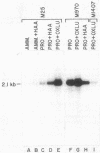
Abstract
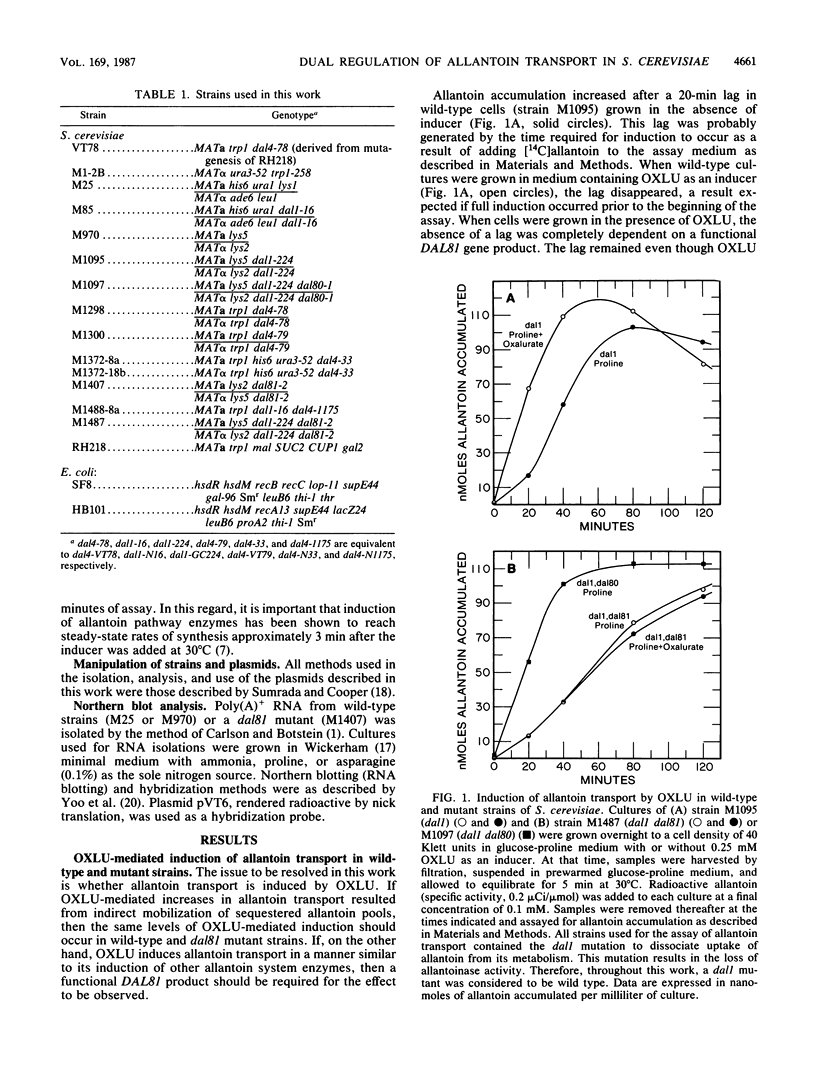
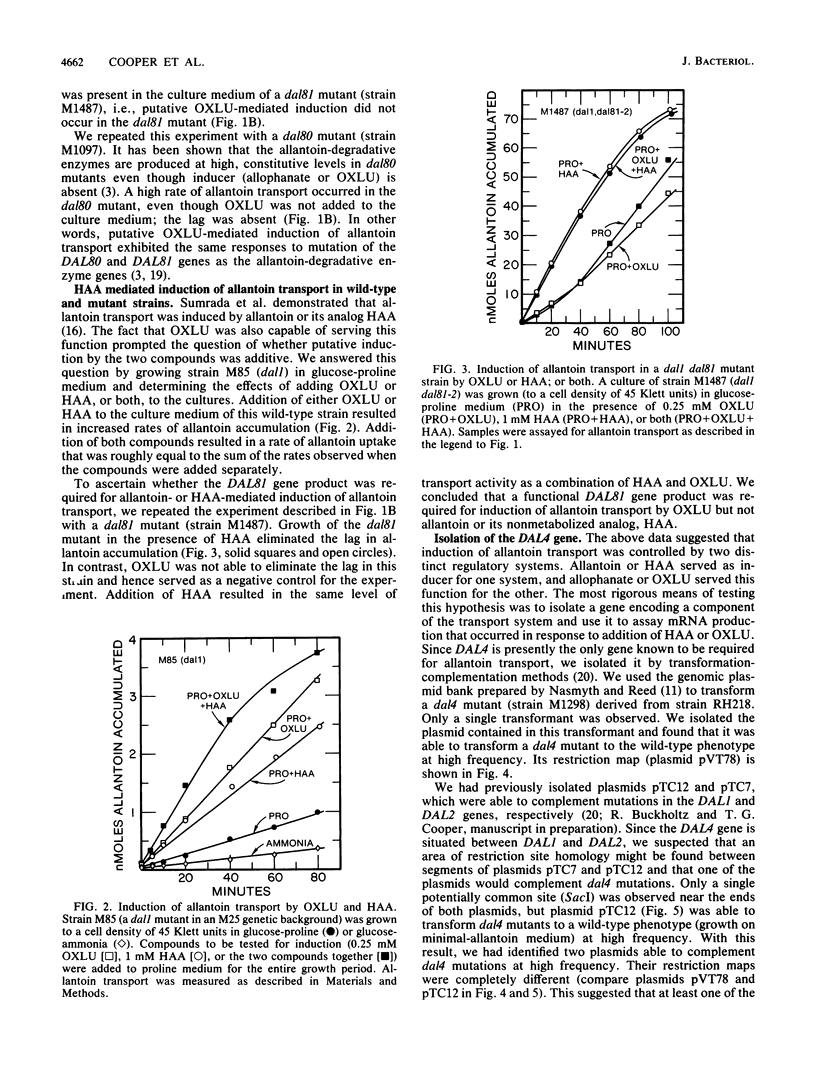
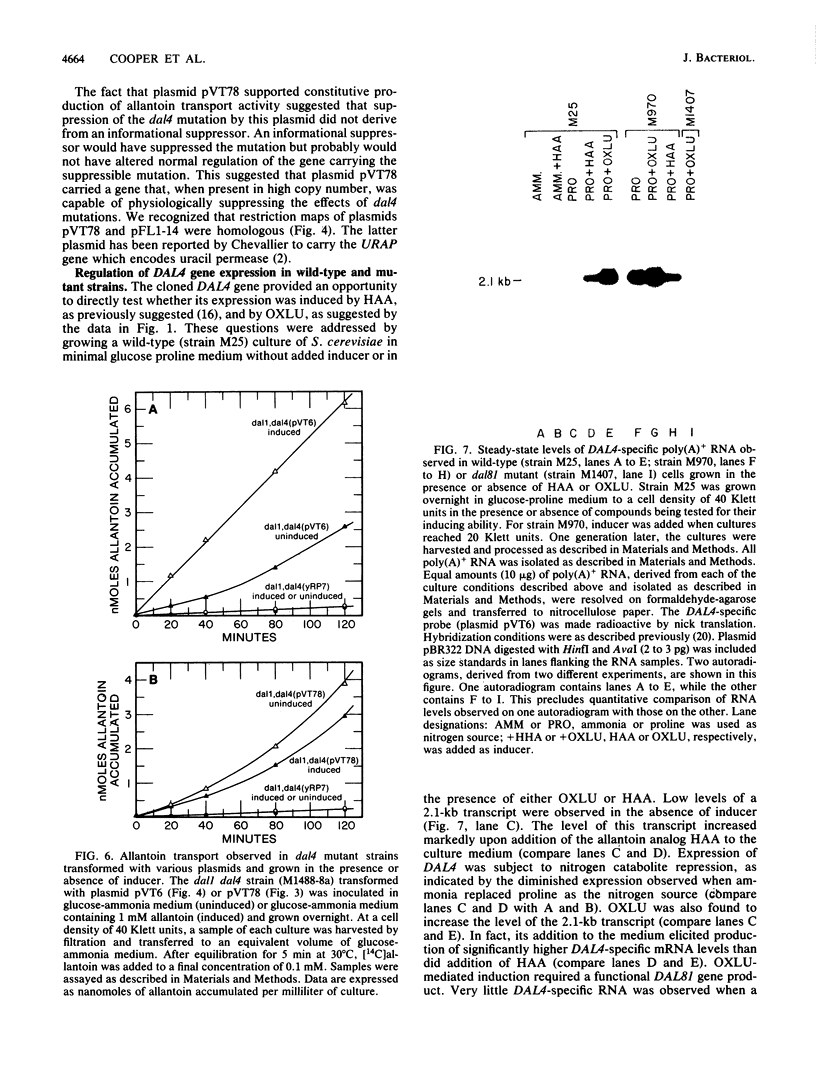
We show that the allantoin transport system of Saccharomyces cerevisiae responds to two induction systems, one mediated by allophanate or its analog oxalurate and the other mediated by allantoin or its analog hydantoin acetate. The effects of the two inducers were additive in strain M85. Like other allantoin pathway genes, oxalurate-mediated induction of allantoin transport required a functional DAL81 gene product. Hydantoin acetate-mediated induction of the system, on the other hand, occurred normally in dal81 mutants. This suggests that induction was not only mediated by two separate inducers, but also involved different regulatory proteins. Induction is probably a transcriptionally regulated process, because addition of hydantoin acetate or oxalurate to the culture medium increased the steady-state levels of mRNA encoded by a gene required for allantoin transport (DAL4).
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Chevallier M. R. Cloning and transcriptional control of a eucaryotic permease gene. Mol Cell Biol. 1982 Aug;2(8):977–984. doi: 10.1128/mcb.2.8.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm G., Cooper T. G. Isolation and characterization of mutants that produce the allantoin-degrading enzymes constitutively in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Sep;2(9):1088–1095. doi: 10.1128/mcb.2.9.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Gorski M., Turoscy V. A cluster of three genes responsible for allantoin degradation in Saccharomyces cerevisiae. Genetics. 1979 Jun;92(2):383–396. doi: 10.1093/genetics/92.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Lawther R. P. Induction of the allantoin degradative enzymes in Saccharomyces cerevisiae by the last intermediate of the pathway. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2340–2344. doi: 10.1073/pnas.70.8.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawther R. P., Cooper T. G. Kinetics of induced and repressed enzyme synthesis in Saccharomyces cerevisiae. J Bacteriol. 1975 Mar;121(3):1064–1073. doi: 10.1128/jb.121.3.1064-1073.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawther R. P., Riemer E., Chojnacki B., Cooper T. G. Clustering of the genes for allantoin degradation in Saccharomyces cerevisiae. J Bacteriol. 1974 Aug;119(2):461–468. doi: 10.1128/jb.119.2.461-468.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz D. The crystal structure of DL-allantoin. Acta Crystallogr. 1965 Nov 10;19(5):726–734. doi: 10.1107/s0365110x65004280. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A., Reed S. I. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2119–2123. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. D. Biochemical Mutants in the Smut Fungus Ustilago Maydis. Genetics. 1949 Sep;34(5):607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Isolation of the CAR1 gene from Saccharomyces cerevisiae and analysis of its expression. Mol Cell Biol. 1982 Dec;2(12):1514–1523. doi: 10.1128/mcb.2.12.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R., Cooper T. G. Allantoin transport in Saccharomyces cerevisiae. J Bacteriol. 1977 Sep;131(3):839–847. doi: 10.1128/jb.131.3.839-847.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R., Cooper T. G. Oxaluric acid: a non-metabolizable inducer of the allantoin degradative enzymes in Saccharomyces cerevisiae. J Bacteriol. 1974 Mar;117(3):1240–1247. doi: 10.1128/jb.117.3.1240-1247.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R., Gorski M., Cooper T. Urea transport-defective strains of Saccharomyces cerevisiae. J Bacteriol. 1976 Mar;125(3):1048–1056. doi: 10.1128/jb.125.3.1048-1056.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R., Zacharski C. A., Turoscy V., Cooper T. G. Induction and inhibition of the allantoin permease in Saccharomyces cerevisiae. J Bacteriol. 1978 Aug;135(2):498–510. doi: 10.1128/jb.135.2.498-510.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turoscy V., Cooper T. G. Pleiotropic control of five eucaryotic genes by multiple regulatory elements. J Bacteriol. 1982 Sep;151(3):1237–1246. doi: 10.1128/jb.151.3.1237-1246.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. S., Genbauffe F. S., Cooper T. G. Identification of the ureidoglycolate hydrolase gene in the DAL gene cluster of Saccharomyces cerevisiae. Mol Cell Biol. 1985 Sep;5(9):2279–2288. doi: 10.1128/mcb.5.9.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharski C. A., Cooper T. G. Metabolite compartmentation in Saccharomyces cerevisiae. J Bacteriol. 1978 Aug;135(2):490–497. doi: 10.1128/jb.135.2.490-497.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]