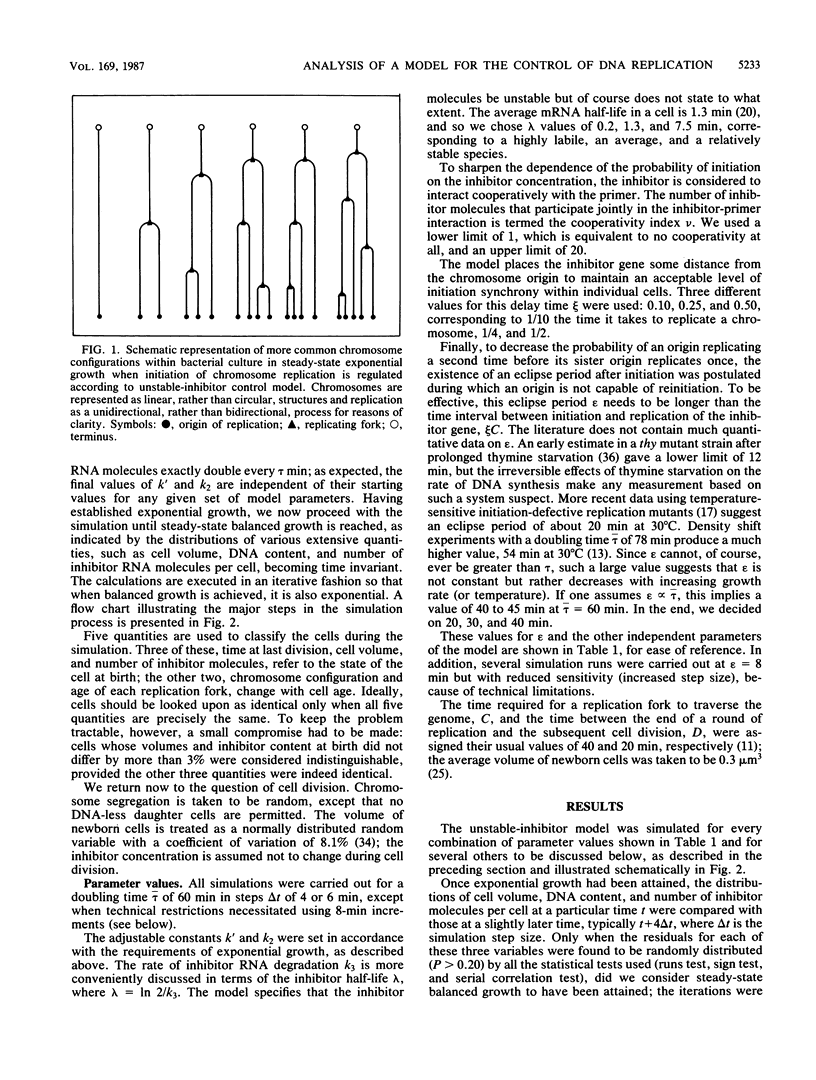
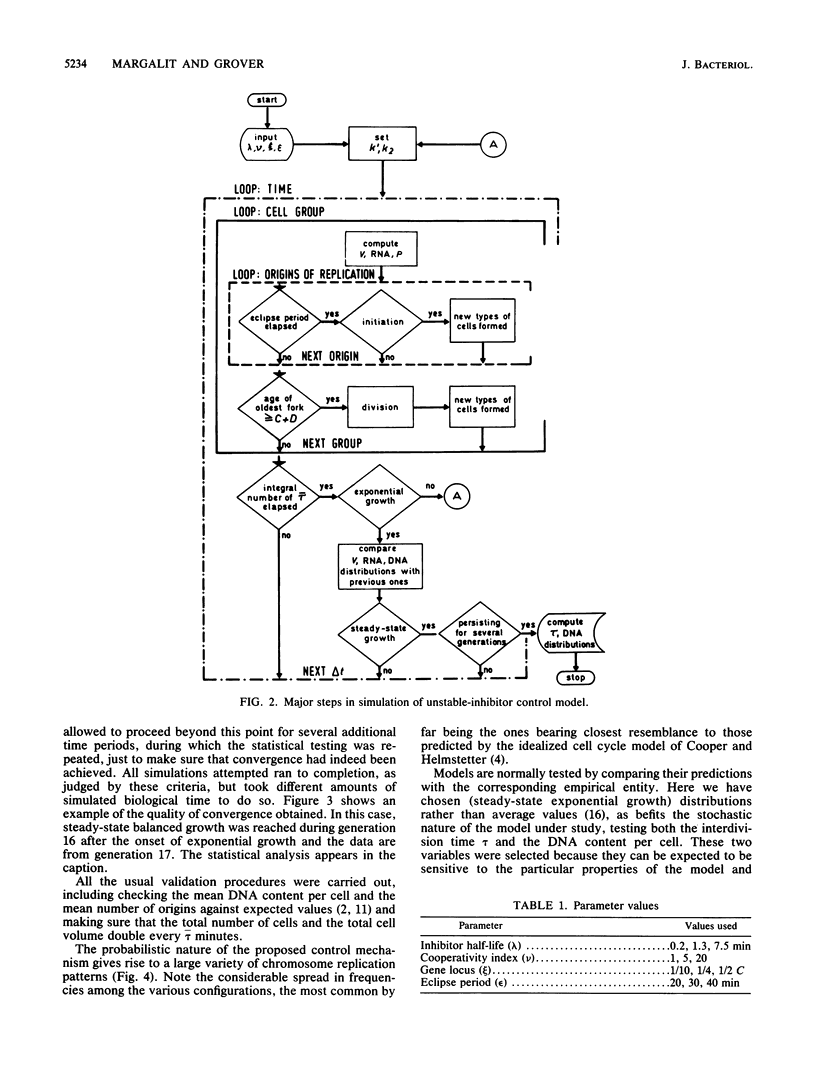
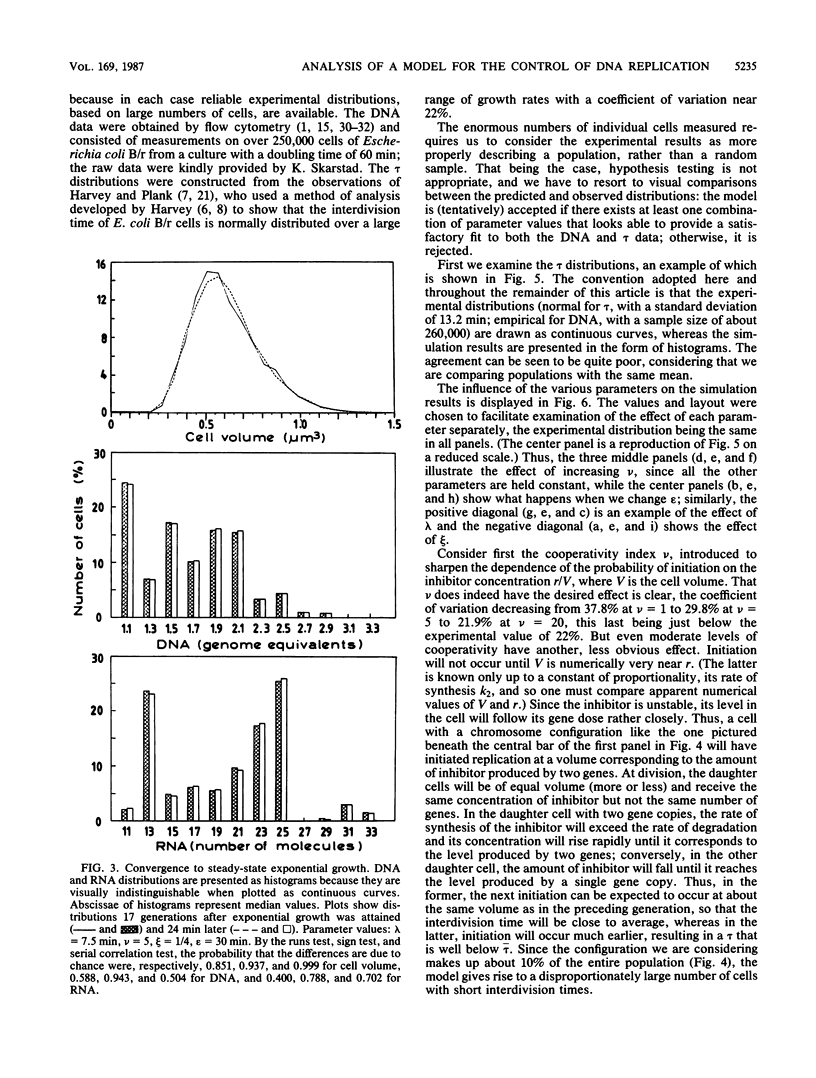
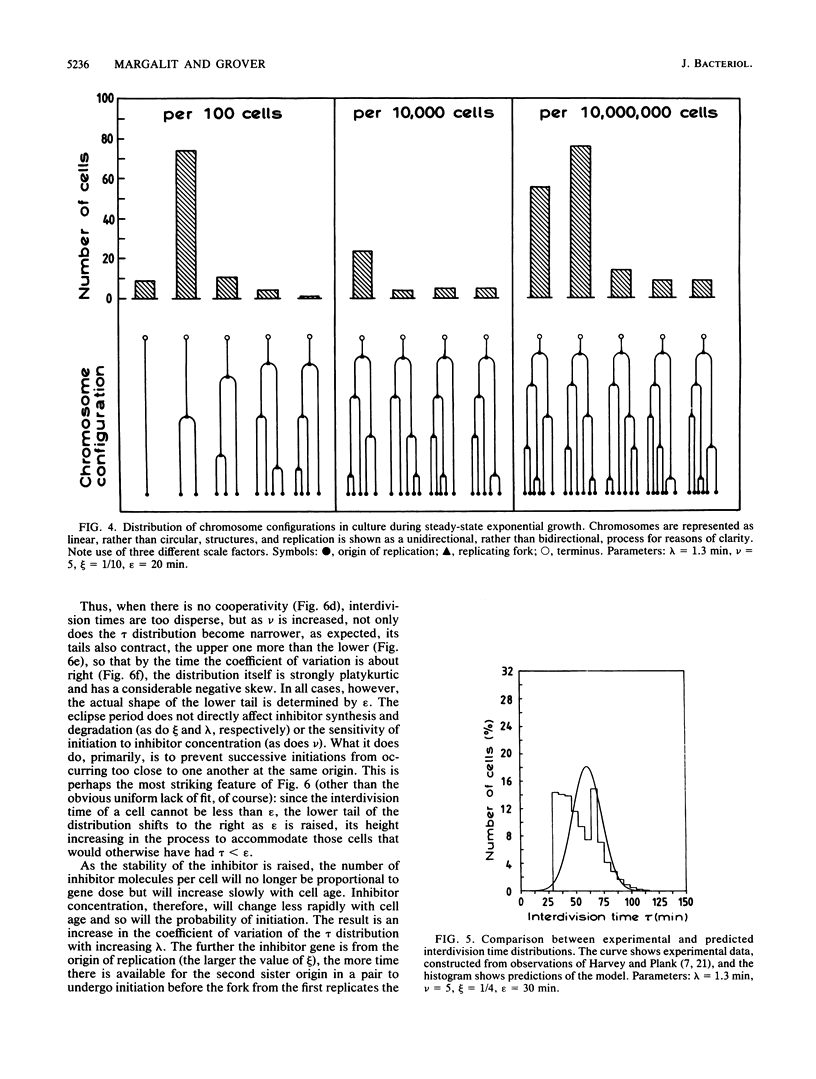
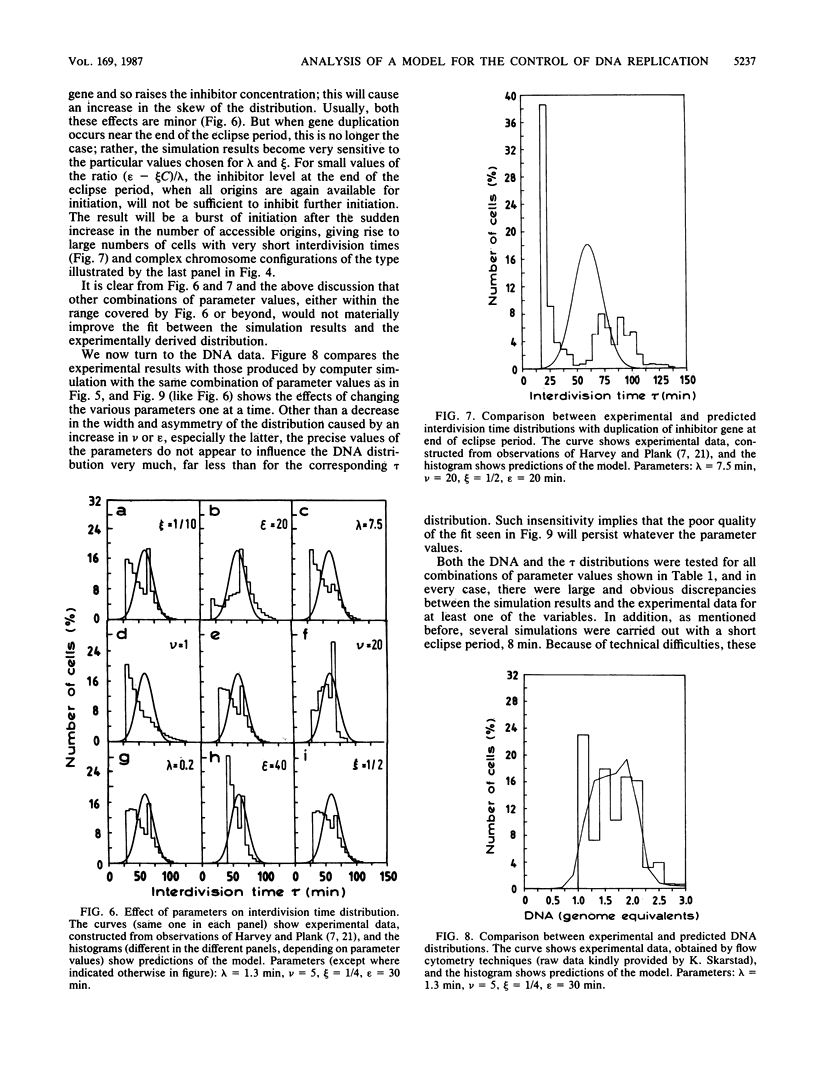
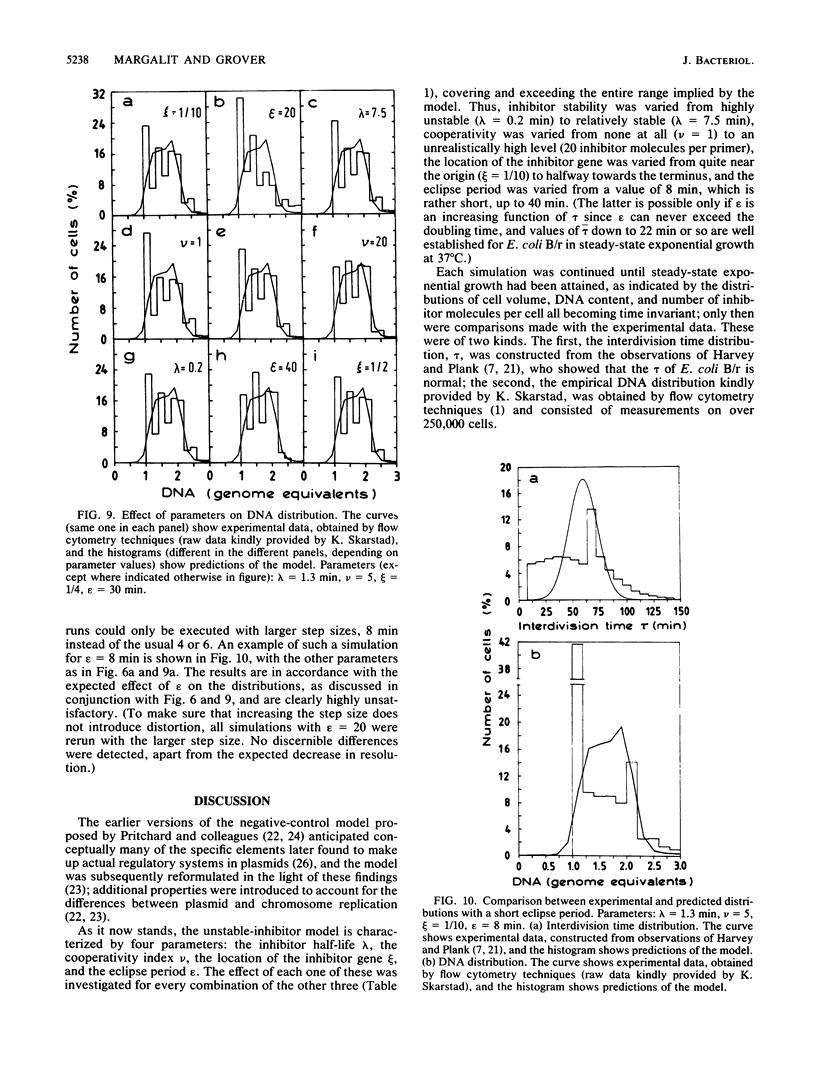
Abstract
This article contains an analysis of a version of the well-known inhibitor-dilution model for the control of initiation of chromosome replication in bacteria. According to this model, an unstable inhibitor interacts with an initiation primer in a hit-and-destroy fashion to prevent successful initiation; both constituents are presumed to be RNA species that are synthesized constitutively. The model further postulates that the inhibitor interacts cooperatively with the primer, that the inhibitor gene is removed some distance from the origin of replication, and that an eclipse period exists during which the chromosome origin is not able to reinitiate. This unstable-inhibitor version is characterized by four parameters: the inhibitor half-life, the cooperativity index, the location of the inhibitor gene, and the eclipse period; computer simulations are used to study the effect of each of these on the DNA and interdivision time distributions in exponentially growing steady-state cultures. In neither case was any combination of parameter values found that could provide even moderately satisfactory agreement between the simulation results and experimental data. From the examples furnished and the associated discussion, it appears that there are none--that no combination of parameter values exists that can reasonably be expected to produce a significantly better fit than those tested. We conclude that the model in its present form cannot be a valid description of chromosome replication control in bacteria. It is pointed out that this does not necessarily apply to negative initiation control models in general, or even to all inhibitor-dilution systems, merely to the particular ColE1-like mechanism considered here. Nevertheless, recent experimental results, which can only be understood in terms of a very high degree of initiation synchrony within individual cells, offer strong evidence against stochastic models of this kind for the control of chromosome replication.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boye E., Steen H. B., Skarstad K. Flow cytometry of bacteria: a promising tool in experimental and clinical microbiology. J Gen Microbiol. 1983 Apr;129(4):973–980. doi: 10.1099/00221287-129-4-973. [DOI] [PubMed] [Google Scholar]
- Churchward G., Bremer H., Young R. Macromolecular composition of bacteria. J Theor Biol. 1982 Feb 7;94(3):651–670. doi: 10.1016/0022-5193(82)90305-8. [DOI] [PubMed] [Google Scholar]
- Churchward G., Estiva E., Bremer H. Growth rate-dependent control of chromosome replication initiation in Escherichia coli. J Bacteriol. 1981 Mar;145(3):1232–1238. doi: 10.1128/jb.145.3.1232-1238.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S., Helmstetter C. E. Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol. 1968 Feb 14;31(3):519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- Donachie W. D. Relationship between cell size and time of initiation of DNA replication. Nature. 1968 Sep 7;219(5158):1077–1079. doi: 10.1038/2191077a0. [DOI] [PubMed] [Google Scholar]
- Harvey J. D. Parameters of the generation time distribution of Escherichia coli B-r. J Gen Microbiol. 1972 Apr;70(1):109–114. doi: 10.1099/00221287-70-1-109. [DOI] [PubMed] [Google Scholar]
- Harvey J. D. Synchronous growth of cells and the generation time distribution. J Gen Microbiol. 1972 Apr;70(1):99–107. doi: 10.1099/00221287-70-1-99. [DOI] [PubMed] [Google Scholar]
- Hashimoto-Gotoh T., Inselburg J. ColE1 plasmid incompatibility: localization and analysis of mutations affecting incompatibility. J Bacteriol. 1979 Aug;139(2):608–619. doi: 10.1128/jb.139.2.608-619.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstetter C., Cooper S., Pierucci O., Revelas E. On the bacterial life sequence. Cold Spring Harb Symp Quant Biol. 1968;33:809–822. doi: 10.1101/sqb.1968.033.01.093. [DOI] [PubMed] [Google Scholar]
- Koppes L. J., Nanninga N. Positive correlation between size at initiation of chromosome replication in Escherichia coli and size at initiation of cell constriction. J Bacteriol. 1980 Jul;143(1):89–99. doi: 10.1128/jb.143.1.89-99.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppes L., Nordström K. Insertion of an R1 plasmid into the origin of replication of the E. coli chromosome: random timing of replication of the hybrid chromosome. Cell. 1986 Jan 17;44(1):117–124. doi: 10.1016/0092-8674(86)90490-3. [DOI] [PubMed] [Google Scholar]
- Lacatena R. M., Cesareni G. Interaction between RNA1 and the primer precursor in the regulation of Co1E1 replication. J Mol Biol. 1983 Nov 5;170(3):635–650. doi: 10.1016/s0022-2836(83)80125-9. [DOI] [PubMed] [Google Scholar]
- Lindmo T., Steen H. B. Characteristics of a simple, high-resolution flow cytometer based o a new flow configuration. Biophys J. 1979 Oct;28(1):33–44. doi: 10.1016/S0006-3495(79)85157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalit H., Rosenberger R. F., Grover N. B. Initiation of DNA replication in bacteria: analysis of an autorepressor control model. J Theor Biol. 1984 Nov 7;111(1):183–199. doi: 10.1016/s0022-5193(84)80204-0. [DOI] [PubMed] [Google Scholar]
- Messer W., Bellekes U., Lother H. Effect of dam methylation on the activity of the E. coli replication origin, oriC. EMBO J. 1985 May;4(5):1327–1332. doi: 10.1002/j.1460-2075.1985.tb03780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K., Molin S., Light J. Control of replication of bacterial plasmids: genetics, molecular biology, and physiology of the plasmid R1 system. Plasmid. 1984 Sep;12(2):71–90. doi: 10.1016/0147-619x(84)90054-4. [DOI] [PubMed] [Google Scholar]
- Pato M. L., Bennett P. M., von Meyenburg K. Messenger ribonucleic acid synthesis and degradation in Escherichia coli during inhibition of translation. J Bacteriol. 1973 Nov;116(2):710–718. doi: 10.1128/jb.116.2.710-718.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank L. D., Harvey J. D. Generation time statistics of Escherichia coli B measured by synchronous culture techniques. J Gen Microbiol. 1979 Nov;115(1):69–77. doi: 10.1099/00221287-115-1-69. [DOI] [PubMed] [Google Scholar]
- Rosenberger R. F., Grover N. B., Zaritsky A., Woldringh C. L. Surface growth in rod-shaped bacteria. J Theor Biol. 1978 Aug 21;73(4):711–721. doi: 10.1016/0022-5193(78)90132-7. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Regulation of plasmid replication. Microbiol Rev. 1984 Mar;48(1):1–23. doi: 10.1016/b978-0-12-048850-6.50006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard H. M., Gelfand D. H., Polisky B. Analysis of a recessive plasmid copy number mutant: evidence for negative control of Col E1 replication. Cell. 1979 Oct;18(2):267–275. doi: 10.1016/0092-8674(79)90046-1. [DOI] [PubMed] [Google Scholar]
- Skarstad K., Boye E., Steen H. B. Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J. 1986 Jul;5(7):1711–1717. doi: 10.1002/j.1460-2075.1986.tb04415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sompayrac L., Maaloe O. Autorepressor model for control of DNA replication. Nat New Biol. 1973 Jan 31;241(109):133–135. doi: 10.1038/newbio241133a0. [DOI] [PubMed] [Google Scholar]
- Steen H. B. A microscope-based flow cytophotometer. Histochem J. 1983 Feb;15(2):147–160. doi: 10.1007/BF01042283. [DOI] [PubMed] [Google Scholar]
- Steen H. B. Further developments of a microscope-based flow cytometer: light scatter detection and excitation intensity compensation. Cytometry. 1980 Jul;1(1):26–31. doi: 10.1002/cyto.990010107. [DOI] [PubMed] [Google Scholar]
- Steen H. B., Lindmo T. Flow cytometry: a high-resolution instrument for everyone. Science. 1979 Apr 27;204(4391):403–404. doi: 10.1126/science.441727. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T. Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6096–6100. doi: 10.1073/pnas.78.10.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueba F. J. On the precision and accuracy achieved by Escherichia coli cells at fission about their middle. Arch Microbiol. 1982 Feb;131(1):55–59. doi: 10.1007/BF00451499. [DOI] [PubMed] [Google Scholar]
- Trueba F. J., Woldringh C. L. Changes in cell diameter during the division cycle of Escherichia coli. J Bacteriol. 1980 Jun;142(3):869–878. doi: 10.1128/jb.142.3.869-878.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaritsky A. Rate stimulation of deoxyribonucleic acid synthesis after inhibition. J Bacteriol. 1975 Jun;122(3):841–846. doi: 10.1128/jb.122.3.841-846.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]


