are not present in GCB DLBCLs (12, 13). Blimp-1 is the master regulator of plasmacytic differentiation that represses directly or indirectly the entire gene expression program of mature B cells (14). Thus, the inactivation of Blimp-1 in roughly one quarter of ABC DLBCL cases provides direct evidence that a block in differentiation is a key event in the pathogenesis of this lymphoma type.
The third subgroup, PMBL, has a characteristic gene expression signature that distinguishes it from GCB DLBCL and ABC DLBCL, but which it shares with the malignant cells of Hodgkin lymphoma (5, 7). PMBL characteristically arises in the mediastinum and, upon pathological examination, a thymic remnant can often be found associated with the tumor mass. This finding suggests that PMBL arises from a rare B cell subpopulation that resides in the thymus (15). PMBLs express some genes in common with GCB DLBCLs, including BCL6, but do not express the ABC DLBCL signature genes and, particularly, do not express genes associated with plasmacytic differentiation (8).
An important event after activation of mature B cells by antigen is Ig class switch recombination (CSR), which can occur within the germinal center or during the extrafollicular activation of B cells. CSR is a recombination and deletion mechanism that juxtaposes a downstream Ig heavy chain (CH) segment to the rearranged VHDHJH segment, thereby switching the Ig isotype of a B cell from IgM to IgG, IgA, or IgE. CSR occurs within the highly repetitive switch regions located 5′ of each constant region (16, 17). The enzyme activation-induced cytidine deaminase (AID) is the B cell–specific factor that is absolutely required for CSR (18–22). In the current understanding of CSR, AID initiates the reaction by creating double-strand DNA breaks in the switch regions. These double-strand DNA breaks recruit and activate DNA damage response proteins, including ataxia-teleangiectasia, histone H2AX, and 53BP1, which coordinate a ligation reaction that uses the enzymatic machinery of the nonhomologous end joining reaction (23–27). CSR is precisely regulated by extracellular signals that influence the choice of which downstream switch regions are involved in the reaction. This regulation is achieved by the production of sterile mRNA transcripts that traverse a switch region and facilitate the recruitment and action of AID (17).
The elaborate controls governing CSR can, however, go awry as indicated by the frequent subversion of CSR to create aberrant switch translocations in multiple myeloma and other lymphoid malignancies (28). Indeed, experimental overexpression of AID can directly promote chromosomal translocations between the Ig heavy chain locus and c-MYC in mouse B lymphocytes (29, 30). Interestingly, AID is one of the few germinal center genes that is more highly expressed in ABC DLBCL than in GCB DLBCL (6), which may be due to the high expression of IRF4 (3, 4, 6), a transcription factor that directly or indirectly increases AID expression (31, 32).
Given the high expression of AID in ABC DLBCL and the ability of AID to promote aberrant class switch translocations, we investigated the nature and extent of CSR events in this lymphoma type in comparison to other subgroups of DLBCL. To this end, we probed the genomic structure of the Ig switch regions by Southern blotting in 22 DLBCL cell lines and 92 DLBCL patient samples, which revealed significant differences in the frequency of CSR between the three subgroups. Unexpectedly, we discovered an abnormality in switch recombination in ABC DLBCLs that led to frequent nonproductive intra-switch CSR, insertion of foreign chromosomal DNA into the Ig locus, and chromosomal translocations involving the switch regions.
RESULTS
Class switch recombination events in subgroups of DLBCL
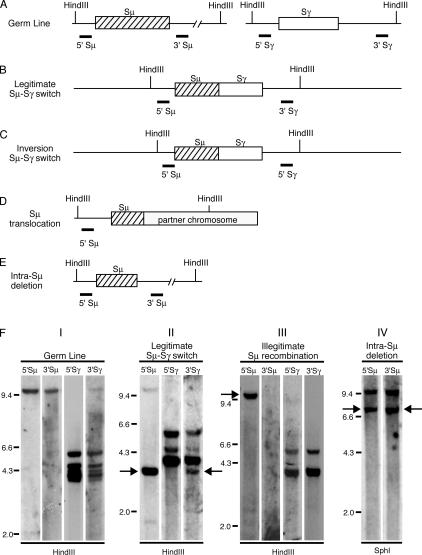
To investigate the frequency and nature of CSR events in tumors of different DLBCL subgroups, we used a Southern blot assay that can detect both legitimate CSR events and illegitimate recombination and deletion events involving the Ig switch regions (28, 33). This assay uses three different pairs of Southern blot probes that hybridize either 5′ or 3′ of the Sμ, Sγ, and Sα switch regions (Fig. 1). With the Sμ probe pairs, a single hybridizing fragment is present in germ line DNA, whereas the Sγ probes hybridize to four germ line fragments representing Sγ1, Sγ2, Sγ3, and Sγ4, and the Sα probes hybridize to two germ line fragments representing Sα1 and Sα2. Genomic DNAs from 22 DLBCL cell lines and 92 DLBCL patient samples were digested with either of two restriction enzymes (HindIII or SphI) and subsequently hybridized with the different probes. A switch region in germ line configuration will be reflected by a restriction fragment that hybridizes with the corresponding 5′ and 3′ switch probes and that is also present in the placental control DNA (Fig. 1, A and F). A legitimate switch event, such as between Sμ and Sγ, creates a restriction fragment that will be detected by the 5′ Sμ and the 3′ Sγ probes (Fig. 1, B and F). Legitimate switch recombinations may also occur between downstream switch regions, such as between Sγ and Sα. Inversion CSR events, such as between Sμ and Sγ, generate a restriction fragment that either hybridizes with the 5′ switch μ and γ probes or with the two 3′ switch probes (Fig. 1 C). An illegitimate switch recombination event is operationally defined by a restriction fragment that hybridizes with only one switch region probe (Fig. 1 F). This may be caused by a chromosomal translocation (Fig. 1 D), an insertion of foreign sequences into a switch region, a polymorphism or mutation of the restriction enzyme binding site, or an atypical deletion within the Ig heavy chain locus. Intra-switch recombination events may occur in which part of a switch region is deleted without resulting in legitimate switch recombination, and these can be detected reliably when they occur in Sμ (Fig. 1, E and F). The human Ig locus contains four Sγ and two Sα regions. CSR can potentially join different Sγ regions or different Sα regions together. Using the Southern blot assay, these events would be impossible to distinguish from intra-Sγ deletions and intra-Sα deletions, respectively.
Figure 1.
Analysis of class switch recombination by Southern blot. (A–E) The position of HindIII restriction sites are shown. Thick lines indicate binding sites of respective probes. (A) Germ line configuration of switch μ and switch γ regions. (B) Legitimate switch μ to γ, detected by 5′ Sμ and 3′ Sγ probe. (C) Inversion switch μ to γ, detected by 5′ Sμ and 5′ Sγ probe. (D) Chromosomal translocation, detected by 5′ Sμ probe. (E) Intra-switch μ deletion, detected by 5′ Sμ and 3′ Sμ probe. (F) Applied restriction enzyme is indicated below lanes. The probes used for hybridization are shown above each lane. Panel I: Restriction digest of placental DNA shows germ line configuration. Panel II: Restriction digest of patient sample 529 genomic DNA. Arrows indicate legitimate switch recombination μ to γ. Panel III: Restriction digest of SUDHL-2 genomic DNA. Arrow indicates illegitimate switch recombination detected by 5′ Sμ probe. Panel IV: Restriction digest of patient sample 1002 genomic DNA. Arrows indicate intra-switch μ deletion.
The analysis of 6 ABC DLBCL and 16 GCB DLBCL cell lines demonstrated differences in the frequency of legitimate switch recombination events (Table S1, available at http://www.jem.org/cgi/content/full/jem.20062041/DC1). Whereas none of the ABC DLBCL cell lines had a germ line configuration of a switch region, four (25%) of the GCB DLBCL lines had only germ line switch regions. 1 ABC DLBCL cell line (17%) had a legitimate switch involving Sμ and Sγ, whereas 11 GCB DLBCL cell lines (69%) had a legitimate switch from Sμ to either Sγ or Sα or from Sα to Sγ. Illegitimate CSR events were detected in half of the ABC DLBCL and GCB DLBCL cell lines.
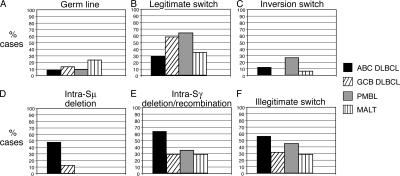
Analysis of 92 primary biopsy samples revealed differences between ABC DLBCL and GCB DLBCL with respect to the nature and extent of CSR (Fig. 2 and Table S2, which is available at http://www.jem.org/cgi/content/full/jem.20062041/DC1). Among 50 ABC DLBCL cases, the switch regions were in germ line configuration in only 4 (8%). Only 15 cases (30%) had a legitimate switch recombination. 28 cases (56%) showed an illegitimate switch recombination event, and 6 cases (12%) had an inversion switch recombination between Sμ and Sγ or Sα (5 cases with inversion switch recombination between Sμ and Sγ, and 1 case with inversion switch recombination between Sμ and Sα). Strikingly, 24 ABC DLBCLs (48%) had intra-Sμ deletion events (Fig. 1, E and F), and 64% of cases showed a deletion/recombination in Sγ.
Figure 2.
Summary of the analysis of class switch recombination in 92 DLBCL and 17 MALT lymphoma patients. (A) Germ line. No significant differences can be observed between the different lymphoma subtypes. (B) Legitimate switch recombination. GCB DLBCL and PMBL show a significantly higher frequency of legitimate switch recombinations compared with ABC DLBCL (ABC DLBCL vs. GCB DLBCL, P = 0.012; ABC DLBCL vs. PMBL, P = 0.035). (C) Inversion switch recombination. ABC DLBCL shows a significantly higher rate of inversion switch recombinations compared with GCB DLBCL (ABC DLBCL vs. GCB DLBCL, P = 0.045). (D) Intra-switch μ deletions. ABC DLBCL shows a significantly higher rate of intra-switch μ deletions compared with GCB DLBCL, PMBL, or MALT (ABC DLBCL vs. GCB DLBCL, P = 0.0012; ABC DLBCL vs. PMBL, P = 0.003; ABC DLBCL vs. MALT, P = 0.0004). (E) Intra-switch γ deletion/recombination. ABC DLBCL shows a significantly higher rate of intra-switch γ deletion/recombination compared with GCB DLBCL and MALT (ABC DLBCL vs. GCB DLBCL, P = 0.002; ABC DLBCL vs. MALT, P = 0.013). (F) Illegitimate switch recombination. ABC DLBCL shows a significantly higher rate of illegitimate switch recombinations compared with GCB DLBCL (ABC DLBCL vs. GCB DLBCL, P = 0.037).
As with ABC DLBCLs, only a small fraction of GCB DLBCLs had switch regions in the germ line configuration (4 out of 31 cases; 13%). However, legitimate CSR events were more common in GCB DLBCLs, occurring in 18 of 31 cases tested (58%) compared with ABC DLBCLs (P = 0.012). Conversely, GCB DLBCLs had lower frequencies of several aberrant class switch events compared with ABC DLBCLs, including illegitimate switch recombination (32%; P = 0.037), inversion switch recombination (0%; P = 0.045), intra-Sμ deletion (13%; P = 0.0012), and intra-Sγ deletion/recombination (29%; P = 0.002).
Of 11 PMBL cases available for analysis, 7 (64%) had legitimate CSR events, which was significantly more than among ABC DLBCLs (P = 0.035). Illegitimate CSR events were detected in five cases (45%), and inversion switch events were detected in three cases (27%). In contrast to ABC DLBCLs, intra-Sμ deletions were not present in any PMBL patient sample analyzed (P = 0.003).
As a control, we investigated CSR events in mucosa-associated lymphoid tissue (MALT) lymphoma because this postgerminal center lymphoma type has been reported to have aberrant CSR events in some cases (34). Like ABC DLBCL, MALT lymphomas had a relatively low frequency of legitimate CSR events (6/17 cases investigated; 35%). However, in contrast to ABC DLBCLs, none of the MALT lymphomas had intra-Sμ deletions (P = 0.0004). MALT lymphomas also had fewer illegitimate CSR events than ABC DLBCLs (29%; P = 0.058) and intra-Sγ deletions/recombinations (29%; P = 0.013).
In summary, ABC DLBCLs had a lower frequency of legitimate class switch recombination events relative to the other lymphoma subtypes and a higher frequency of intra-Sμ deletion, intra-Sγ deletion/recombination events as well as illegitimate switch recombination events. Intra-Sα deletion/recombination events were detected, but there were no significant differences in frequency of these events among the lymphoma subtypes (Table S2).
Expression of AID in lymphoma subsets
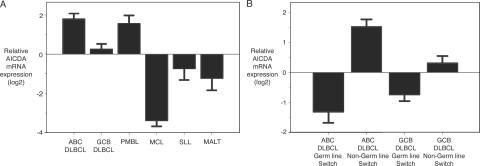
AID mRNA levels were assessed in 245 primary lymphoma specimens with the use of Affymetrix oligonucleotide microarrays (Fig. 3 A). Highest levels of AID expression were observed in ABC DLBCL and PMBL, and these lymphoma types had, respectively, threefold and 2.5-fold higher expression than GCB DLBCL (ABC DLBCL vs. GCB DLBCL, P = 0.00006; PMBL vs. GCB DLBCL, P = 0.01). The pre-germinal center lymphoma types, mantle cell lymphoma and small lymphocytic lymphoma, both had low levels of AID expression compared with any of the DLBCL subgroups as did the post-germinal center entity, MALT lymphoma (ABC DLBCL vs. mantle cell lymphoma, P = 2.2 × 10−16; ABC DLBCL vs. small lymphocytic lymphoma, P = 0.0003; ABC DLBCL vs. MALT, P = 2.16 × 10−5).
Figure 3.
Relative AID mRNA expression in different lymphoma subtypes. (A) ABC DLBCLs and PMBLs express the highest level of AID mRNA. (B) Comparison of AID mRNA expression in ABC DLBCL and GCB DLBCL germ line cases versus non-germ line cases. Non-germ line cases express higher AID mRNA compared with germ line cases.
Having identified a small subset of DLBCLs in which the Ig switch regions were in germ line configuration, we investigated whether these lymphomas might have lower levels of AID expression. Indeed, among ABC DLBCLs, cases with germ line switch regions had eightfold lower expression compared with non-germ line cases (Fig. 3 B; P = 0.0012). Likewise, those GCB DLBCLs with germ line switch regions had lower AID expression than their non-germ line counterparts (P = 0.09).
Structure of intra-Sμ deletions
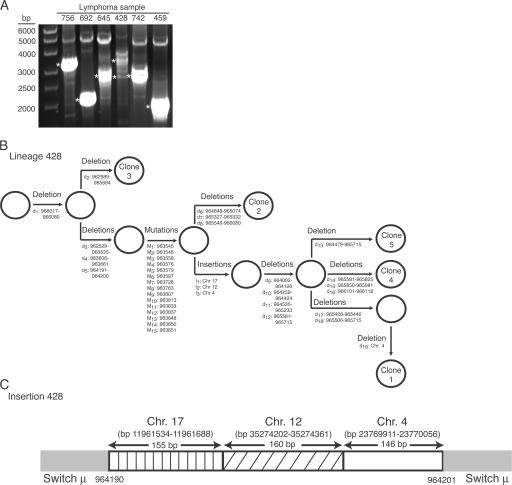
To characterize intra-Sμ deletions further, we amplified the Sμ regions from four ABC DLBCL cases (cases 756, 692, 645, and 428) that had putative intra-Sμ deletions based on the Southern blot assay. All intra-Sμ deletions were confirmed by PCR (Fig. 4 A). Unexpectedly, however, case 428, which had a single intra-Sμ deletion band using the Southern blot assay, yielded a cluster of bands ranging from ∼2.7 to ∼3.9 kb in size after PCR for the Sμ region. This finding suggested that there might be intra-clonal variation in the structure of the intra-Sμ deletions.
Figure 4.
Analysis of intra-switch μ deletions. (A) PCR amplification of intra-switch μ deletions (ethidium bromide–stained 0.8% agarose gel). Asterisks indicate cloned PCR bands for sequence analysis. (B) Lineage model of aberrations in ABC DLBCL patient sample 428. d, deletion; M, mutation; I, insertion. Open circles represent putative clonal precursors. Numbers refer to GenBank accession number NG_001019.4. (C) Sequence analysis of DNA insertions into Sμ in ABC DLBCL case 428. bp numbers refer to accession number Hs17_10875 for chromosome 17, accession number Hs12_29578 for chromosome 12, and accession number Hs4_16510 for chromosome 4.
To examine this possibility, we cloned the amplified intra-Sμ deletion bands of the four ABC DLBCL samples and determined the sequence of three to five clones per case. The molecular clones derived from a single case were remarkably diverse, showing 1–19 separate deletion events within Sμ (Table I, Fig. 4 B, and Fig. S1, which is available at http://www.jem.org/cgi/content/full/jem.20062041/DC1). Even in cases in which the Sμ PCR product appeared to be a single band, the clones obtained were diverse in sequence.
Table I.
Sequence analysis of Sμ deletions in ABC DLBCL and GCB DLBCL patient samples
| Sample | DLBCL subtype |
No. of Sμ deletions |
No. of somatic mutationsa |
No. of duplications |
|---|---|---|---|---|
| 428 | ABC DLBCL | 19 | n.a. | 0 |
| 645 | ABC DLBCL | 13 | n.a. | 3 |
| 692 | ABC DLBCL | 9 | n.a. | 2 |
| 756 | ABC DLBCL | 5 | 3 | 0 |
| 459 | GCB DLBCL | 11 | 3 | 0 |
| 742 | GCB DLBCL | 6 | 6 | 1 |
n.a., not available, as investigated 250-bp segment partly or completely deleted.
Somatic mutations determined in 250-bp segment at 5′ end of Sμ.
We next examined the 5′ end of the amplified Sμ region in detail for the presence of somatic mutations. From each case, we obtained molecular clones that had a large number of somatic mutations (Table I, Fig. 4 B, and Fig. S1). These mutations occurred most frequently at RGYW motifs, strongly suggesting that they were caused by AID (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20062041/DC1). In some instances, different molecular clones from the same case had distinct somatic mutations whereas other molecular clones had no mutations.
As a control, we performed a similar sequence analysis of the Sμ region in two ABC DLBCL cases in which Sμ was in germ line configuration by Southern blot analysis. As expected, no deletion was detected in the amplified Sμ regions from these cases by PCR. Further, these two cases had significantly fewer somatic mutations in Sμ than ABC DLBCL cases with intra-Sμ deletions.
In addition to deletions and mutations, sequence analysis revealed several duplications of Sμ segments of up to 40 bp in length (Table I and Fig. S1). Unexpectedly, we also detected insertions of short DNA segments derived from other chromosomes into the Sμ region of one of the four ABC DLBCL cases analyzed (Fig. 4, B and C). These insertions, which varied in length from 146 to 160 bp, were derived from chromosomes 17, 12, and 4 and were arranged in tandem with no gaps between the insertions (Fig. 4 C). These segments were situated within a Sμ deletion of 10 bp. Interestingly, one of the three clones analyzed from this case had a 19-bp deletion within the inserted chromosome 4 sequence (Fig. 4 B). This finding suggests that AID is able to create breaks and deletions in ectopic sequences that are inserted into the Sμ region.
For each of the four cases, we were able to organize all of the Sμ deletions, mutations, duplications, and insertions into lineage models of the stepwise accumulation of these events in individual molecular clones (Fig. 4 B and Fig. S1). The fact that such lineage diagrams can be drawn is consistent with ongoing molecular alterations of the Sμ regions within the malignant clones. In three of four cases, the first abnormality that occurred was one or several internal deletions. Subsequently, each clone underwent several steps of mutations, internal tandem duplications, and insertions of DNA from other chromosomes. Finally, most of the clones sustained one or more Sμ deletions. Thus, multiple molecular mechanisms that perturb Sμ structure were ongoing and interspersed in time after malignant transformation of a B cell into ABC DLBCL.
Although intra-Sμ deletions were much less frequent in GCB DLBCLs than in ABC DLBCLs (Fig. 2 D), we investigated whether ongoing intra-clonal Sμ alterations could also occur in GCB DLBCL. To this end, we amplified the Sμ regions of two GCB DLBCL patient samples (cases 742 and 459) that had intra-Sμ deletions by the Southern blot assay. In both cases, intra-Sμ deletions were confirmed by PCR (Fig. 4 A). Sequence analysis revealed clonal heterogeneity in both cases, with 6–11 deletions per case, various somatic mutations, and a duplication of 194 bp in GCB DLBCL case 742 (Table I). Thus, GCB DLBCL and ABC DLBCL can apparently use similar aberrant CSR mechanisms, albeit with different frequencies.
Switch translocations in DLBCL
As assessed by the Southern blotting, ABC DLBCL had the highest frequency of illegitimate switch recombination events (Fig. 2). To determine whether these illegitimate events might represent chromosomal translocations involving the switch regions, we first performed fluorescence in situ hybridization (FISH) using CH and VH probes to determine the presence of IgH translocations. We investigated 10 DLBCL cell lines (2 ABC DLBCL cell lines: OCI-Ly10, and SUDHL2, and 8 GCB DLBCL cell lines: NUDHL-1, NUDUL-1, OCI-Ly1, OCI-Ly2, OCI-Ly-8, SUDHL-4, Toledo, and RCK8) that had an illegitimate switch recombination phenotype by Southern blotting. As a control, we analyzed the ABC DLBCL cell line U2932 that did not show an illegitimate switch recombination. IgH translocations were detected by FISH in all 10 DLBCL cell lines with the illegitimate CSR phenotype, but not in U2932, demonstrating that the majority of illegitimate CSR events represent chromosomal translocations.
We next investigated whether two oncogenes of importance in DLBCL, MYC and BCL6, were involved in illegitimate switch translocations. FISH analyses of DLBCL samples were performed using “break-apart” probes spanning the MYC and BCL6 loci, revealing MYC disruptions in 7% of cases analyzed (n = 87; reference 35) and BCL6 disruptions in 19% of cases analyzed (n = 133; unpublished data). Some of these positive cases were investigated further using long-distance PCR assays to detect MYC or BCL6 translocations into the Sμ or Sγ regions. These assays revealed an MYC translocation into the Sγ region in one ABC DLBCL case and a BCL6 translocation into the Sμ region in another (Fig. 5 A). In cell lines with illegitimate switch CSR events, long-distance PCR demonstrated a MYC-Sγ translocation in the GCB DLBCL cell line NUDUL-1 (Fig. 5 A).
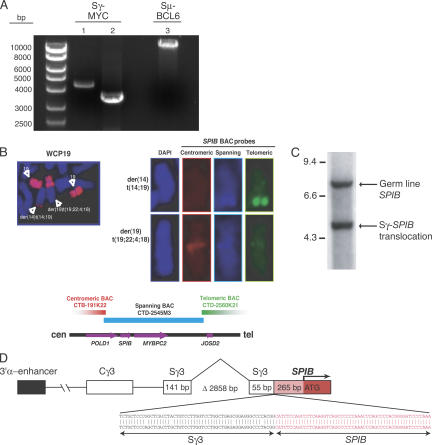
Figure 5.
Switch translocations in DLBCL. (A) PCR for detection of BCL6/MYC switch translocations (ethidium bromide–stained 0.8% agarose gel). Lane 1, Sγ-MYC translocation in ABC DLBCL case 1000; lane 2, Sγ-MYC translocation in GCB DLBCL cell line NUDUL-1; lane 3, Sμ-BCL6 translocation in ABC DLBCL case 709. (B) Whole chromosome painting of chromosome 19 detects t(14; 19) and t(19; 22; 4; 18). Hybridization of t(14; 19) with spanning and telomeric BAC SPIB probes. No hybridization with centromeric BAC probe. Hybridization of t(19; 22; 4; 18) with centromeric and spanning BAC SPIB probe, no hybridization with telomeric probe. (C) Detection of switch γ-SPIB translocation by Southern blot in OCI-Ly3. (D) Sequence analysis of switch γ-SPIB translocation. Δ, deletion in Sγ.
Additionally, we investigated whether a t(14;19) translocation in the ABC DLBCL cell line OCI-Ly3 might be the result of aberrant switch recombination. FISH using a whole chromosome 19 painting probe revealed that OCI-Ly3 cells show a t(14;19) as well as a complicated translocation involving chromosomes 19, 22, 4, and 18 (unpublished data; Fig. 5 B). A variety of BAC probes from chromosome 19 were used in FISH analyses to narrow the region of chromosome 19 involved in the translocations (unpublished data). Ultimately, three BAC probes in the vicinity of the SPIB gene were identified that clustered around the translocation breakpoint (Fig. 5 B). The t(14;19) derivative chromosome hybridized with BAC probes spanning or telomeric to the SPIB gene, but not with a BAC probe centromeric to SPIB. This suggested that the translocation breakpoint was 5′ of the transcriptional start site of SPIB. In contrast, the t(19; 22; 4; 18) hybridized with the centromeric BAC probe and weakly with the spanning BAC probe (Fig. 5 B).
We next used Southern blot hybridization to confirm the SPIB translocation that was detected by FISH. With OCI-Ly3 cells, a 332-bp probe binding ∼200 bp 3′ of the start codon of SPIB hybridized to one germ line fragment and to a second smaller fragment (Fig. 5 C), whereas only the germ line fragment was observed in other lymphoma cell lines (unpublished data). Finally, we were able to amplify the translocation breakpoint using PCR primers derived from the Sγ region and from the 5′ end of SPIB. Sequence analysis confirmed that the breakpoint occurred 265 bp 5′ of the SPIB transcriptional start site and joined SPIB with the Sγ3 region (Fig. 5 D). Additionally, the translocated Sγ3 region sustained a second deletion of 2,858 bp, demonstrating that the processes responsible for intra-switch deletion and switch translocation can target the same Ig allele (Fig. 5 D). As a consequence of this translocation, the intact SPIB coding region was placed in proximity to the Ig 3′α enhancer (Fig. 5 D), resulting in a high level of SPIB mRNA expression compared with most other DLBCL biopsy samples and cell lines (not depicted).
DISCUSSION
Using a Southern blot assay, we have discovered significant differences in the frequency and character of CSR among three molecular subtypes of DLBCL. Unexpectedly, almost half of the ABC DLBCLs had internal deletions in Sμ, events that were present rarely among GCB DLBCLs (13%) and absent among PMBLs and MALT lymphomas. Furthermore, 64% of ABC DLBCL cases had abnormal CSR events involving Sγ regions, which occurred much less commonly in GCB DLBCL (29%), PMBL (36%), and MALT lymphoma (29%). In normal B cells, intra-Sμ deletion occurs in only 12.5–18% of cells (36, 37), and intra-Sγ deletion/recombination events are extremely rare (37). Therefore, the high frequency of both intra-Sμ and intra-Sγ deletion/recombination events in ABC DLBCL strongly suggests that CSR is functionally impaired in this lymphoma type, potentially predisposing this lymphoma type to chromosomal translocations. Conversely, legitimate CSR was significantly less common in ABC DLBCL than GCB DLBCL or PMBL. Consistent with this finding, ABC DLBCLs express IgM mRNA at significantly higher levels than do GCB DLBCLs (6).
Constitutive overexpression of AID seems a likely contributor to the aberrant CSR in ABC DLBCL. Normal B lymphocytes transcriptionally activate AID upon stimulation by antigen or mitogens, and this up-regulation requires the activity of the transcription factor IRF4 (31, 32). ABC DLBCLs have high expression of IRF4 due to constitutive NF-κB pathway activation (3, 4, 38, 39), which may account for the high AID expression in ABC DLBCL despite the fact that most other germinal center B cell signature genes are down-regulated (6). Indeed, of all lymphoma types studied, AID mRNA is highest in ABC DLBCL (Fig. 3) and, consequently, AID protein is more highly expressed in ABC DLBCL than in GCB DLBCL (40). Interestingly, the few ABC DLBCLs that lacked AID expression were those that had a germ line configuration of the Ig switch regions. This result is consistent with the hypothesis that AID action is required for the aberrant CSR that characterizes the majority of ABC DLBCLs.
In normal B cell responses to antigen, AID expression is limited to a short window during which the B cell is diversifying its IgV regions in the germinal center reaction. AID expression is normally extinguished during plasmacytic differentiations, thereby limiting the exposure of B cells to this potentially mutagenic enzyme. In contrast, constitutive AID expression in ABC DLBCL may expose the genome of this tumor type to repeated mutation and DNA strand breaks. Sequence analysis of the intra-Sμ deletions in ABC DLBCL revealed that AID was constitutively active in the malignant clone over many generations. These Sμ regions contained as many as 19 independent deletions that were interspersed with somatic mutations targeted at AID hot spots. This remarkably high frequency of switch region deletions is unprecedented in analyses of switch regions from normal mouse B cells. These deletions and mutations could be organized into lineage diagrams that imply that AID-dependent single-stranded and double-stranded breaks were being generated over many generations of the ABC DLBCL clones. GCB DLBCLs had aberrant CSR events less commonly, but in two patient samples we identified ongoing deletions, mutations, and duplications in Sμ. GCB DLBCLs expressed AID mRNA at levels that were lower than in ABC DLBCL but still higher than in mantle cell lymphoma, small lymphocytic lymphoma, and MALT lymphoma (Fig. 3 A). Because AID is likely to participate in the Sμ abnormalities, the level of AID in GCB DLBCL is apparently sufficient for aberrant CSR. Nonetheless, the significantly higher AID expression in ABC DLBCL may contribute to the higher frequency of Sμ deletions in this DLBCL subtype.
However, none of the PMBL samples showed intra-Sμ deletions despite having AID expression levels that were comparable to those in ABC DLBCLs. Therefore, it is unlikely that high AID expression is the only molecular abnormality that contributes to aberrant CSR in ABC DLBCL. The fact that intra-switch deletions predominated over legitimate switch recombination suggests that ABC DLBCLs may have defects in the process of “synapsis” that brings two different switch regions together before recombination (17). Genetic deficiency in three DNA repair proteins, H2AX, 53BP1, and ataxia-teleangiectasia, reduces legitimate CSR in mouse B cells while allowing intra-Sμ deletions to occur (25, 27, 37, 41, 42), leading to the hypothesis that these proteins may facilitate the formation of a protein lattice that physically links switch regions that have sustained AID-induced double-stranded breaks. Thus, it is possible that ABC DLBCLs in contrast to PMBL may have a genetic or functional deficiency in one of these DNA repair proteins.
One of the most unanticipated findings in the present study was the insertion of foreign DNA segments within the Sμ region of one ABC DLBCL case, which may reflect a dysregulation of CSR that predisposes this lymphoma type to chromosomal translocations. The inserted DNA segments originated from three chromosomes and were ligated in tandem without homologous sequences at the junctions, suggesting that they were joined by the NHEJ mechanism. These insertions have not been reported in switch regions from normal B cells (37), further supporting the hypothesis that CSR is dysregulated in ABC DLBCL. However, it has been published that in one case of MALT lymphoma, a 336-bp segment from the ARNT2 gene on chromosome 15 was inserted into Sμ, suggesting that this aberrant CSR mechanism also occurs in other lymphoma types (34). The molecular mechanism responsible for the blunt end insertion of foreign DNA into switch region remains unknown.
We speculate that the extraordinary frequency of intra-switch deletions and the insertions of ectopic DNA into Sμ in ABC DLBCLs may predispose these lymphomas to chromosomal translocations. In the Sγ3-SPIB translocation in the OCI-Ly3 cell line, the translocated switch region had also sustained an intra-switch deletion, raising the possibility that these events might be mechanistically linked. By the Southern blot assay, 56% of ABC DLBCLs had illegitimate CSR events, and these events were less frequent in the other DLBCL subtypes and in MALT lymphoma. At least some of these illegitimate CSR events represent chromosomal translocations as we were able to identify illegitimate switch translocations involving the BCL6, MYC, and SPIB genes in ABC DLBCLs. Also, all cell lines that had illegitimate CSR events in the Southern blot assay had IgH translocations that were demonstrable by FISH. AID is a likely candidate to mediate these chromosomal translocations because it has been shown to be required for spontaneous MYC/IgH translocations that occur in IL-6-transgenic (29), 53BP1−/− (30), and H2AX −/− (43) mice. Compared with resting blood mature B cells, ABC DLBCLs have 26-fold higher AID mRNA expression (unpublished data). In this regard, it is notable that ectopic overexpression of AID in mouse B cells by ∼10-fold was sufficient to cause chromosomal translocations linking the MYC locus to the IgH locus (30). Thus, it is certainly plausible that the high expression of AID in ABC DLBCL contributes to the higher frequency of intra-Sμ deletion and illegitimate CSR in this lymphoma type. AID could contribute to an initiating chromosomal translocation event involving a potent oncogene such as MYC and BCL6 or could act later in malignant transformation to generate translocations that drive clonal progression. However, because PMBL and ABC DLBCL express AID mRNA at approximately equivalent levels, it is likely that ABC DLBCLs have additional mechanisms that deregulate CSR given the fact that this lymphoma type has higher intra-Sμ and intra-Sγ deletions and higher illegitimate CSR events than PMBL.
The novel switch translocation involving the SPIB gene that we identified in the ABC cell line OCI-Ly3 highlights the potential biological importance of the switch translocations in ABC DLBCL. SPIB is an Ets family transcription factor that is expressed exclusively in mature B cells, T cell progenitors, and plasmacytoid dendritic cells (44, 45). SPIB is required for full B cell receptor signaling and T cell–dependent antibody responses (46). SPIB is central to the germinal center reaction because germinal centers in SPIB-deficient mice are smaller in size, persist for a shorter duration after immunization, and contain more apoptotic cells than those in wild-type animals (46). The importance of SPIB as a regulator of mature B cell function is also emphasized by the fact that it is directly repressed by Blimp-1 during plasmacytic differentiation (14). Interestingly, SPIB is significantly more highly expressed in ABC DLBCLs than in GCB DLBCLs (4). This fact together with the translocation of SPIB in the ABC DLBCL cell line OCI-Ly3 suggests that SPIB may play an important role in the pathophysiology of this lymphoma subtype. More generally, we believe that the present study indicates that a comprehensive identification of switch translocations in ABC DLBCL is likely to uncover additional oncogenic pathways in this lymphoma type.
MATERIALS AND METHODS
Patient samples.
Tumor biopsy specimens, which have previously been analyzed by gene expression profiling (4, 5), were obtained before treatment from 92 patients with DLBCL and 17 patients with MALT lymphoma. All samples were studied according to a protocol approved by the NCI Institutional Review Board.
Culture of lymphoma cell lines.
Lymphoma cell lines were cultured in RPMI, 10% FBS, except for OCI-Ly1, OCI-Ly2, OCI-Ly3, OCI-Ly4, OCI-Ly7, OCI-Ly8, and OCI-Ly10, which were cultured in Iscove's modified Dulbecco medium with 20% human plasma.
Southern blot analysis of switch regions.
Genomic DNA from cell lines and patient samples was extracted with the DNeasy Tissue kit (QIAGEN) according to the manufacturer's instructions. 10 μg of genomic DNA was digested with appropriate restriction enzymes, fractionated on a 0.8% agarose gel, denatured with a buffer consisting of 0.5 N NaOH, 0.6 M NaCl, neutralized with 1 M Tris-HCL/0.6 M NaCL, and transferred with 20× SSC to nylon membranes (PerkinElmer). Two radiolabeled probes were created by PCR for each Ig switch region, binding either 5′ or 3′ of the switch region (Fig. 1) as described previously (28, 33). Filters were hybridized overnight with the radiolabeled probes at 42°C in 1 M NaCl, 50 mM Tris-HCL, pH 7.4, 40% formamide, 10% dextran sulfate, 1% SDS, and 100 μg/ml salmon sperm DNA. The filters were subsequently washed in 2× SSC/0.1% SDS at room temperature and then in 0.1× SSC/0.1% SDS at 52–60°C, followed by exposure to XAR films (Kodak) at –70°C with an intensifying screen.
The Chi-squared test was used for statistical analysis of the difference in CSR events between lymphoma types.
Amplification and sequencing of aberrant Sμ regions MYC, BCL6, and SPIB.
The Sμ region was amplified by PCR using primers binding 5′ and 3′ outside of Sμ (M-F1, GGAGGGGATGCTCCGGGAAGGTGG; M-R1, CGAGGCAGCCAACGGCCACGC). The PCR was performed with TaKaRa La Taq polymerase (Takara Bio, Inc.) using the following conditions: 94°C for 1 min, followed by 33 cycles of denaturation, 30 s at 94°C; annealing, 30 s at 68°C; extension, 6 min at 72°C; and a final extension for 10 min at 72°C. The PCR products were visualized by electrophoresis on an 0.8% agarose gel and ethidium bromide staining. PCR products were cloned into the Topo XL cloning vector (Invitrogen) and sequenced (BigDye sequencing system; Applied Biosystems).
The PCR reactions for detection of translocations involving the Sμ and Sγ region and MYC or BCL6 were performed as described previously (47). The PCR reactions to confirm a translocation involving the Sγ region and SPIB were performed applying one of the SPIB primers (GGGGCCCTCACTTACTGTG or AACCAGAAGGGGCAAGAGAC) and one of the Sγ primers (GACCAGTGGACACTGTTCTCAGATGG or CCTCCAAGGCCCTTTTCTTCTGTG; reference 48) using the following conditions: 95°C for 10 min, followed by 40 cycles of denaturation, 30 s at 95°C; annealing, 30 s at 65°C; extension, 3 min at 72°C, and final extension for 10 min at 72°C. The PCR products were visualized by electrophoresis on an 0.8% agarose gel with ethidium bromide staining, and the PCR product was subsequently sequenced.
FISH.
FISH for detection of IgH translocations with CH BAC and VH cosmid probes was performed as described previously (49). For the analysis of SPIB translocation, BAC clones CTB-191K22, CTD-2545M3, and CTD-2560K21 as well as a whole chromosome painting probe for chromosome 19 were used. Isolation and labeling of BAC DNA and FISH were performed as described previously (50). MYC and BCL6 translocations were detected using break-apart probes (Vysis probes; Abbott).
Southern blot analysis of SPIB.
10 μg of genomic DNA was digested with SphI, and Southern blotting was performed as described above. A radiolabeled 332-bp probe binding ∼200 bp 3′ of the SPIB start codon was created by PCR, and hybridization, washing, and exposure was performed as described above.
Gene expression profiling.
RNA was extracted from the biopsy specimens as described previously (3) and was profiled for gene expression with the use of “Lymphochip” DNA microarrays (51) or Affymetrix U133A and U133B oligonucleotide microarrays. The primary gene expression profiling data are available from the Gene Expression Omnibus of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/geo) through GEO accession number GSE4732.
Online supplemental material.
Table S1 and S2 contain the complete results of the analysis of class switch recombination by Southern blot in DLBCL cell lines and DLBCL and MALT patient samples. Fig. S1 shows lineage models of Sμ alterations in three ABC DLBCL patient samples (cases 756, 692, and 645) and two GCB DLBCL patient samples (cases 742 and 459). Fig. S2 displays sequence analysis of the 5′ end of Sμ deletion in ABC DLBCL case 756. The online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20062041/DC1.
Acknowledgments
We wish to thank Claudia Becher for expert technical assistance.
This research was supported by the Intramural Research Program of the NIH, NCI, Center for Cancer Research, and an NCI Director's Challenge grant (UO1-CA84967). G. Lenz was also supported by a research grant of the German Research Foundation (DFG), and R. Siebert was supported by the Deutsche Krebshilfe and the Kinder-Krebs-Initiative Buchholz Holm-Seppensen. This project was performed under the auspices of the Lymphoma/Leukemia Molecular Profiling Project of the NCI.
The authors have no conflicting financial interests.
Abbreviations used: ABC, activated B cell–like; AID, activation-induced cytidine deaminase; CSR, class switch recombination; DLBCL, diffuse large B cell lymphoma; FISH, fluorescence in situ hybridization; GCB, germinal center B cell–like; MALT, mucosa-associated lymphoid tissue; PMBL, primary mediastinal B cell lymphoma.
References
- 1. 1997. A clinical evaluation of the International Lymphoma Study Group classification of non-Hodgkin's lymphoma. The Non-Hodgkin's Lymphoma Classification Project. Blood. 89:3909–3918. [PubMed] [Google Scholar]
- 2.Coiffier, B. 200 1. Diffuse large cell lymphoma. Curr. Opin. Oncol. 13:325–334. [DOI] [PubMed] [Google Scholar]
- 3.Alizadeh, A.A., M.B. Eisen, R.E. Davis, C. Ma, I.S. Lossos, A. Rosenwald, J.C. Boldrick, H. Sabet, T. Tran, X. Yu, et al. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 403:503–511. [DOI] [PubMed] [Google Scholar]
- 4.Rosenwald, A., G. Wright, W.C. Chan, J.M. Connors, E. Campo, R.I. Fisher, R.D. Gascoyne, H.K. Muller-Hermelink, E.B. Smeland, J.M. Giltnane, et al. 2002. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 346:1937–1947. [DOI] [PubMed] [Google Scholar]
- 5.Rosenwald, A., G. Wright, K. Leroy, X. Yu, P. Gaulard, R.D. Gascoyne, W.C. Chan, T. Zhao, C. Haioun, T.C. Greiner, et al. 2003. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J. Exp. Med. 198:851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright, G., B. Tan, A. Rosenwald, E.H. Hurt, A. Wiestner, and L.M. Staudt. 2003. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. USA. 100:9991–9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savage, K.J., S. Monti, J.L. Kutok, G. Cattoretti, D. Neuberg, L. De Leval, P. Kurtin, P. Dal Cin, C. Ladd, F. Feuerhake, et al. 2003. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 102:3871–3879. [DOI] [PubMed] [Google Scholar]
- 8.Staudt, L.M., and S. Dave. 2005. The biology of human lymphoid malignancies revealed by gene expression profiling. Adv. Immunol. 87:163–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phan, R.T., and R. Dalla-Favera. 2004. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 432:635–639. [DOI] [PubMed] [Google Scholar]
- 10.Shaffer, A.L., X. Yu, Y. He, J. Boldrick, E.P. Chan, and L.M. Staudt. 2000. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 13:199–212. [DOI] [PubMed] [Google Scholar]
- 11.Shaffer, A.L., M. Shapiro-Shelef, N.N. Iwakoshi, A.-H. Lee, S.-B. Qian, H. Zhao, X. Yu, L. Yang, B.K. Tan, A. Rosenwald, et al. 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 21:81–93. [DOI] [PubMed] [Google Scholar]
- 12.Tam, W., M. Gomez, A. Chadburn, J.W. Lee, W.C. Chan, and D.M. Knowles. 2006. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood. 107:4090–4100. [DOI] [PubMed] [Google Scholar]
- 13.Pasqualucci, L., M. Compagno, J. Houldsworth, S. Monti, A. Grunn, S.V. Nandula, J.C. Aster, V.V. Murty, M.A. Shipp, and R. Dalla-Favera. 2006. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J. Exp. Med. 203:311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaffer, A.L., K.I. Lin, T.C. Kuo, X. Yu, E.M. Hurt, A. Rosenwald, J.M. Giltnane, L. Yang, H. Zhao, K. Calame, and L.M. Staudt. 2002. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 17:51–62. [DOI] [PubMed] [Google Scholar]
- 15.Copie-Bergman, C., A. Plonquet, M.A. Alonso, M.L. Boulland, J. Marquet, M. Divine, P. Moller, K. Leroy, and P. Gaulard. 2002. MAL expression in lymphoid cells: further evidence for MAL as a distinct molecular marker of primary mediastinal large B-cell lymphomas. Mod. Pathol. 15:1172–1180. [DOI] [PubMed] [Google Scholar]
- 16.Manis, J.P., M. Tian, and F.W. Alt. 2002. Mechanism and control of class-switch recombination. Trends Immunol. 23:31–39. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhuri, J., and F.W. Alt. 2004. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 4:541–552. [DOI] [PubMed] [Google Scholar]
- 18.Muramatsu, M., V.S. Sankaranand, S. Anant, M. Sugai, K. Kinoshita, N.O. Davidson, and T. Honjo. 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274:18470–18476. [DOI] [PubMed] [Google Scholar]
- 19.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 20.Longerich, S., U. Basu, F. Alt, and U. Storb. 2006. AID in somatic hypermutation and class switch recombination. Curr. Opin. Immunol. 18:164–174. [DOI] [PubMed] [Google Scholar]
- 21.Arakawa, H., J. Hauschild, and J.M. Buerstedde. 2002. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 295:1301–1306. [DOI] [PubMed] [Google Scholar]
- 22.Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Gennery, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 102:565–575. [DOI] [PubMed] [Google Scholar]
- 23.Honjo, T., H. Nagaoka, R. Shinkura, and M. Muramatsu. 2005. AID to overcome the limitations of genomic information. Nat. Immunol. 6:655–661. [DOI] [PubMed] [Google Scholar]
- 24.Petersen, S., R. Casellas, B. Reina-San-Martin, H.T. Chen, M.J. Difilippantonio, P.C. Wilson, L. Hanitsch, A. Celeste, M. Muramatsu, D.R. Pilch, et al. 2001. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 414:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reina-San-Martin, B., H.T. Chen, A. Nussenzweig, and M.C. Nussenzweig. 2004. ATM is required for efficient recombination between immunoglobulin switch regions. J. Exp. Med. 200:1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schrader, C.E., E.K. Linehan, S.N. Mochegova, R.T. Woodland, and J. Stavnezer. 2005. Inducible DNA breaks in Ig S regions are dependent on AID and UNG. J. Exp. Med. 202:561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manis, J.P., J.C. Morales, Z. Xia, J.L. Kutok, F.W. Alt, and P.B. Carpenter. 2004. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat. Immunol. 5:481–487. [DOI] [PubMed] [Google Scholar]
- 28.Bergsagel, P.L., M. Chesi, E. Nardini, L.A. Brents, S.L. Kirby, and W.M. Kuehl. 1996. Promiscuous translocations into immunoglobulin heavy chain switch regions in multiple myeloma. Proc. Natl. Acad. Sci. USA. 93:13931–13936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramiro, A.R., M. Jankovic, T. Eisenreich, S. Difilippantonio, S. Chen-Kiang, M. Muramatsu, T. Honjo, A. Nussenzweig, and M.C. Nussenzweig. 2004. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 118:431–438. [DOI] [PubMed] [Google Scholar]
- 30.Ramiro, A.R., M. Jankovic, E. Callen, S. Difilippantonio, H.T. Chen, K.M. McBride, T.R. Eisenreich, J. Chen, R.A. Dickins, S.W. Lowe, et al. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 440:105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sciammas, R., A.L. Shaffer, J.H. Schatz, H. Zhao, L.M. Staudt, and H. Singh. 2006. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 25:225–236. [DOI] [PubMed] [Google Scholar]
- 32.Klein, U., S. Casola, G. Cattoretti, Q. Shen, M. Lia, T. Mo, T. Ludwig, K. Rajewsky, and R. Dalla-Favera. 2006. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 7:773–782. [DOI] [PubMed] [Google Scholar]
- 33.Chesi, M., P.L. Bergsagel, L.A. Brents, C.M. Smith, D.S. Gerhard, and W.M. Kuehl. 1996. Dysregulation of cyclin D1 by translocation into an IgH gamma switch region in two multiple myeloma cell lines. Blood. 88:674–681. [PubMed] [Google Scholar]
- 34.Nardini, E., A. Aiello, R. Giardini, M.I. Colnaghi, S. Menard, and A. Balsari. 2000. Detection of aberrant isotype switch recombination in low-grade and high-grade gastric MALT lymphomas. Blood. 95:1032–1038. [PubMed] [Google Scholar]
- 35.Dave, S.S., K. Fu, G.W. Wright, L.T. Lam, P. Kluin, E.J. Boerma, T.C. Greiner, D.D. Weisenburger, A. Rosenwald, G. Ott, et al. 2006. Molecular diagnosis of Burkitt's lymphoma. N. Engl. J. Med. 354:2431–2442. [DOI] [PubMed] [Google Scholar]
- 36.Dudley, D.D., J.P. Manis, A.A. Zarrin, L. Kaylor, M. Tian, and F.W. Alt. 2002. Internal IgH class switch region deletions are position-independent and enhanced by AID expression. Proc. Natl. Acad. Sci. USA. 99:9984–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reina-San-Martin, B., S. Difilippantonio, L. Hanitsch, R.F. Masilamani, A. Nussenzweig, and M.C. Nussenzweig. 2003. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 197:1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis, R.E., K.D. Brown, U. Siebenlist, and L.M. Staudt. 2001. Constitutive nuclear factor κB activity is required for survival of activated B cell–like diffuse large B cell lymphoma cells. J. Exp. Med. 194:1861–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lam, L.T., R.E. Davis, J. Pierce, M. Hepperle, Y. Xu, M. Hottelet, Y. Nong, D. Wen, J. Adams, L. Dang, and L.M. Staudt. 2005. Small molecule inhibitors of IkB-kinase are selectively toxic for subgroups of diffuse large B cell lymphoma defined by gene expression profiling. Clin. Cancer Res. 11:28–40. [PubMed] [Google Scholar]
- 40.Pasqualucci, L., R. Guglielmino, J. Houldsworth, J. Mohr, S. Aoufouchi, R. Polakiewicz, R.S. Chaganti, and R. Dalla-Favera. 2004. Expression of the AID protein in normal and neoplastic B cells. Blood. 104:3318–3325. [DOI] [PubMed] [Google Scholar]
- 41.Lumsden, J.M., T. McCarty, L.K. Petiniot, R. Shen, C. Barlow, T.A. Wynn, H.C. Morse III, P.J. Gearhart, A. Wynshaw-Boris, E.E. Max, and R.J. Hodes. 2004. Immunoglobulin class switch recombination is impaired in Atm-deficient mice. J. Exp. Med. 200:1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reina-San-Martin, B., J. Chen, A. Nussenzweig, and M.C. Nussenzweig. 2007. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1(−/−) B cells. Eur. J. Immunol. 37:235–239. [DOI] [PubMed] [Google Scholar]
- 43.Franco, S., M. Gostissa, S. Zha, D.B. Lombard, M.M. Murphy, A.A. Zarrin, C. Yan, S. Tepsuporn, J.C. Morales, M.M. Adams, et al. 2006. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell. 21:201–214. [DOI] [PubMed] [Google Scholar]
- 44.Schotte, R., M.C. Rissoan, N. Bendriss-Vermare, J.M. Bridon, T. Duhen, K. Weijer, F. Briere, and H. Spits. 2003. The transcription factor Spi-B is expressed in plasmacytoid DC precursors and inhibits T-, B-, and NK-cell development. Blood. 101:1015–1023. [DOI] [PubMed] [Google Scholar]
- 45.Su, G.H., H.S. Ip, B.S. Cobb, M.M. Lu, H.M. Chen, and M.C. Simon. 1996. The Ets protein Spi-B is expressed exclusively in B cells and T cells during development. J. Exp. Med. 184:203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su, G.H., H.M. Chen, N. Muthusamy, L.A. Garrett-Sinha, D. Baunoch, D.G. Tenen, and M.C. Simon. 1997. Defective B cell receptor-mediated responses in mice lacking the Ets protein, Spi-B. EMBO J. 16:7118–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akasaka, T., M. Muramatsu, H. Ohno, I. Miura, E. Tatsumi, S. Fukuhara, T. Mori, and M. Okuma. 1996. Application of long-distance polymerase chain reaction to detection of junctional sequences created by chromosomal translocation in mature B-cell neoplasms. Blood. 88:985–994. [PubMed] [Google Scholar]
- 48.Sonoki, T., T.G. Willis, D.G. Oscier, E.L. Karran, R. Siebert, and M.J. Dyer. 2004. Rapid amplification of immunoglobulin heavy chain switch (IGHS) translocation breakpoints using long-distance inverse PCR. Leukemia. 18:2026–2031. [DOI] [PubMed] [Google Scholar]
- 49.Shou, Y., M.L. Martelli, A. Gabrea, Y. Qi, L.A. Brents, A. Roschke, G. Dewald, I.R. Kirsch, P.L. Bergsagel, and W.M. Kuehl. 2000. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc. Natl. Acad. Sci. USA. 97:228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin-Subero, J.I., I. Chudoba, L. Harder, S. Gesk, W. Grote, F.J. Novo, M.J. Calasanz, and R. Siebert. 2002. Multicolor-FICTION: expanding the possibilities of combined morphologic, immunophenotypic, and genetic single cell analyses. Am. J. Pathol. 161:413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alizadeh, A., M. Eisen, R.E. Davis, C. Ma, H. Sabet, T. Tran, J. Powell, L. Yang, G. Marti, T. Moore, et al. 1999. The lymphochip: a specialized cDNA microarray for the genomic-scale analysis of gene expression in normal and malignant lymphocytes. Cold Spring Harb. Symp. Quant. Biol. 64:71–78. [DOI] [PubMed] [Google Scholar]