Abstract
RanGAP1 is the GTPase-activating protein for Ran, a small ras-like GTPase involved in regulating nucleocytoplasmic transport. In vertebrates, RanGAP1 is present in two forms: one that is cytoplasmic, and another that is concentrated at the cytoplasmic fibers of nuclear pore complexes (NPCs). The NPC-associated form of RanGAP1 is covalently modified by the small ubiquitin-like protein, SUMO-1, and we have recently proposed that SUMO-1 modification functions to target RanGAP1 to the NPC. Here, we identify the domain of RanGAP1 that specifies SUMO-1 modification and demonstrate that mutations in this domain that inhibit modification also inhibit targeting to the NPC. Targeting of a heterologous protein to the NPC depended on determinants specifying SUMO-1 modification and also on additional determinants in the COOH-terminal domain of RanGAP1. SUMO-1 modification and these additional determinants were found to specify interaction between the COOH-terminal domain of RanGAP1 and a region of the nucleoporin, Nup358, between Ran-binding domains three and four. Together, these findings indicate that SUMO-1 modification targets RanGAP1 to the NPC by exposing, or creating, a Nup358 binding site in the COOH-terminal domain of RanGAP1. Surprisingly, the COOH-terminal domain of RanGAP1 was also found to harbor a nuclear localization signal. This nuclear localization signal, and the presence of nine leucine-rich nuclear export signal motifs, suggests that RanGAP1 may shuttle between the nucleus and the cytoplasm.
Posttranslational protein modifications are required for a variety of cell functions, regulating protein interactions, enzymatic activity, subcellular localization, and stability. Ubiquitination is a posttranslational modification that involves the covalent attachment of ubiquitin (Ub),1 itself a 76 amino acid protein, to lysine residues of targeted substrates (for review see Wilkinson, 1995; Hochstrasser, 1996). Ub can be considered to be a posttranslationally added signal that targets its substrates to specific fates. Among other factors, different metabolic fates can depend on the number of Ub molecules conjugated to a particular substrate, with mono-ubiquitinated proteins being relatively stable (such as histone H2A; Goldknopf and Busch, 1977), and multiubiquitinated proteins being relatively unstable. Covalent attachment of Ub to intracellular proteins has effects on a wide range of cell functions, including gene expression, cell division, DNA repair, programmed cell death, peroxisome biogenesis, mitochondrial protein import, and ribosome assembly. While the precise mechanisms underlying the roles of Ub in many of these processes are not fully understood, the best characterized function of the Ub signal is to target proteins for ATP-dependent proteolysis by the 26S proteasome (Wilkinson, 1995; Hochstrasser, 1996). Targeting by Ub is mediated by direct interactions between 26S proteasome subunits and Ub itself (Deveraux et al., 1994).
It has long been suspected that Ub modification may have other consequences for targeted substrates, and recently ubiquitination was shown to function as a signal for ligand-induced receptor endocytosis and lysosomal targeting (Hicke and Riezman, 1996; Strous et al., 1996), as well as in activation of the IκBα protein kinase (Chen et al., 1996). The question of whether there may be related ubiquitin systems using novel, ubiquitin-like proteins as modifiers has also been raised. The first example of such a system was described for the interferon inducible protein, UCRP (ubiquitin cross-reactive protein), a 15-kD protein containing two Ub-related domains that are 43 and 62% homologous to Ub, respectively (Haas et al., 1987). UCRP is covalently ligated to a heterogeneous set of proteins by a pathway that is parallel to, but also distinct from, ubiquitination (Narasimhan et al., 1996). Like Ub, UCRP also appears to function as a posttranslationally added signal, targeting modified substrates to intermediate filaments in the cytoplasm (Loeb and Haas, 1994).
Recently, a novel family of Ub-like proteins, termed small ubiquitin-like modifiers (SUMOs), has been described that, while sharing only 18% identity with Ub, also appear to be processed and covalently ligated to protein substrates by mechanisms similar to ubiquitination (Boddy et al., 1996; Mannen et al., 1996; Matunis et al., 1996; Okura et al., 1996; Shen et al., 1996a ; Mahajan et al., 1997). The first member of this family of proteins, SUMO-1 (previously called GMP1, PIC1, UBL1, and Sentrin), was identified as a covalent modification of the Ran GTPase-activating protein, RanGAP1 (Matunis et al., 1996; Mahajan et al., 1997). SUMO-1 was also identified in two-hybrid screens with the Fas/APO-1 receptor (Okura et al., 1996), PML (Boddy et al., 1996), and Rad51 and Rad52 (Shen et al., 1996a ), although it remains to be determined whether SUMO-1 is covalently ligated to these proteins. While RanGAP1 remains the only confirmed substrate for SUMO-1 modification, SUMO-1 is conjugated to a limited number of predominantly nuclear, high molecular mass proteins (Matunis et al., 1996; Kamitani et al., 1997; Mahajan et al., 1997). As predicted from conserved elements in its primary sequence, reactions involved in SUMO-1 processing and ligation parallel ubiquitination. First, SUMO-1 contains a COOH-terminal extension of four amino acids (HSTV) that is proteolytically removed to expose the mature COOH terminus ending with a double glycine (Kamitani et al., 1997). This double glycine is invariant in all known Ub and SUMO proteins. Secondly, the exposed COOH-terminal glycine residue is essential for subsequent conjugation reactions, similar to the COOH-terminal glycine of Ub (Kamitani et al., 1997). And finally, SUMO-1 modification is reversible, as is ubiquitination (Matunis et al., 1996; Mahajan et al., 1997). Unlike Ub, SUMO-1 does not appear to form polymeric chains (at least when attached to RanGAP1), and as will be discussed below, SUMO-1 modification has novel functions distinct from those of ubiquitination.
Saccharomyces cerevisiae contains a single SUMO protein, encoded by the SMT3 gene, which is 40% identical to SUMO-1 (Meluh and Koshland, 1995). The SMT3 gene was first identified in a genetic screen as a suppressor of MIF2, a centromere-associated protein (Meluh and Koshland, 1995). Smt3p also functions as a Ub-like modifier, and considerable progress has recently been made in characterizing its processing and conjugation reactions. After proteolytic processing, Smt3p is activated by a heterodimeric enzyme consisting of Uba2p, a protein homologous to the COOH terminus of Ub-activating enzymes (E1s), and Aos1p, a protein homologous to the NH2 terminus of E1s (Johnson et al., 1997). The second step in the pathway, Smt3p conjugation, is mediated by Ubc9p, a protein homologous to Ub-conjugating enzymes, or E2s (Johnson and Blobel, 1997). While the enzymes involved in conjugating the vertebrate SUMOs have not been identified to date, it can be anticipated that they will be homologous to those required for Smt3p.
As indicated, SUMO-1 was identified as a covalent modification of the Ran GTPase-activating protein, RanGAP1 (Matunis et al., 1996; Mahajan et al., 1997). Ran is a small nuclear ras-like GTPase required for the bidirectional transport of proteins and ribonucleoproteins across the nuclear pore complex (NPC) (Melchior et al., 1993; Moore and Blobel, 1993; for review see Rush et al., 1996). RanGAP1 is the only known GTPase-activating protein for Ran (Bischoff et al., 1994, 1995a ,b), and its subcellular localization is, therefore, an important indicator of where RanGTP hydrolysis is required during transport through the NPC (for recent reviews of nuclear transport see Corbett and Silver, 1997; Nigg, 1997). In vertebrate cells, unmodified RanGAP1 is present in the cytoplasm and SUMO-1–modified RanGAP1 is associated with the cytoplasmic fibers of NPCs (Matunis et al., 1996; Mahajan et al., 1997). This observation led us to propose that SUMO-1 modification functions to target RanGAP1 to the NPC. Here, we provide evidence for this proposal by demonstrating that mutations in RanGAP1 that block SUMO-1 modification also prevent RanGAP1 from localizing at the NPC. We have also investigated the mechanism by which SUMO-1 modification targets RanGAP1 to the NPC. Our results indicate that SUMO-1 modification exposes, or creates, a binding site in the COOH-terminal domain of RanGAP1 that binds to a region in the COOH terminus of Nup358, a nucleoporin associated with the cytoplasmic fibers of the NPC (Wu et. al., 1995; Yokoyama et al., 1995). Finally, we have identified a nuclear localization signal in the COOH-terminal domain of RanGAP1, raising the interesting possibility that RanGAP1 may shuttle between the nucleus and the cytoplasm.
Materials and Methods
Plasmid Constructions
The cDNA coding for mouse RanGAP1 was kindly provided by Dr. Mark Rush (New York University, New York; Ren et al., 1995). The expression vector coding for myc-tagged, wild-type RanGAP1 was constructed as follows: a synthetic 5′ PCR primer complementary to the NH2 terminus of RanGAP1 and containing an EcoRI site was used in conjunction with a 3′ primer complementary to the COOH terminus and containing a NotI site to prime PCR using the mouse RanGAP1 cDNA as template. The amplified fragment was digested with EcoRI and NotI and ligated into EcoRI-NotI–digested pcDNA3-mycPK, replacing the insert coding for pyruvate kinase. pcDNA3-mycPK was kindly provided by Dr. Haru Siomi (University of Pennsylvania, Philadelphia, PA) and contains an insert cloned into the EcoRI and NotI sites coding for pyruvate kinase, and an insert cloned into the BstXI and EcoRI sites that codes for the sequence MEQKLISEEDL in frame with pyruvate kinase (Siomi and Dreyfuss, 1995). This sequence is the epitope recognized by the anti-myc monoclonal antibody, 9E10 (Evan et al., 1985). COOH- and NH2-terminal deletions of RanGAP1 were generated using a similar strategy, using PCR primers complementary to appropriate coding sequences. Point mutations in the COOH terminus of RanGAP1 were generated using the Altered Sites II Mammalian Mutagenesis System (Promega Corp., Madison, WI). The cDNA coding for myc-tagged RanGAP1 was excised from pcDNA3 using KpnI and NotI and ligated into KpnI-NotI–digested pAlter-Max. Oligonucleotide-directed point mutations were generated according to instructions provided by the manufacturer. Vectors for expressing pyruvate kinase–RanGAP1 fusion proteins were synthesized as follows: 5′ primers complementary to the appropriate coding sequences of RanGAP1 and containing EcoRI sites were used in conjunction with 3′ primers complementary to the COOH terminus of RanGAP1 and containing XhoI sites to prime PCR using mouse RanGAP1 cDNA as template. The amplified fragments were digested with EcoRI and XhoI and ligated into EcoRI-XhoI–digested pcDNA1-mycPK. pcDNA1-mycPK was kindly provided by Dr. Matthew Michael (University of San Diego, San Diego, CA) and contains an insert coding for myc-tagged pyruvate kinase cloned into the BamHI and EcoRI sites of pcDNA1 (Michael et al., 1995). The expression vector for the SUMO-1–pyruvate kinase fusion protein was made by PCR amplification of a fragment of SUMO-1 coding for amino acids 1–95. The resulting fragment, containing a BamHI site at the 5′-end and a BstXI site at the 3′-end, was digested and cloned into the BamHI and BstXI sites of pcDNA3-mycPK.
Transfections and Immunofluorescence Localization
HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Cells were grown on glass coverslips in 35-mm dishes and transfected with 1 μg of plasmid using lipofectamine (GIBCO-BRL, Gaithersburg, MD), as described by the manufacturer. 36 h after transfection, the cells were washed twice with PBS, fixed for 30 min at room temperature with 2% formaldehyde in PBS, and permeabilized with −20°C acetone for 3 min. After rinsing three times with PBS, the cells were incubated with the anti-myc monoclonal antibody, 9E10 (diluted in PBS containing 2% BSA), for 1 h at room temperature, followed by three more washes with PBS. Cells were subsequently incubated with fluorescein-conjugated goat anti–mouse (diluted in PBS containing 2% BSA) for 30 min at room temperature. After three washes with PBS, coverslips were mounted in buffer containing 80% glycerol, 50 mM Tris-HCl, pH 8.0, and 0.1% p-phenylenediamine and analyzed with a Zeis Axiophot fluorescence microscope (Thornwood, NY).
Gel Electrophoresis and Immunoblot Analysis
HeLa cells transfected as described above were lysed in SDS sample buffer 36 h after transfection. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membrane as previously described (Dreyfuss et al., 1984). Membranes were blocked in 5% nonfat dry milk in PBST (PBS containing 0.1% Tween-20), followed by incubation with monoclonal antibody 9E10 (diluted 1:1,000 in PBST containing 2% BSA). Antibodies were detected using luminol-based chemiluminescence.
Expression and Purification of Recombinant Proteins
DNA fragments coding for specific regions of Nup358 were generated by PCR amplification using appropriate complementary primers. These fragments were cloned into pGEX-2TL (pGEX-2T with a modified multiple cloning site; generously provided by Dr. Hui Ge, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD), and the plasmids were transformed into DH5α cells. Protein expression was induced with 0.1 mM IPTG, and proteins were purified by affinity chromatography using glutathione Sepharose 4B as described by the manufacturer (Pharmacia Biotech Inc., Piscataway, NJ).
In Vitro Protein Binding Assay
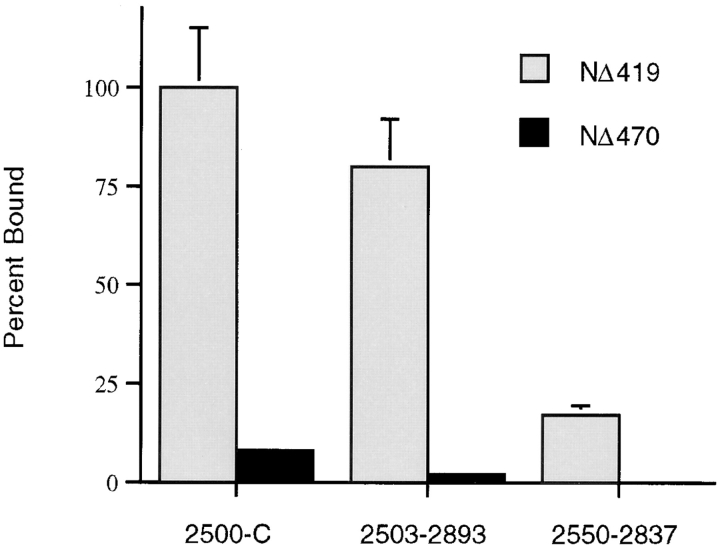
RanGAP1 substrates were expressed in rabbit reticulocyte transcription/ translation extracts in the presence of [35S]methionine as described by the manufacturer (Promega Corp.). Incorporation of radioactive label was determined by TCA precipitation followed by filter binding and counting in a liquid scintillation counter. Binding assays were performed as follows: 1 μg of each purified protein was diluted into 100 μl of PBS and bound to the wells of a microtiter plate overnight at 4°C. The wells were subsequently blocked with 150 μl of assay buffer (2% BSA, 20 mM Hepes-KOH, pH 7.3, 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 0.05% Tween-20) for 1 h at room temperature. In vitro translated RanGAP1 substrates were diluted into 100 μl of assay buffer and incubated with the bound proteins for 1 h at room temperature. After five washes with 150 μl of assay buffer, proteins were eluted with SDS sample buffer. Quantification of NΔ419 and NΔ470 binding was as follows: assays were performed as described above in triplicate, loading equivalent amounts of NΔ419 and NΔ470 to the assays (based on radioactive incorporation during in vitro translation). Radioactive counts bound in each binding assay were determined using a liquid scintillation counter, and counts bound to glutathione-S-transferase (GST) were subtracted from each assay as background. NΔ419 binding to Nup358 fragment 2500-C was arbitrarily set at 100%, and percentages bound in all other assays were calculated relative to this reaction. Approximately 20% of the SUMO-1–modified NΔ419 added to the assays was bound by 2500-C. 2500-C is estimated to be at ∼200-fold molar excess over SUMO-1–modified NΔ419.
Results
Lysine 526 Is Required for SUMO-1 Modification of RanGAP1
To identify elements of RanGAP1 required for SUMO-1 modification, we developed a rapid and simple in vitro assay using rabbit reticulocyte transcription and translation extracts. When a cDNA coding for myc-tagged RanGAP1 was transcribed and translated in an vitro extract, two products were observed, migrating with relative molecular masses of 75 and 95 kD (Fig. 1, lane 1). Both products appeared as doublets, possibly as a result of initiation of translation from an internal methionine. Their 20-kD difference suggested that the larger product represented SUMO-1–modified RanGAP1, which was confirmed by immunopurification with antibodies specific for SUMO-1 (data not shown). These results indicate that rabbit reticulocyte extracts contain all of the necessary factors for SUMO-1 modification of RanGAP1.
Figure 1.
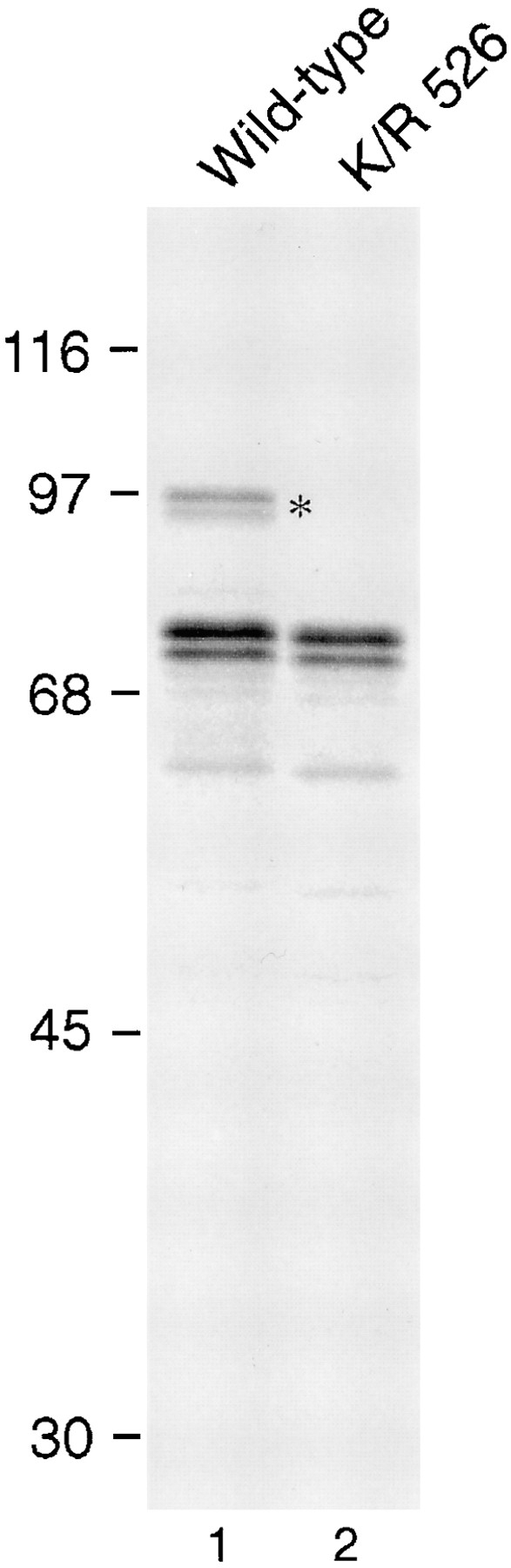
Lysine 526 is essential for SUMO-1 modification of RanGAP1 (A) Myc-tagged wild-type RanGAP1 (lane 1) and myc-tagged RanGAP1 containing a single lysine to arginine substitution at amino acid 526 (lane 2) were transcribed and translated in vitro in the presence of [35S]methionine and analyzed by SDS-PAGE. Molecular mass standards are indicated on the left and an asterisk denotes SUMO-1–modified wild-type RanGAP1.
We used this in vitro assay to analyze a series of NH2- and COOH-terminal deletions of RanGAP1 and found that the COOH-terminal domain of RanGAP1 is essential for SUMO-1 modification (data not shown; also see Fig. 3 B). To identify specific amino acid residues required for SUMO-1 modification, lysines in the COOH terminus of RanGAP1 were systematically mutated to arginine, based on the assumption that SUMO-1 conjugation would occur via a lysine, similar to ubiquitination (Wilkison, 1995; Hochstrasser, 1996). Substitution of arginine for lysine residues at positions 567, 555, 532, and 530 had no effect on RanGAP1 modification in vitro (data not shown). However, when lysine 526 was mutated to arginine, RanGAP1 was no longer modified by SUMO-1 (Fig. 1, lane 2). This finding identifies lysine 526 of RanGAP1 as essential for modification and implicates this residue as the acceptor for SUMO-1 conjugation.
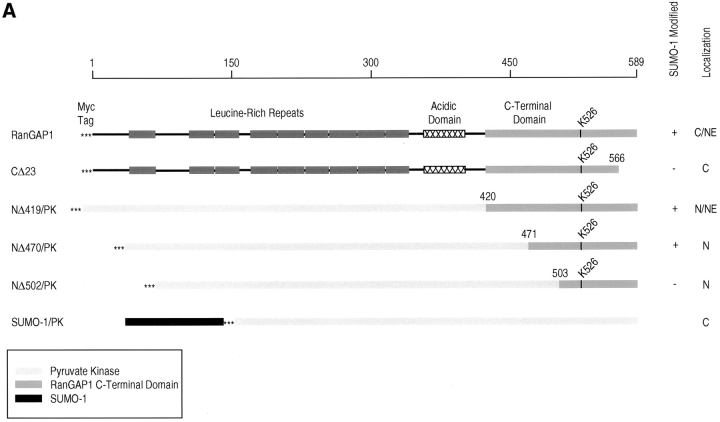
Figure 3.
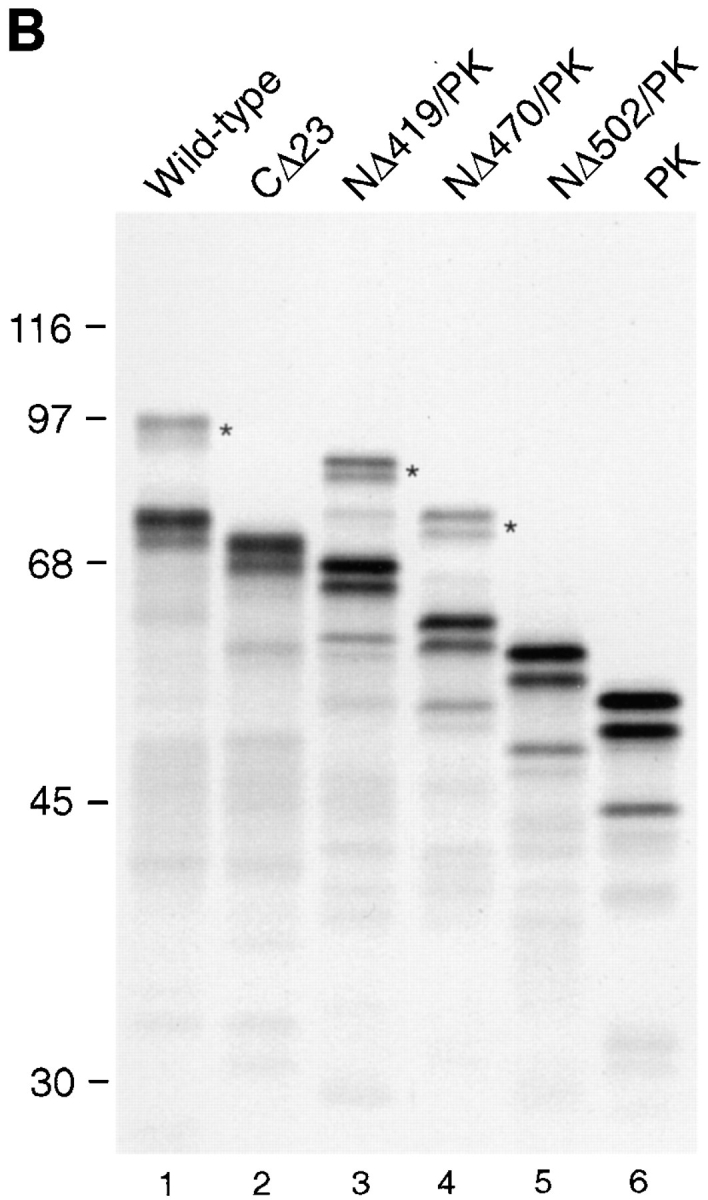
Determinants for SUMO-1 modification reside in the COOH-terminal domain of RanGAP1. (A) Schematic representations of RanGAP1 and pyruvate kinase fusion proteins. Asterisks indicate the presence of a myc epitope tag at the NH2 terminus of each protein. Leucine-rich repeats in RanGAP1 are indicated by dark-shaded boxes, the acidic domain by a hatched box, and the COOH-terminal domain by a light-shaded box. Pyruvate kinase is represented by very light-shaded boxes, and SUMO-1 by a black box. Results of in vitro SUMO-1 modification and immunolocalization of the transiently expressed proteins are indicated on the right. C, cytoplasm; NE, nuclear envelope; N, nucleus. (B) Wild-type RanGAP1 (lane 1), the COOH-terminal deletion mutant CD23 (lane 2), the pyruvate kinase fusion proteins NΔ419/PK (lane 3), NΔ470/PK (lane 4) and NΔ502/PK (lane 5), and pyruvate kinase (lane 6) were transcribed and translated in rabbit reticulocyte extracts in the presence of [35S]methionine. Reactions were separated by SDS-PAGE and analyzed by autoradiography. Molecular mass standards are indicated on the left and asterisks indicate SUMO-1–modified substrates.
SUMO-1 Modification Is Required for Localization of RanGAP1 at the NPC
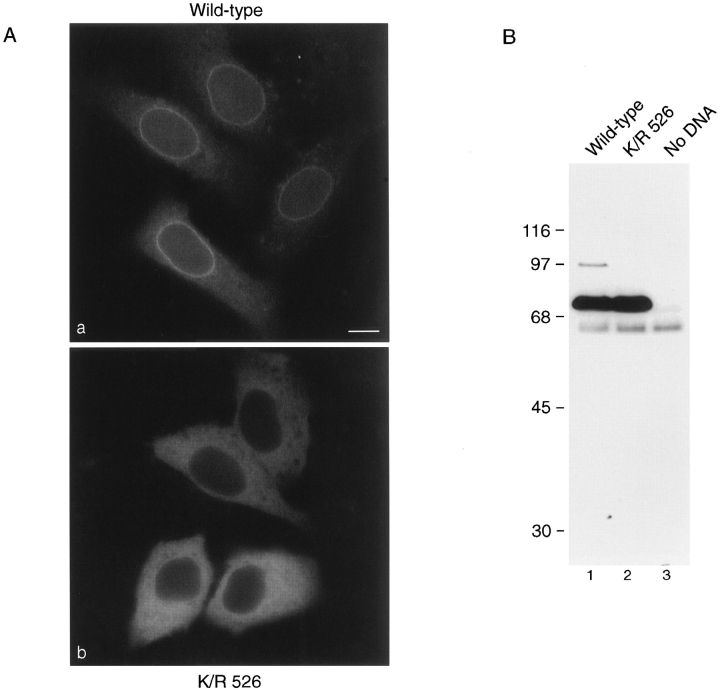
To analyze the effects of SUMO-1 modification on RanGAP1 localization, we next transfected cells with plasmids coding for either wild-type RanGAP1 or for mutant RanGAP1 with the lysine to arginine substitution at residue 526 (K/R 526). Both proteins were designed to contain a myc epitope tag at their NH2 terminus, which was used for immunolocalization and for immunoblot analysis. Transiently expressed wild-type protein showed a pattern of localization similar to the endogenous RanGAP1; it was detected both in the cytoplasm and concentrated at the nuclear envelope (Fig. 2 A, a). Discontinuous rim staining at the equatorial plane of the nuclear envelope, and punctate staining on the surface indicated association with NPCs. In cells expressing increasing amounts of RanGAP1, NPC labeling remained constant while the cytoplasmic signal increased, indicating that NPC binding sites were saturated. In contrast to wild-type RanGAP1, the K/R 526 mutant form of RanGAP1 was confined strictly to the cytoplasm and showed no evidence of association with the nuclear envelope or NPCs even at the lowest levels of expression (Fig. 2 A, b). No signal was detected in untransfected cells (data not shown).
Figure 2.
SUMO-1 modification is required for localization of RanGAP1 at the NPC. (A) HeLa cells were transfected with plasmids expressing myc-tagged wild-type RanGAP1 (a) or myc-tagged RanGAP1 containing a single lysine to arginine substitution at amino acid 526 (b). Subcellular localizations were determined 36 h after transfection by indirect immunofluorescence using the anti-myc monoclonal antibody, 9E10. (B) Cells were lysed in SDS sample buffer 36 h after transfection and analyzed by immunoblot analysis with the anti-myc monoclonal antibody, 9E10. Bar, 10 μm.
To determine whether the transiently expressed forms of RanGAP1 were modified in vivo as predicted, immunoblot analysis was performed on lysates prepared from transfected cells. Consistent with the in vitro translation assays, modified RanGAP1 was detected in cells transfected with wild-type RanGAP1 (Fig. 2 B, lane 1), but not in cells expressing RanGAP1 with the K/R 526 mutation (Fig. 2 B, lane 2). The relative ratio of modified to unmodified RanGAP1 was very low, indicating that SUMO-1 conjugation to RanGAP1 is closely regulated. These results confirm that lysine 526 is required for SUMO-1 modification of RanGAP1 in vivo, and they provide further evidence that SUMO-1 modification is required for RanGAP1 localization at the NPC.
The COOH-terminal Domain of RanGAP1 Specifies SUMO-1 Modification and NPC Localization
We next designed a series of constructs to assay for the minimal sequence in RanGAP1 required to specify SUMO-1 modification (Fig. 3 A). Either RanGAP1 with COOH-terminal deletions or pyruvate kinase fused to regions of the COOH-terminal domain of RanGAP1 were assayed for SUMO-1 modification using the in vitro assay described above. When RanGAP1 lacking the COOH-terminal 23 amino acids (CΔ23) was translated in rabbit reticulocyte lysate, modified RanGAP1 was not detected, indicating that the extreme COOH terminus of RanGAP1 is required for SUMO-1 modification (Fig. 3 B, lane 2). Pyruvate kinase fused with amino acids 420–589 of RanGAP1 (NΔ419/PK), as well as amino acids 471–589 (NΔ470/PK), were modified with efficiencies similar to wild-type RanGAP1 (Fig. 3 B, lanes 1, 3, and 4), indicating that the first 470 amino acids of RanGAP1 could be deleted without affecting SUMO-1 modification. When pyruvate kinase was fused to amino acids 503–589 of RanGAP1 (NΔ502/PK), however, no modification was detected, indicating that amino acid residues between 471 and 502 are also required for SUMO-1 modification (Fig. 3 B, lane 5). Together, these data indicate that SUMO-1 modification of RanGAP1 requires a domain extending from amino acid 470 to the COOH terminus.
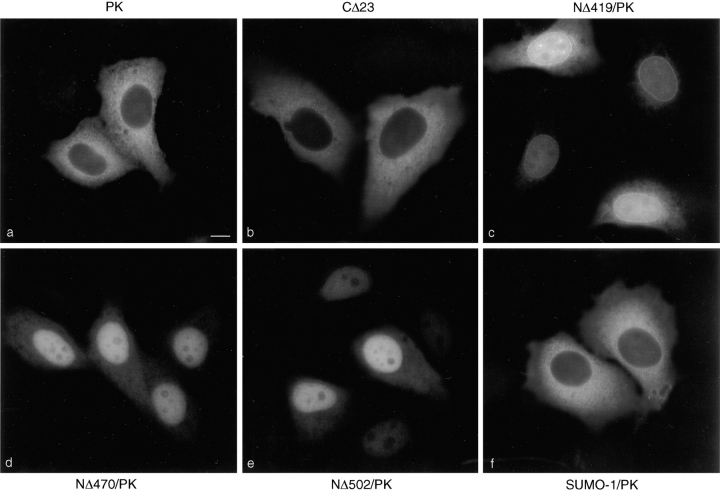
To investigate the determinants specifying NPC localization further, the constructs described above were transfected into HeLa cells, and the transiently expressed proteins were localized by indirect immunofluorescence. As evident in Fig. 4 a, pyruvate kinase was localized in the cytoplasm and showed no detectable nuclear envelope staining. CΔ23, which is not a substrate for SUMO-1 modification, was also strictly cytosolic (Fig. 4 b). In contrast, NΔ419/PK was concentrated at the nuclear envelope in a pattern consistent with NPC binding and also in the nucleus (Fig. 4 c). NΔ470/PK was also localized to the nucleus but showed no evidence of NPC binding, despite the fact that it is predicted to be modified by SUMO-1 (Fig. 4 d). NΔ502/PK showed a similar intranuclear localization and no NPC staining (Fig. 4 e). These results indicate that SUMO-1 modification is required, but not sufficient, for RanGAP1 localization at the NPC. This conclusion is further supported by the finding that pyruvate kinase, expressed as a fusion protein with SUMO-1, was strictly cytosolic (Fig. 4 f). The difference in localization between NΔ419 and NΔ470 indicate that the 50 amino acid residues between 419 and 470 are also required for localization of RanGAP1 at the NPC, in addition to SUMO-1 modification.
Figure 4.
Pyruvate kinase is targeted to the NPC by SUMO-1 modification and by additional determinants in the COOH-terminal domain of RanGAP1. Plasmids coding for the proteins illustrated in Fig. 3 A were transfected into HeLa cells, and the transiently expressed proteins we immunolocalized with the anti-myc monoclonal antibody, 9E10. Representative micrographs of the observed immunolocalizations are presented. Bar, 10 μm.
To determine whether the proteins transiently expressed in vivo were modified as expected, lysates from transfected cells were analyzed by immunoblot analysis using an antibody to the myc epitope tag. In cells expressing NΔ419/PK and NΔ470/PK, SUMO-1–modified forms of the proteins were detected, with approximately half of the proteins present in the modified form (Fig. 5, lanes 2 and 3). This ratio of modified to unmodified protein was noticeably higher than that observed with wild-type RanGAP1, where a relatively small amount of modified protein was detected (see Fig. 2, lane 1). Surprisingly, NΔ502/PK was also modified in vivo, contrary to the in vitro translation result, with the ratio of modified to unmodified forms again being ∼50% (Fig. 5, lane 4). SUMO-1 modification of the transiently expressed fusion proteins was confirmed by immunoblot analysis with an antibody specific for SUMO-1 (data not shown).
Figure 5.
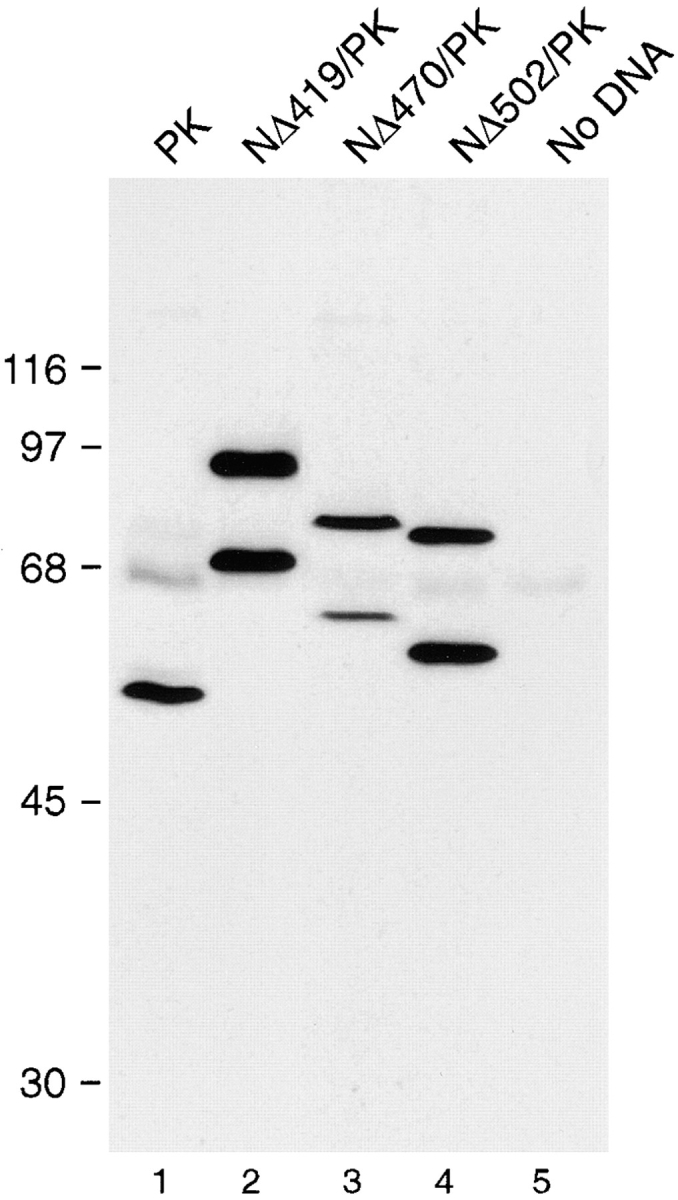
RanGAP1/PK fusion proteins are SUMO-1 modified in vivo. Cells transfected as in Fig. 4 were lysed in SDS sample buffer and analyzed by immunoblot analysis with the anti-myc monoclonal antibody, 9E10. In cells transfected with native pyruvate kinase, a single specific band migrating at 50 kD is detected (lane 1). The band migrating at 65 kD is nonspecific and detected in cells transfected with no DNA (lane 5). Two forms of the fusion proteins NΔ419/ PK (lane 2), NΔ470/PK (lane 3), and NΔ502/PK (lane 4) are detected, the lower molecular mass forms corresponding to unmodified protein, the higher molecular mass forms corresponding to SUMO-1–modified protein. Molecular mass standards are indicated on the left.
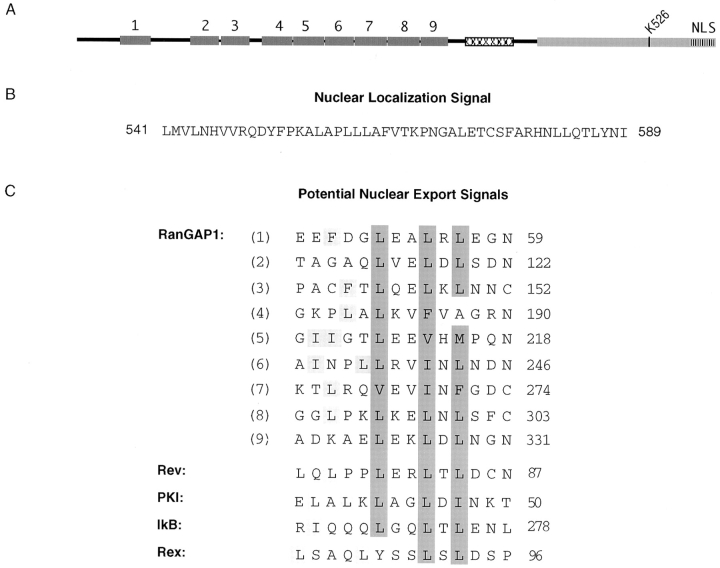
RanGAP1 Contains a Functional Nuclear Localization Signal and Nine Putative Nuclear Export Signals
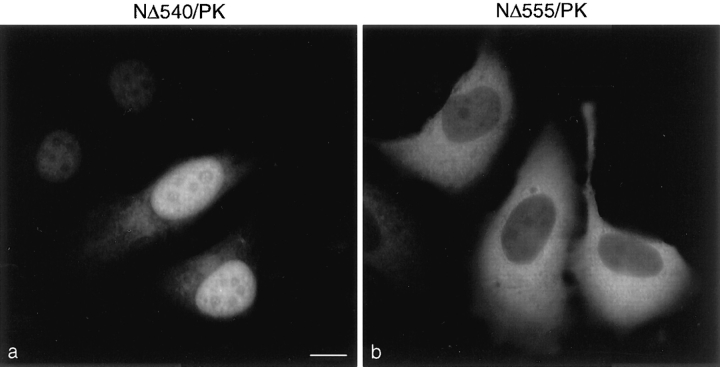
The localization of the pyruvate kinase fusion proteins (Fig. 4, c–e) indicated that the COOH terminus of RanGAP1 contains a nuclear localization signal (NLS). This was further mapped using additional pyruvate kinase fusion proteins. When fused to amino acids 541–589 of RanGAP1 (NΔ540/PK), pyruvate kinase was again targeted to the nucleus (Fig. 6 a). However, when fused with amino acids 556–589 of RanGAP1 (NΔ555/PK), pyruvate kinase was detected only in the cytosol (Fig. 6 b). These results indicated that amino acids 541–589 of RanGAP1 can function as an NLS. A database search with the NLS of RanGAP1 (shown in Fig. 7 B) revealed no significant homologies with other proteins and no homologies to other known NLSs. However, the nine amino-terminal leucine-rich repeats of RanGAP1 were each found to have striking homology with the leucine-rich nuclear export sequence motif (Fig. 7 C), first described for the HIV REV protein and the cAMP-dependent protein kinase inhibitor (Fischer et al., 1995; Wen et al., 1995). The presence of potential nuclear import and export signals raises the interesting possibility that RanGAP1 may shuttle between the nucleus and the cytoplasm.
Figure 6.
The COOH-terminal domain of RanGAP1 contains a nuclear localization signal. The pyruvate kinase fusion proteins NΔ540/PK (a) and NΔ555/PK (b) were transiently expressed in HeLa cells and localized with the anti-myc monoclonal antibody, 9E10.
Figure 7.
RanGAP1 contains nine potential nuclear export signals in addition to a nuclear localization signal. (A) Schematic representation of RanGAP1 indicating the positions of the nine NH2-terminal nuclear export motifs and the COOH-terminal nuclear localization signal. (B) Amino acid sequence of the nuclear localization signal in the COOH-terminal domain of RanGAP1. (C) Amino acid sequences and alignments of the nine putative nuclear export motifs in the NH2-terminal domain of RanGAP1 and the nuclear export motifs of Rev (Fischer et al., 1995), PKI (Wen et al., 1995), IκB, and Rex (Fritz and Green, 1996). Hydrophobic residues comprising the core of the motif are highlighted in dark gray, and hydrophobic residues outside of the core are highlighted in light gray.
SUMO-1–modified RanGAP1 Binds to a COOH-terminal Region of Nup358
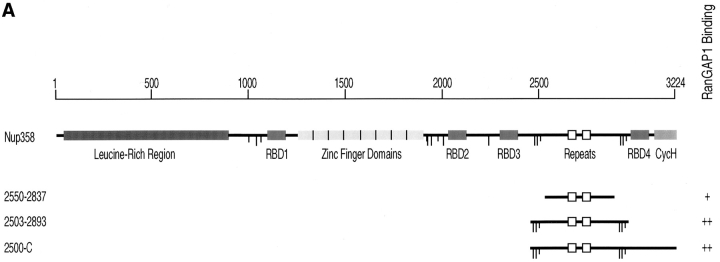
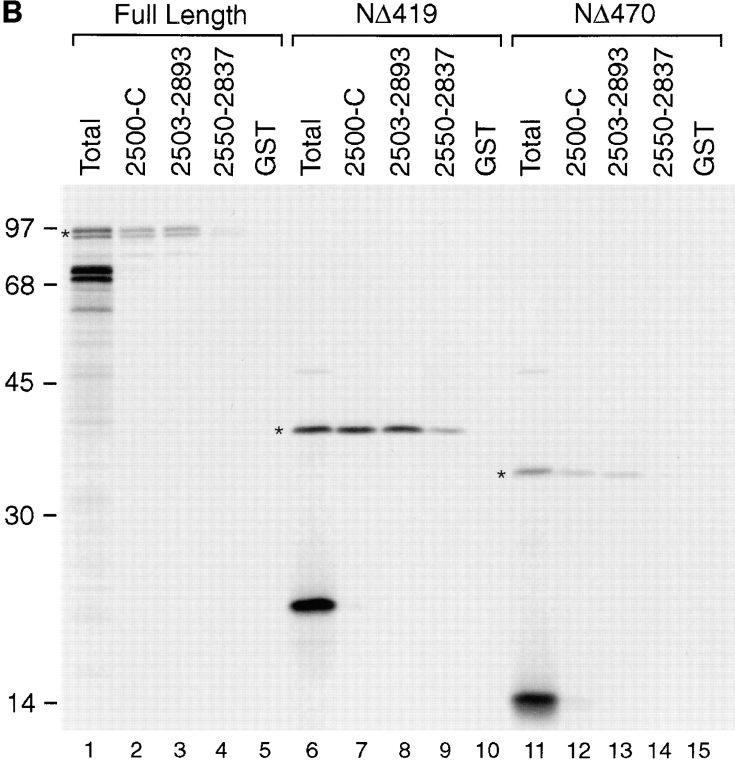
SUMO-1–modified RanGAP1 interacts with Nup358 (Mahajan et al., 1997; Saitoh et al., 1997), a nucleoporin that is a component of the cytoplasmic fibers of the NPC (Wu et al., 1995; Yokoyama et al., 1995). To characterize this interaction in more detail, we used an in vitro binding assay using bacterially expressed regions of Nup358 and radiolabeled RanGAP1 produced in rabbit reticulocyte extracts. Purified GST fusion proteins corresponding to various domains of Nup358 were bound to the wells of a microtiter plate and incubated with in vitro–translated RanGAP1. After incubation, the wells were washed, and bound proteins were eluted with SDS sample buffer. No binding of either modified or unmodified RanGAP1 was detected to regions of Nup358 corresponding to the NH2-terminal leucine-rich region, the Ran-binding domains, or the cyclophilin homologous domain (data not shown). RanGAP1 binding was, however, detected with a region of Nup358 extending from amino acid 2500 to the COOH terminus (Fig. 8 B, lane 2). Binding was specific for SUMO-1–modified RanGAP1, with no unmodified RanGAP1 being detected. The specificity of the binding was further demonstrated by the absence of interactions with GST (Fig. 8 B, lane 5). The region of Nup358 from amino acid 2500 to the COOH terminus contains two Ran-binding domains (domains three and four) separated by a 470–amino acid segment, and the cyclophilin homology domain. The 470– amino acid segment between the two Ran-binding domains contains direct repeats of ∼40 amino acids (Yokoyama et al., 1995). Because our results showed that the Ran-binding domains and cyclophilin homology domain did not interact with RanGAP1, we assayed more specifically for binding to the region separating Ran-binding domains three and four. This domain is flanked by clusters of FXFG repeats, and fragments with (amino acids 2503–2893) and without (amino acids 2550–2837) these repeats were tested in the binding assay. SUMO-1–modified RanGAP1 bound specifically to both of these fragments (Fig. 8 B, lanes 3 and 4). However, interaction with the fragment lacking FXFG repeats was of lower affinity relative to the fragment containing FXFG repeats, based on the four- to fivefold reduction in observed binding (see Fig. 9). Again, unmodified RanGAP1 did not bind to these fragments.
Figure 8.
SUMO-1–modified RanGAP1 binds to a COOH-terminal region of Nup358, between Ran-binding domains three and four. (A) Schematic representation of Nup358. RBD, Ran-binding domain; CycH, cyclophilin homologous domain. Long vertical lines indicate FXFG repeats, and short vertical lines indicate FG repeats. Fragments that were found to bind modified RanGAP1 are indicated, as well as a summary of their binding activity (+, weak binding; ++, strong binding). Internal direct repeats in the RanGAP1-binding domain are indicated by open boxes. (B) Full-length RanGAP1 (lane 1) and COOH-terminal regions of RanGAP1 from amino acids 420–589 (NΔ419; lane 6) and 471– 589 (NΔ470; lane 11) were translated in vitro in the presence of [35S]methionine. Translated proteins were assayed for binding to the domains of Nup358 indicated in A, as described in Materials and Methods. Molecular mass standards are indicated on the left, and asterisks denote SUMO-1–modified proteins.
Figure 9.
NΔ419 and NΔ470 have different affinities for the COOH-terminal region of Nup358 between Ran-binding domains three and four. Binding assays and quantification were performed as described in Material and Methods. Mean values are shown, with error bars indicating variance observed between triplicate experiments.
Based on the in vivo NPC targeting data, it was anticipated that the COOH terminus of RanGAP1, from amino acid 420 to 589 (NΔ419), would be sufficient for interaction with Nup358. We therefore translated this domain in vitro (without pyruvate kinase) and assayed for its ability to bind to the regions of Nup358 found to interact with full-length, modified RanGAP1. Both SUMO-1–modified and unmodified forms of NΔ419 were produced when translated in vitro (Fig. 8 B, lane 6), and as observed with full-length RanGAP1, the modified protein bound to the region of Nup358 between Ran-binding domains three and four, whereas the unmodified protein did not (Fig. 8 B, lanes 7–9). Similar to full-length RanGAP1, NΔ419 had a lower affinity for the domain lacking FXFG repeats, based on a fivefold reduction in binding compared with the domain containing these repeats (Fig. 8 B, lanes 8 and 9; Fig. 9). We next assayed for binding with the region of RanGAP1 between amino acids 471 and 589 (NΔ470), which is SUMO-1 modified but does not have NPC targeting activity in vivo. NΔ470 also showed the same binding specificity as full-length RanGAP1 and NΔ419, with only the SUMO-1–modified form binding to the region of Nup358 between Ran-binding domains three and four (Fig. 8 B, lanes 12–14). However, when their relative binding properties were compared, it was found that NΔ470 had a much lower affinity for Nup358, as evidenced by a six- to sevenfold reduction in the observed binding compared with NΔ419 (Fig. 9). These results are consistent with the in vivo localization data and indicate that the region of RanGAP1 between amino acids 420 and 470 stabilizes the interaction between RanGAP1 and the COOH terminus of Nup358. As previously reported, competition with free SUMO-1 had little if any effect on the binding of SUMO-1–modified RanGAP1 to Nup358 (data not shown; Mahajan et al., 1997).
Discussion
We have further characterized the ubiquitin-related protein, SUMO-1, and its effects on RanGAP1 localization. Our results, based on several independent lines of analysis, indicate that SUMO-1 modification is required for the association of RanGAP1 with the NPC. First, we have previously shown by cellular fractionation and immunoelectron microscopy that unmodified RanGAP1 is strictly cytoplasmic, whereas SUMO-1–modified RanGAP1 is primarily associated with the NPC. While this result was in itself suggestive, SUMO-1 modification as a cause for RanGAP1 association with the NPC, or an effect of RanGAP1 association with the NPC, could not be distinguished. Here, we have mapped the domain in RanGAP1 specifying SUMO-1 modification and show that a single amino acid substitution in this domain that prevents SUMO-1 modification also prevents NPC targeting. In addition, we have demonstrated that the binding of RanGAP1 to a COOH-terminal domain in Nup358 is positively regulated by SUMO-1 modification. Together, these data indicate that SUMO-1 modification functions to target RanGAP1 to the NPC through regulation of its interaction with Nup358.
Our results indicate that SUMO-1 modification is not the only element required for targeting RanGAP1 to the NPC. This conclusion is based on the observations that neither direct fusion of SUMO-1 to pyruvate kinase, nor fusion of the minimal domain of RanGAP1 that specifies SUMO-1 modification of pyruvate kinase, is sufficient to target RanGAP1 to the NPC. This result is not completely unexpected, as RanGAP1 is the only SUMO-1 modified substrate associated with the NPC, the remaining substrates being located primarily in the nucleus (Matunis et al., 1996; Mahajan et al., 1997). Differences in the subcellular localization of the constructs NΔ419/PK and NΔ470/PK indicate that the 50 amino acids between residues 420 and 470 of RanGAP1 are also required for NPC targeting. In vitro binding studies with these regions of RanGAP1 demonstrate that this 50–amino acid domain is required for efficient binding to Nup358. When expressed as a fusion protein with pyruvate kinase, this 50–amino acid domain (and several larger domains) was not sufficient to mediate Nup358 binding or NPC localization (data not shown). This result suggests that SUMO-1 is either required to form part of the Nup358 binding site or that the binding site overlaps with elements that inhibit binding in the absence of modification. Based on these observations, we propose that SUMO-1 functions to target RanGAP1 to the NPC by exposing, or possibly creating, a Nup358 binding site. In this capacity, SUMO-1 is different from Ub and UCRP, which appear to mediate the interactions between their substrates and their intracellular targets directly (Deveraux et al., 1994; Loeb et al., 1994). Rather, SUMO-1 modification of RanGAP1 appears to be more analogous to a phosphorylation event that exposes a masked signal sequence.
The most significant mutation affecting RanGAP1 modification was a lysine to arginine substitution at residue 526. The effect of this single conservative substitution on RanGAP1 strongly implicates lysine 526 as the acceptor for SUMO-1 ligation, although protein sequence analysis of modified RanGAP1 will be required for definitive proof. In addition to lysine 526, deletion analysis indicated that a region from amino acid 470 to the COOH terminus is required for SUMO-1 modification in vitro. (In vivo results suggest that this region may actually be smaller.) This region is likely to contain elements required for recognition of RanGAP1 by the SUMO-1–conjugating enzyme (E2) and possibly SUMO-1 ligase (E3), if one is required. Alignment of the region between amino acids 470 and the COOH terminus of RanGAP1 with sequences in the protein data bases revealed a surprising homology with the Ran-binding motifs of the yeast nucleoporins, Nup2 and Nup36 (data not shown; Loeb et al., 1993; Noguchi et al., 1997). Whether this region in RanGAP1 binds Ran, and whether yeast Nup2 and Nup36 are substrates for SUMO-1 modification, remain to be determined. The yeast homologue of RanGAP1, Rna1p (Traglia et al., 1989; Bischoff et al., 1995a ), lacks the entire COOH-terminal domain found in vertebrate RanGAP1 and is therefore not likely to be a substrate for SUMO-1 modification.
While other substrates for SUMO-1 have not yet been confirmed, a number have been implicated in both yeast and vertebrates based on genetic analysis and two-hybrid screens. The yeast homologue of SUMO-1, Smt3p, was first identified as a suppressor of a temperature-sensitive allele of MIF2, which codes for a centromere binding protein (Meluh and Koshland, 1995). In addition, SUMO-1 was found to interact with the death domain of the Fas/ APO-1 receptor, which is involved in apoptosis (Okura et al., 1996), and with Rad51 and Rad52, proteins involved in DNA recombination and repair (Shen et al., 1996a ). Finally, SUMO-1 was also identified in a two-hybrid screen with PML, a protein normally localized to discrete nuclear foci, called PML nuclear bodies (Boddy et al., 1996). PML fusion with the retinoic acid receptor, as a result of chromosomal translocation, leads to a redistribution of PML throughout the nucleus and acute promyelocytic leukemia. The functional relevance of the interactions involving each of these proteins, and whether they are substrates for SUMO-1 modification, remains to be determined.
A significant body of evidence now indicates that the E2 enzyme for SUMO-1 conjugation is Ubc9p (Seufert et al., 1995), including biochemical and genetic evidence that Ubc9p is essential for Smt3p modification in yeast (Johnson and Blobel, 1997). In addition, vertebrate Ubc9p has been found to interact with SUMO-1 by two-hybrid analysis (Shen et al., 1996b ) and to be present in a complex with Nup358 and modified RanGAP1 in Xenopus egg extracts (Saitoh et al., 1997). The association of Ubc9p with Nup358 raises the interesting question of where RanGAP1 is modified in vivo. Immunoblot analysis of whole cell extracts indicates that approximately half of the endogenous RanGAP1 is modified by SUMO-1 and that most, if not all, of this is associated with the NPC (Matunis et al., 1996; Mahajan et al., 1997). In this study, we found that overexpression of RanGAP1 leads to a relatively small fraction of RanGAP1 being modified and localized to the NPC, with the majority of the protein accumulating unmodified and in the cytoplasm. These observations suggest that SUMO-1 modification is sensitive to the association of RanGAP1 with the NPC and can be explained by a number of models. First, modified RanGAP1 in the cytoplasm may negatively regulate SUMO-1–conjugating enzymes once sites at the NPC are saturated. Alternatively, unmodified RanGAP1 may associate with the NPC transiently, being modified after this association by Nup358-bound Ubc9p. Modification, as we have shown, would allow RanGAP1 to form a stable complex with Nup358. A third possibility is that RanGAP1 could be modified by SUMO-1 in the cytoplasm, with the modification being rapidly removed by a SUMO-1 demodifying activity. According to this model, interaction with Nup358 would stabilize modified RanGAP1 by preventing removal of SUMO-1. Better characterization and subcellular localization of the SUMO-1 conjugating and demodifying enzymes will be required to test these models. Interestingly, we also observed that SUMO-1 modification of several nuclear pyruvate kinase fusion proteins (NΔ419, NΔ470, and NΔ502) appeared to be less strictly regulated compared with modification of cytoplasmic RanGAP1. This finding suggests that there may be differences in the regulation of SUMO-1 modification in different subcellular compartments. The modification of NΔ502 in vivo, but not in vitro, could also be indicative of SUMO-1 conjugating activities with different specificities.
Our data indicate that the COOH-terminal domain of RanGAP1 also contains a novel nuclear localization signal. This NLS was mapped to amino acids 541–589 by virtue of its ability to target pyruvate kinase to the nucleus, although a minimal domain remains to be defined. Similar fusions with pyruvate kinase have been used to identify the NLSs of a number of proteins, including the SV-40 large T antigen (Kalderon et al., 1984), p53 (Dang and Lee, 1989), and hnRNP A1 (Siomi and Dreyfuss, 1995). At this time, we have no evidence that the NLS in RanGAP1 actually functions to target RanGAP1 to the nucleus, and immunofluorescence data actually indicates that there is little if any intranuclear RanGAP1 (Matunis et al., 1996; Mahajan et al., 1997). However, the presence of nine potential nuclear export signals in its NH2 terminus raises the interesting possibility that RanGAP1 may shuttle between the nucleus and the cytoplasm. Consistent with this possibility, we previously found ∼10% of the NPC-associated RanGAP1 to be located on the nucleoplasmic side of the pore complex (Matunis et al., 1996). RanGAP1 could conceivably shuttle to regulate RanGTP hydrolysis on the nucleoplasmic side of the NPC, where RanGTP has been proposed to bind to karyopherin β1 (importin β) to release import substrates into the nucleoplasm (Görlich et al., 1996). It appears almost certain that RanGTP hydrolysis will occur at the cytoplasmic filaments of the NPC, where we have localized SUMO-1–modified RanGAP1. We have mapped the binding site for RanGAP1 to a region on Nup358 that is flanked by Ran-binding domains. The Ran-binding domains in Nup358 are homologous to the Ran-binding domain of RanBP1 (Coutavas et al., 1993), and like RanBP1, these domains costimulate GTP hydrolysis by Ran in the presence of RanGAP1 (Beddow et al., 1995). RanGTP hydrolysis at the cytoplasmic filaments likely serves the dual purpose of preventing free RanGTP from entering the cytoplasm, where it could prematurely dissociate karyopherin/substrate complexes (Rexach and Blobel, 1995), and providing RanGDP, which has been proposed to be the active form of Ran required for nuclear import (Nehrbass and Blobel, 1995; Görlich et al., 1996; Weis et al., 1996). RanGTP hydrolysis could also be involved in recycling RanGTP-bound karyopherin β1 (Floer et al., 1997), and the nuclear export receptor, CRM1 (Fornerod et al., 1997; Stade et al., 1997). We found that the FXFG and FG repeats affected RanGAP1 binding, suggesting that RanGAP1 may have some affinity for these repeats, or that there may be interactions between RanGAP1 and other factors bound to these repeats (Moroianu et al., 1995; Radu et al., 1995). (Reticulocyte extracts contain, among other factors, karyopherin α and β1 [Adam and Adam, 1994].) It remains to be tested whether the longer, direct repeats in the RanGAP1 binding site on Nup358 are involved in the interaction with RanGAP1. It is conceivable that each of these repeats serves as a binding site for a single molecule of RanGAP1, making it possible for two molecules of RanGAP1 to bind to a single molecule of Nup358. The binding assays that we have performed to date do not allow us to address this issue.
In conclusion, our findings indicate that SUMO-1 modification exposes, or creates, a binding site on RanGAP1 that enables it to interact with Nup358. A similar function has not been described previously for Ub or any other Ub-like protein and demonstrates the versatility of this type of posttranslational protein modification. Other effects of SUMO-1 modification on RanGAP1 have not been identified but could possibly include regulation of its GTPase-activating activity or its interactions with additional proteins. It is apparent that SUMO-1 modification will have functions other than that described here for RanGAP1, given that the majority of SUMO-1–modified substrates are not localized at the NPC. Understanding these additional functions awaits identification of other SUMO-1 substrates.
Acknowledgments
We thank members of the Blobel lab, especially Elias Coutavas, Beatriz Fontoura, and Erica Johnson, for helpful discussions and critical reading of the manuscript. We are also grateful to Dr. Haruhiko Siomi and Dr. Matthew Michael for generously providing us with Myc-tagged expression vectors, and Dr. Hui Ge for providing us with a GST expression vector.
This work was supported by the Howard Hughes Medical Institute. M.J. Matunis is an American Cancer Society-Amgen Fellow (PF-4195), J. Wu is supported by a National Institutes of Health Fellowship (1F32GM16758-01).
Abbreviations used in this paper
- GST
glutathione-S-transferase
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- SUMO-1
small ubiquitin-like modifier 1
- Ub
ubiquitin
- UCRP
ubiquitin cross-reactive protein
Footnotes
Address all correspondence to Michael Matunis, Laboratory of Cell Biology, Howard Hughes Medical Institute, The Rockefeller University, New York, NY 10021. Tel.: (212) 327-8101. Fax: (212) 327-7880. E-mail: matunim@rockvax.rockefeller.edu
References
- Adam EJH, Adam SA. Identification of cytosolic factors required for nuclear location sequence–mediated binding to the nuclear envelope. J Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddow AL, Richards SA, Orem NR, Macara IG. The Ran/TC4 GTPase-binding domain: identification by expression cloning and characterization of a conserved sequence motif. Proc Natl Acad Sci USA. 1995;92:3328–3332. doi: 10.1073/pnas.92.8.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc Natl Acad Sci USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Kempf T, Hermes I, Ponstingl H. Human RanGTPase-activating protein RanGAP1 is a homologue of yeast Rna1p involved in mRNA processing and transport. Proc Natl Acad Sci USA. 1995a;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Smirnova E, Dong W, Ponstingl H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO (Eur Mol Biol Organ) J. 1995b;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, Howe K, Etkin LD, Solomon E, Freemont PS. PIC1, a novel ubiquitin-like protein that interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of IκBα by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. Nucleocytoplasmic transport of macromolecules. Microb Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutavas E, Ren M, Oppenheim JD, D'Eustachio P, Rush MG. Characterization of proteins that interact with the cell-cycle regulatory protein Ran/TC4. Nature. 1993;366:585–587. doi: 10.1038/366585a0. [DOI] [PubMed] [Google Scholar]
- Dang CV, Lee WM. Identification of the human c-myc protein nuclear translocation signal. Mol Cell Biol. 1988;8:4048–4054. doi: 10.1128/mcb.8.10.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7071. [PubMed] [Google Scholar]
- Dreyfuss G, Adam SA, Choi YD. Physical change in cytoplasmic messenger ribonucleoproteins in cells treated with inhibitors of mRNA transcription. Mol Cell Biol. 1984;4:415–423. doi: 10.1128/mcb.4.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular TNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Floer M, Blobel G, Rexach M. Disassembly of RanGTP-karyopherin β complex, an intermediate in nuclear protein import. J Biol Chem. 1997;272:19538–19546. doi: 10.1074/jbc.272.31.19538. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fritz CC, Green MR. HIV Rev uses a conserved cellular protein export pathway for the nucleocytoplasmic transport of viral RNAs. Curr Biol. 1996;6:848–854. doi: 10.1016/s0960-9822(02)00608-5. [DOI] [PubMed] [Google Scholar]
- Goldknopf IL, Busch H. Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate protein A24. Proc Natl Acad Sci USA. 1977;74:864–868. doi: 10.1073/pnas.74.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Panté N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear import. EMBO (Eur Mol Biol Organ) J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Haas AL, Ahrens P, Bright PM, Ankel H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J Biol Chem. 1987;262:11315–11323. [PubMed] [Google Scholar]
- Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Ubc9 is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO (Eur Mol Biol Organ) J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kamitani T, Nguyen HP, Yeh ET. Preferential modification of nuclear proteins by a novel ubiquitin-like molecule. J Biol Chem. 1997;272:14001–14004. doi: 10.1074/jbc.272.22.14001. [DOI] [PubMed] [Google Scholar]
- Loeb JDJ, Davis LI, Fink GR. Nup2, a novel yeast nucleoporin, has functional overlap with other proteins of the nuclear pore complex. Mol Biol Cell. 1993;4:209–222. doi: 10.1091/mbc.4.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb KR, Haas AL. Conjugates of ubiquitin cross-reactive protein distribute in a cytoskeletal pattern. Mol Cell Biol. 1994;14:8408–8419. doi: 10.1128/mcb.14.12.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Mannen H, Tseng H-M, Cho C, Li SS-L. Cloning and expression of human homolog HSMT3 to yeast SMT3 suppressor of MIF2 mutations in a centromere protein gene. Biochem Biophys Res Commun. 1996;222:178–180. doi: 10.1006/bbrc.1996.0717. [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase–activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiaeencodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin α1β and α2β heterodimers: α1 or α2 subunit binds nuclear localization signal and β subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan J, Potter JL, Haas AL. Conjugation of the 15-kD interferon-induced ubiquitin homolog is distinct from that of ubiquitin. J Biol Chem. 1996;271:324–330. doi: 10.1074/jbc.271.1.324. [DOI] [PubMed] [Google Scholar]
- Nehrbass U, Blobel G. Role of the nuclear transport factor p10 in nuclear import. Science. 1996;272:120–122. doi: 10.1126/science.272.5258.120. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Science. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Noguchi E, Hayashi N, Nakashima N, Nishimoto T. Yrb2p, a Nup2p-related yeast protein, has a functional overlap with Rna1p, a yeast Ran-GTPase-activating protein. Mol Cell Biol. 1997;17:2235–2246. doi: 10.1128/mcb.17.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okura T, Gong L, Kamitani T, Wada T, Okura I, Wei CF, Chang HM, Yeh ET. Protection against Fas/APO-1- and tumor necrosis factor-mediated cell death by a novel protein, sentrin. J Immunol. 1996;157:4277–4281. [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Ren M, Villamarin A, Shih A, Cautavas E, Moore MS, LoCurcio M, Clarke V, Oppenheim JD, D'Eustachio P, Rush MG. Separate domains of the Ran GTPase interact with different factors to regulate nuclear protein import and RNA processing. Mol Cell Biol. 1995;15:2117–2124. doi: 10.1128/mcb.15.4.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Rush MG, Drivas G, D'Eustachio P. The small GTPase Ran: how much does it run? . BioEssays. 1996;18:103–112. doi: 10.1002/bies.950180206. [DOI] [PubMed] [Google Scholar]
- Saitoh H, Pu R, Cavenagh M, Dasso M. RanBP2 associates with Ubc9 and a modified form of RanGAP1. Proc Natl Acad Sci USA. 1997;94:3736–3741. doi: 10.1073/pnas.94.8.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- Shen Z, Pardington-Purtymun PE, Comeaux JC, Moyzis RK, Chen DJ. UBL1, a human ubiquitin-like protein associating with human RAD51/RAD52 proteins. Genomics. 1996a;36:271–279. doi: 10.1006/geno.1996.0462. [DOI] [PubMed] [Google Scholar]
- Shen Z, Pardington-Purtymun PE, Comeaux JC, Moyzis RK, Chen DJ. Associations of UBE21 with RAD52, UBL1, p53, and RAD51 proteins in a yeast two-hybrid system. Genomics. 1996b;37:183–186. doi: 10.1006/geno.1996.0540. [DOI] [PubMed] [Google Scholar]
- Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weiss K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Strous GJ, Kerkhof PV, Govers R, Ciechanover A, Schwartz AL. The ubiquitin conjugation system is required for ligand-induced endocytosis and degradation of the growth hormone receptor. EMBO (Eur Mol Biol Organ) J. 1996;15:3806–3812. [PMC free article] [PubMed] [Google Scholar]
- Traglia HM, Atkinson NS, Hopper AK. Structural and functional analyses of Saccharomyces cerevisiae wild-type and mutant RNA1genes. Mol Cell Biol. 1989;9:2989–2999. doi: 10.1128/mcb.9.7.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K, Dingwall C, Lamond AI. Characterization of the nuclear protein import mechanism using Ran mutants with altered nucleotide binding specificities. EMBO (Eur Mol Biol Organ) J. 1996;15:7120–7128. [PMC free article] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- Wilkinson KD. Roles of ubiquitinylation in proteolysis and cellular regulation. Annu Rev Nutri. 1995;15:161–189. doi: 10.1146/annurev.nu.15.070195.001113. [DOI] [PubMed] [Google Scholar]
- Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, ran-gtp binding sites, zinc fingers, a cyclophilin a homologous domain, and a leucine-rich region. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, et al. A giant nucleopore protein that binds ran/tc4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]