Abstract
Metals play a variety of roles in biological processes, and hence their presence in a protein structure can yield vital functional information. Because the residues that coordinate a metal often undergo conformational changes upon binding, detection of binding sites based on simple geometric criteria in proteins without bound metal is difficult. However, aspects of the physicochemical environment around a metal binding site are often conserved even when this structural rearrangement occurs. We have developed a Bayesian classifier using known zinc binding sites as positive training examples and nonmetal binding regions that nonetheless contain residues frequently observed in zinc sites as negative training examples. In order to allow variation in the exact positions of atoms, we average a variety of biochemical and biophysical properties in six concentric spherical shells around the site of interest. At a specificity of 99.8%, this method achieves 75.5% sensitivity in unbound proteins at a positive predictive value of 73.6%. We also test its accuracy on predicted protein structures obtained by homology modeling using templates with 30%–50% sequence identity to the target sequences. At a specificity of 99.8%, we correctly identify at least one zinc binding site in 65.5% of modeled proteins. Thus, in many cases, our model is accurate enough to identify metal binding sites in proteins of unknown structure for which no high sequence identity homologs of known structure exist. Both the source code and a Web interface are available to the public at http://feature.stanford.edu/metals.
Keywords: zinc, functional annotation, metal binding sites, function prediction, structural bioinformatics
A large percentage of proteins require metals to stabilize their structures or to carry out their functions. Zinc, one of the most common of these metals, is associated with proteins of a wide variety of biological roles. A recent study estimates that 40% of zinc binding proteins in the human proteome are transcription factors; the remaining 60% are primarily enzymes and proteins involved in ion transport (Andreini et al. 2006). Zinc has a number of chemical properties that give rise to its diverse biological function (Vallee and Auld 1990). Because its d-shell is filled, it does not undergo oxidation or reduction reactions; this offers a level of stability in biological environments whose redox potentials may fluctuate. Its ability to take on different coordination numbers and several types of ligating residues may allow proteins to tune their binding sites' affinities and functions.
The roles of structural zinc sites vary. Zinc fingers, which comprise the largest class of transcription factors in the human genome, are structurally stable only in the presence of zinc (Tupler et al. 2001). In cases such as a repressor protein involved in the bacterial response to heavy metal toxicity, zinc binding acts as a conformational switch by inducing a structural rearrangement that affects the protein's activity (Gaither and Eide 2001; Hantke 2001; Eicken et al. 2003). Zinc can also participate in the stabilization of quaternary structure, as is the case for a voltage-gated potassium channel in which two different monomers contribute residues to a binding site (Bixby et al. 1999).
Zinc's wide range of functional and structural roles makes the ability to detect its binding sites important in functional annotation efforts. One of the most ubiquitous zinc binding motifs is the C2H2 zinc finger motif, first identified in transcription factor IIIA (Ginsberg et al. 1984; Brown et al. 1985). Modern sequence motif databases such as Prosite contain patterns for a variety of zinc binding domains that can be used to infer the existence of a binding site in a protein sequence (Hulo et al. 2006). The Pfam database of hidden Markov models, which are built from multiple sequence alignments of protein families, may also recognize a zinc binding domain if its sequence is similar enough to members of a known family (Bateman et al. 2004).
More complex machine learning methods developed in recent years attempt to predict zinc and other metal binding sites directly from sequence. Instead of relying only on sequence comparison, Lin et al. (2005) map each amino acid to biochemical and biophysical features and then use a neural network to identify likely coordinating residues. Rather than predicting exact binding sites, a method using support vector machines operates at a global level to identify zinc binding proteins, and shows that hydrophobicity, predicted solvent accessibility, and the polar or nonpolar nature of the protein's residues are particularly useful for predicting metal binding activity from sequence (Lin et al. 2006). Passerini et al. (2006, 2007) take advantage of the observation that the probability that a residue ligates a metal increases if it is close in sequence to another ligating residue, though they note that prediction accuracy is significantly higher for cysteine residues than for the other amino acids that typically coordinate zinc. Though many of these methods are reasonably successful at locating zinc binding sites in proteins, it should be possible to achieve even higher degrees of accuracy when structural information is available since local structural features are often conserved even when the amino acid sequence diverges.
A number of structural genomics projects are pursuing the goal of solving protein structures whose folds are likely to be unique. As a result, these efforts have increased the number of protein structures available whose functions are unknown (Todd et al. 2005; Chandonia and Brenner 2006); methods that use this three-dimensional information therefore have an important role to play in the post-genomic era. Because even methods based on high levels of sequence homology or on strong matches to annotated sequence motifs can produce errors (Bork and Bairoch 1996; Brenner 1999; Palmer et al. 1999; Gerlt and Babbitt 2000), structure- and sequence-based methods complement one another in functional annotation efforts. In addition, an atomic level understanding of metal binding sites will aid efforts in protein engineering and structure prediction (Banci et al. 1999; Arnesano et al. 2002; Bertini et al. 2002).
Early structure-based methods for predicting sites likely to be of functional importance mapped evolutionary conservation at the sequence level onto a protein's three-dimensional structure. Though patches of conserved residues tend to be involved in catalytic activity or in ligand binding, this type of approach does not directly assign specific functions to these sites (Karlin and Zhu 1996; Zhu and Karlin 1996; Aloy et al. 2001; Lichtarge and Sowa 2002; Landau et al. 2005). Fold-X uses empirical force field calculations to analyze local regions of protein structures and can discriminate between different metal binding sites, though typically in the context of detailed structural refinement of predictions made through other means (Schymkowitz et al. 2005).
In order to assign function to specific sites, some methods define structural templates based on known active site or ligand binding site geometry and use them to search new protein structures for sites similar to the templates. JESS takes templates that consist of atoms with conserved pairwise distances, such as might be observed in a metal binding site with well-defined coordination geometry, and finds sites that match within some tolerance (Barker and Thornton 2003). Deng et al. (2006) use a graph theoretical approach to search for oxygen atoms whose geometric arrangement can accommodate calcium binding. Both Fetrow and Skolnick's fuzzy functional forms and Russell's templates encode not only the geometry of an active site or binding site but also residue identities (Fetrow and Skolnick 1998; Russell 1998).
Since producing templates of known function typically requires manual effort, several groups have developed methods for automatically breaking up proteins into potentially functional templates and using them to search a query protein for matches. If one of these “reverse templates” comes from a protein of known function, one may be able to make a functional inference about proteins it matches (Jambon et al. 2003; Laskowski et al. 2005a,b). Alternatively, Arakaki et al. (2004) suggest an automated method for developing templates from residues annotated as members of functional sites in the Swiss-Prot database of protein sequences. An intriguing approach by Dudev and Lim (2007) translates a protein sequence into an alphabet based on local backbone dihedral angles and searches for one-dimensional magnesium binding motifs in this new space.
Metal binding sites are defined not only by their coordination geometry but also by the second shell atoms that contribute to their stabilities and affinities (Marino and Regan 1999). To account for the observation that the hydrophilic coordination sphere of a metal ion typically occurs within a shell of hydrophobic atoms (Yamashita et al. 1990), Gregory et al. (1993) discard template matches in regions of low hydrophobic contrast. However, this approach still does not account for the possibility that the geometry of the metal binding site could differ significantly in the presence and absence of metals. A recent survey concluded that metal binding site geometry differs between the holo (bound) and apo (unbound) states in more than 40% of cases (Babor et al. 2005). Residue identities may also be misleading in cases where backbone atoms coordinate the metal since these residues are not as strongly conserved as are those that contribute side-chain atoms to the binding site (Kasampalidis et al. 2007). An ideal method for metal binding site detection must therefore account for both conformational changes and residue substitutions in the binding site, and in order to benefit areas of research such as protein structure prediction and protein engineering, it must provide a description of conserved biochemical and biophysical properties both in and around the binding site itself.
MetSite uses PSI-BLAST position-specific scoring matrix scores (Altschul et al. 1997), secondary structure, solvent accessibility, and pairwise Cβ distances of residues near metal binding sites to train a neural network (Sodhi et al. 2004). The use of these structural features takes three-dimensional conformation into account but allows for variation in exact side-chain placements. This method achieves a sensitivity of 47.8% among zinc binding sites at a false-positive rate of 5% on a data set in which no protein is structurally similar to one used to train the neural net. However, the use of a neural network does not necessarily allow for biologically relevant interpretation of the classifier.
We have previously reported a rapid, general purpose method for finding functional sites in proteins and RNA (Bagley and Altman 1995, 1996; Wei and Altman 1998, 2003; Banatao et al. 2003). FEATURE examines a variety of physicochemical properties in concentric radial shells, which typically cover a radius of 7.5 Å around the site center, and then identifies features that are over- or underrepresented with respect to negative training examples. The 7.5 Å spherical environment combined with Bayesian learning allows FEATURE to capture important residues or features that may be somewhat distant from the site of interest without including spurious information. Averaging features in the radial shells conserves some geometric information without requiring proteins to recapitulate the exact arrangements of atoms observed in training sites. The resulting model is orientation independent, which facilitates rapid scanning of large databases. The use of a local environment allows for recognition of functional similarities even in the absence of global sequence or structural homology. Here, we extend the method to incorporate prior information on the nature of zinc binding sites and show strong performance in apo proteins even when conformational changes occur upon metal binding. We furthermore demonstrate that FEATURE is capable of recognizing metal binding sites in predicted structures produced through homology modeling, which makes it an attractive tool for functional annotation of proteins of both known and unknown structure.
Results
The zinc model produced by FEATURE captures not only the coordination geometry of binding sites but also the biochemical and biophysical properties of the surrounding region. The algorithm determines whether each property examined by FEATURE is abundant or deficient in zinc binding sites with respect to negative training examples. This produces a “fingerprint” of zinc binding sites (Fig. 1). As aspartates, histidines, and cysteines commonly coordinate zincs, they are abundant in the second shell (1.25–2.5 Å from the zinc). These distances are consistent with observed coordination geometries in zinc binding proteins (Harding 2001). As expected, the second shell contains more negative charge and less positive charge than the negative training sites due to the presence of the coordinating atoms. The principle of hydrophobic contrast predicts that the polar atoms that surround a metal site will themselves be surrounded by hydrophobic atoms (Yamashita et al. 1990), and this is indeed observed as an abundance of aliphatic and aromatic carbons in the third FEATURE shell (2.5–3.75 Å). The solvent accessibility in the third and fourth shells (2.5–5 Å) is low, most likely due to the presence of structural zincs in the data set, which tend to be buried. In addition, although many catalytic zincs are solvent exposed, the side chains of their coordinating residues may nonetheless be buried (Alberts et al. 1998). The increase in solvent accessibility in the last two shells (5–7.5 Å) may indicate that while many zinc sites are buried, they are closer to the surface of the protein than to the hydrophobic core.
Figure 1.
Abundant and deficient physicochemical properties around zinc binding sites. The distributions of positive and negative training set scores were compared using the Mann-Whitney rank sum test. Red and green squares indicate properties that are significantly more abundant or scarce in the positive training examples than in the negative examples, respectively (P ≤ 0.01).
Performance on holo proteins
The 349 zinc binding proteins in our nonredundant training set yielded 131 unique combinations of coordinating atoms. Of these, 35% contain at least one cysteine, 24% contain at least two cysteines, and 31% have at least one water molecule. Embedding each of the 100 zinc binding proteins in our holo test set in a 1 Å cubic grid produced an average of 137,251 grid points to be scored per protein. This grid did not include points that have less than one nonsolvent atom within the 7.5 Å environment examined by FEATURE. Filtering out grid points that do not have enough residues nearby to create a plausible first coordination shell reduced the number of points to be scanned by 93%. This filtering, which requires only a partial match to a known coordination environment, is described in more detail in the Methods section. Though we remove any zinc ions before scanning a protein for binding sites, the regular geometry of the coordinating residues makes this classification task much easier than in apo proteins.
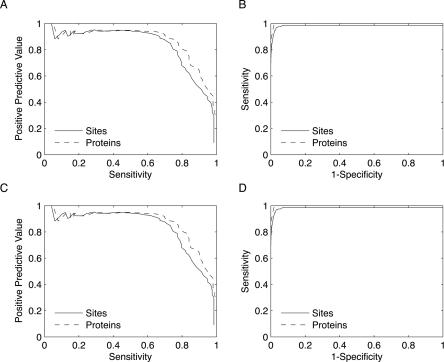
The receiver-operating characteristic (ROC) and precision-recall curves in Figure 2, A and B, demonstrate that FEATURE achieves high sensitivity, specificity, and positive predictive value in holo proteins. At a specificity of 99.8%, the model has a positive predictive value of 75.2% and recognizes 77.8% of binding sites. Because some proteins have multiple binding sites that are related to one another by symmetry, we also report the sensitivity with respect to proteins. At the 99.8% specificity cutoff, we identify at least one binding site in 84.0% of proteins. We consider every high scoring hit within 5 Å of one of a zinc's coordinating residues to be a true positive and every other high scoring hit to be a false positive. As binding sites for many metals share similar characteristics, at this stage we expect that the model may in some cases be unable to differentiate between zinc and other metals. Hence, in the handful of cases in which a protein contains another known metal binding site, we ignored any grid point within 5 Å of its coordinating residues. If we instead consider hits near other occupied metal binding sites to be false positives, we observe a small decrease in the positive predictive value from 75.2% to 72.8%. However, the high scoring hits near other metal binding sites are still informative, as they suggest that a metal with chemical characteristics similar to zinc may bind. Unless otherwise noted, we therefore exclude grid points close to an occupied binding site for a metal other than zinc.
Figure 2.
Model performance on an independent holo test set. (A) As demonstrated in these ROC curves, the zinc model attains high sensitivity and specificity on a holo test set regardless of whether sensitivity is calculated with respect to binding sites (solid line) or proteins (dashed line). (B) At a positive predictive value of 75%, the model recognizes 77.8% of zinc sites (solid) and at least one site in 84% of proteins (dashed). (C, D) When true positives are defined as hits within 5 Å of at least two coordinating residues, the specificities, sensitivities, and positive predictive values are relatively unaffected.
The above definition of a true positive guarantees that any such hit will be close to at least one coordinating residue. In some settings, however, one might desire a criterion that is more sensitive to the exact location of the binding site. We therefore analyzed our results with an alternative definition of a true positive that requires a hit to be within 5 Å of at least two coordinating residues rather than one. This more stringent requirement will, for example, classify a high score occurring in the outskirts of the binding site as a false positive. At the 99.8% specificity score cutoff used above, the positive predictive value drops very slightly to 74.7% while the specificity and the sensitivities with respect to sites and proteins remain unchanged (Fig. 2C,D). Therefore, unless otherwise noted, we use the definition of a true positive that requires proximity to only one coordinating residue.
The proteins in the training set cover 83 different enzyme classification (EC) numbers representing all six EC classes, and the test set contains proteins with 23 EC numbers not seen in the training set. Although the majority of zincs in the training set are coordinated by at least one cysteine residue, the model does identify many test set sites with no cysteines. Thus, the zinc model recognizes binding sites in a disparate set of proteins, and can extrapolate beyond types of sites it has already seen to identify new ones located in proteins with entirely different functions.
Performance on apo proteins
The model's performance in holo proteins indicates that it is able to detect zinc binding sites when the side chains are ideally arranged to bind the metal. In an apo protein structure, however, a binding site's side chains are more likely to take on alternate conformations, making the site harder to detect. These cases are of greater biological relevance since side chain movements may reveal information regarding the mechanism of binding. To obtain a local RMSD for each binding site, we superimposed each of the holo site's coordinating atoms and their residues' α-carbons onto the corresponding atoms in the apo site. The average RMSD of the coordinating atoms among the 81 zinc binding sites in our data set of 51 nonredundant apo proteins is 1.36 ± 1.28 Å.
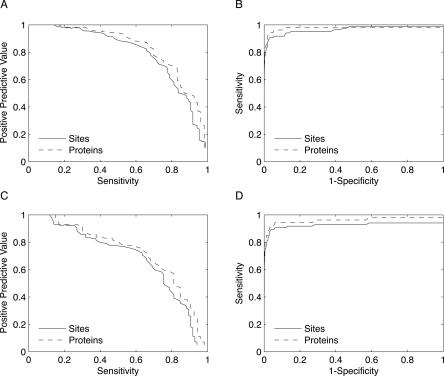
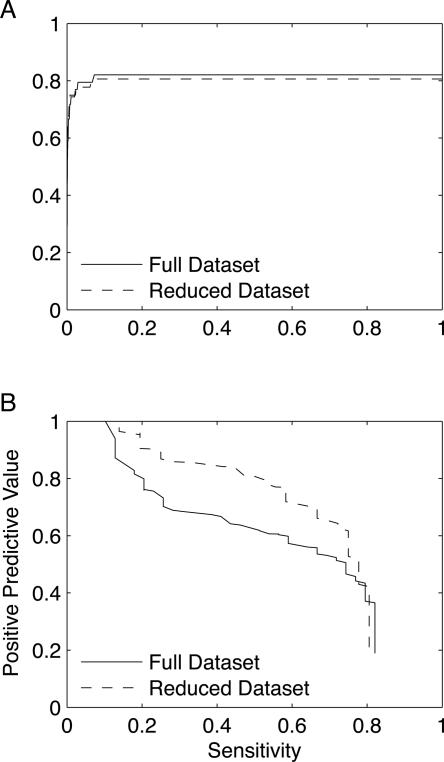
Using the 99.8% specificity score cutoff from the holo test set and the definition of a true positive that requires proximity to one coordinating residue, the zinc model achieves a positive predictive value of 73.6% on a nonredundant data set of 51 apo proteins (Fig. 3A). Its sensitivities with respect to sites and proteins are 71.8% and 75.5%, respectively, and its specificity is 99.7% (Fig. 3B). Hence, the performance of the model on apo proteins is nearly identical to the performance on holo proteins. If we consider grid points in close proximity to an occupied binding site for another metal to be negative rather than positive points, the positive predictive value decreases to 67.2%.
Figure 3.
Model performance on an independent apo test set. (A) A ROC curve assessing the zinc model's performance on the apo test set indicates that the performance is nearly as strong in apo proteins as in holo proteins (Fig. 2). At the 99.8% specificity score threshold determined from the holo test set, the zinc model attains sensitivities of 71.8% and 75.5% with respect to binding sites (solid line) and proteins (dashed line), respectively. (B) At the same score threshold, the positive predictive value is 73.6%, which is nearly as high as for the holo test set. (C, D) Because metal binding often involves a conformational change, defining true positives as hits within 5 Å of at least two residues reduces the model's positive predictive value and sensitivities.
Since structural rearrangements of the binding site are common in apo proteins (Babor et al. 2005), we do not expect that each high scoring hit will necessarily be in close proximity to two coordinating residues. Using this more stringent definition of a true positive, the positive predictive value therefore decreases to 62.3%, and the sensitivities with respect to sites and proteins drop to 69.4% and 73.5%, respectively (Fig. 3C,D). This is not surprising since in some cases the second closest residue to a high scoring hit in an apo protein will have undergone a conformational change.
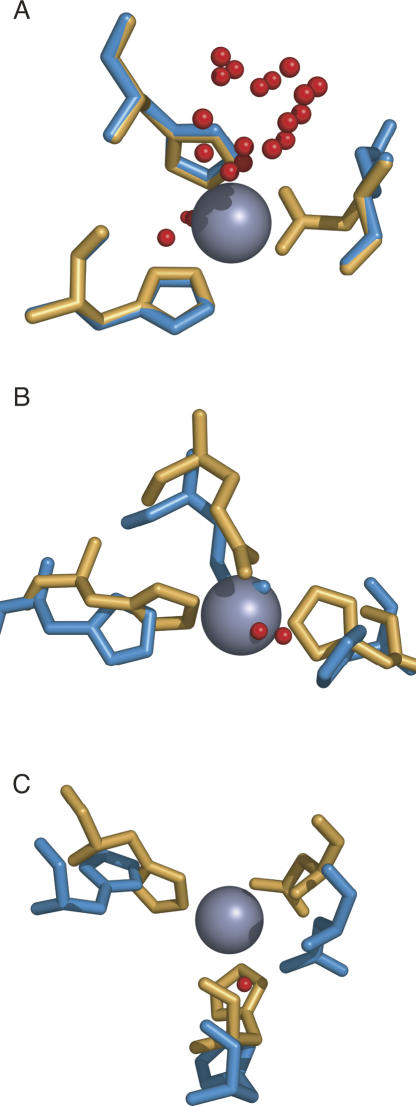
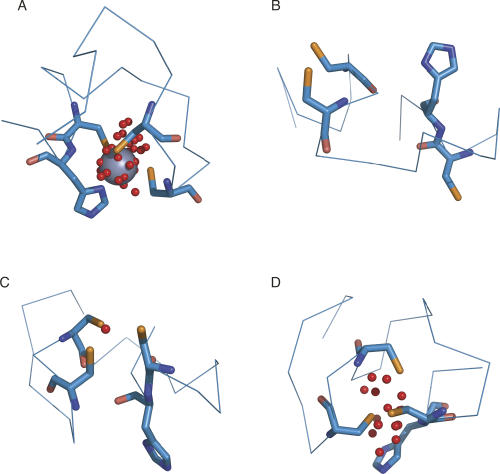
While the probability of detecting a binding site decreases somewhat as the RMSD between the coordinating atoms in the holo and apo structures increases, we identified a number of cases in which conformational changes had taken place and yet FEATURE is still able to find the binding site (Fig. 4). In some cases, only one of the coordinating residues' positions differs (Fig. 4A), while other sites undergo much more significant changes (Fig. 4B,C). The red spheres in Figure 4 denote locations of FEATURE hits whose scores meet or exceed the 99.8% specificity score threshold established for the holo test data set.
Figure 4.
FEATURE detects zinc binding sites that undergo conformational changes. The zinc binding sites in the test set undergo varying degrees of conformational changes upon binding of metal. Holo proteins are shown in orange, apo proteins are in blue, and red spheres represent points with FEATURE hits above the 99.8% specificity threshold. (A) In some cases, only one coordinating residue moves significantly (holo: PDB identifier 1s0e; apo: PDB identifier 1s0g). (B, C) In other cases, multiple coordinating residues are displaced from their positions in the holo protein (holo: 1hp7 [B] and 1ty2 [C]; apo: 1kct [B] and 1ty0 [C]).
Distinguishing between zinc and calcium binding sites
As zinc's chemical properties differ somewhat from those of alkaline earth metals, we sought to distinguish zinc and calcium binding sites by training a second FEATURE model using their binding sites as positive and negative training examples, respectively. We designate this new model the “Zn vs. Ca” model. We apply our original model followed by the Zn vs. Ca model, and require that a hit surpass the 99.8% specificity score cutoff for the original model and a second score cutoff for the new model. This process allows us to assess whether regions initially identified as zinc binding sites are in fact calcium sites. While some features that discriminate zinc from calcium are easily interpreted, such as the greater propensity for histidines and cysteines to coordinate zinc as compared to calcium, others are more subtle, such as differences in typical atom densities in various shells. Nearly every amino acid other than histidine and cysteine is more prevalent in the second through fourth shells of calcium binding sites than in zinc sites; this is likely a reflection of the fact that backbone oxygens coordinate calcium ions more commonly than they do zinc. The complete physicochemical fingerprint of the Zn vs. Ca model is available at http://helix-web.stanford.edu/pubs/zinc.
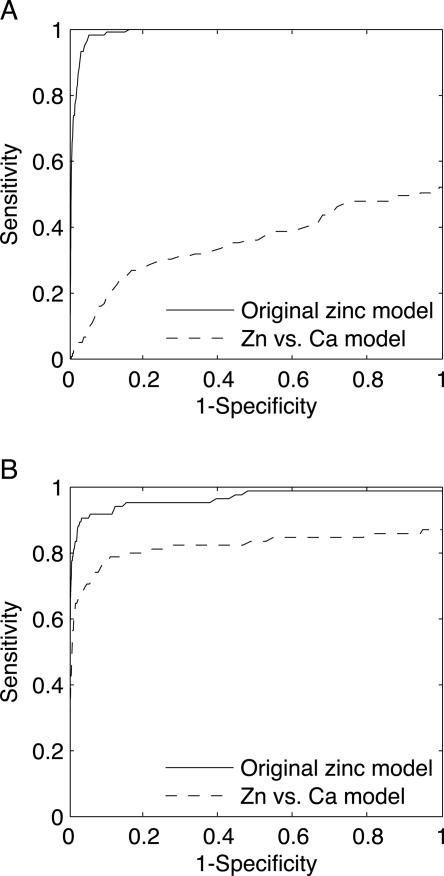
Grid scans of an independent test set of 58 calcium binding proteins against the original zinc model yielded a sensitivity of 51.3% with respect to calcium binding sites, indicating that the model does indeed overlap with a biochemical description of calcium binding sites. In order to assess FEATURE's ability to screen out calcium sites, we scanned all of the high scoring points again with the Zn vs. Ca model and computed a ROC curve for varying score thresholds (Fig. 5A). At a Zn vs. Ca model score cutoff that discards only eight of the 127 zinc sites in the holo test that were detected by the original model, the sensitivity with respect to calcium sites drops to 10.1%. This fivefold decrease in the number of calcium sites confused for zinc indicates that the sequential application of the original zinc model followed by the Zn vs. Ca model allows for discrimination between zinc and calcium binding sites. In order to ensure further that the Zn vs. Ca model does not accidentally filter out zinc sites as well, we applied it to the zinc apo test set (Fig. 5B). This increased the positive predictive value from 73.6% to 76.4% and screened out only one zinc site, while the specificity decreased slightly to 95.6%.
Figure 5.
A zinc model trained against calcium binding sites discriminates between zinc and calcium. (A) We computed a ROC curve for the original zinc model for a test set consisting of calcium binding proteins (solid line). In this case, we consider calcium binding sites to be positive sites. The original zinc model recognizes approximately half of the calcium binding sites at the 99.8% specificity score threshold. When we scan hits above this score threshold with a second model trained against calcium sites (dashed line), we detect far fewer calcium binding sites, indicating that the sequential application of the original model followed by the Zn vs. Ca model results in discrimination between zinc and calcium. (B) Applying this process to the zinc apo test set reduces the sensitivity with respect to zinc binding sites at high specificities only mildly. At the 99.8% specificity score threshold determined from the holo test set, the original model's sensitivity is 71.8% (solid line); this decreases to 70.6% after applying the second model (dashed line). All sensitivities are calculated with respect to binding sites.
Detecting binding sites in modeled structures
Since FEATURE was designed to tolerate conformational changes in binding sites, we tested our zinc model's ability to find binding sites in predicted protein structures, whose side-chain positions are not likely to be accurate. The performance on modeled structures also provides information about how accurate these structures must be in order to allow prediction of binding and active sites. We identified 29 proteins in the holo test set that have 30%–50% sequence identity to a nonmetal binding template structure in the Protein Data Bank (PDB) (Berman et al. 2000), and then modeled the holo test set structures from their templates using the MODELLER program (Marti-Renom et al. 2000) as described in the Methods section. The average holo/template pairwise sequence identity of 35.9 ± 5.2% makes this a reasonably difficult homology modeling exercise. Though the upper limit of 50% sequence identity may seem high, only six of the 29 proteins exceeded 40% identity. Furthermore, 26 of the 127 residues that coordinate zincs are located in gapped regions of the BLAST alignments of the holo and template proteins (Altschul et al. 1997), and 73 differ in sequence between the holo and template proteins. Only 10 of these latter 73 residues are replaced by residues likely to coordinate a zinc ion (e.g., histidine, cysteine, aspartic acid, or glutamic acid). These local differences in the zinc binding sites between the holo proteins and their templates contribute to the difficulty in recognizing zinc binding sites in the modeled structures.
We generated structural diversity by building 10 different models for each of the holo proteins and scanned each for zinc binding sites. In order to recognize the existence of a site, we required that FEATURE detect it in only one of the 10 models. In the case of the C-terminal zinc binding domain of the SecA ATPase (PDB identifier 1sx1), the zinc binding site is well formed in only a small percentage of the models (Fig. 6). We quantified the structural similarity of each modeled zinc site to its counterpart in the true holo structure by superimposing the zinc's coordinating atoms and their residues' α-carbons in the modeled site onto the corresponding atoms from the holo structure. The average local RMSD for the coordinating atoms over all 390 modeled zinc binding sites is 7.8 ± 9.4 Å. The average RMSD for binding sites below the median value is 2.4 ± 1.3 Å.
Figure 6.
Multiple homology models improve the probability of detecting a zinc binding site. MODELLER produced models of varying quality for the site in the C-terminal zinc binding domain of the SecA ATPase (PDB identifier 1sx1). (A) The zinc atom in the holo structure is coordinated by three cysteine residues and one histidine. (B) In many models, the residues that coordinate the zinc were separated in space and FEATURE detected no hits. (C, D) In other models, the binding site was cohesive enough to be detected by FEATURE. We observed no cases among the 10 models in which the histidine was not to some degree rotated away from the binding site. Red spheres indicate FEATURE hits above the 99.8% specificity threshold.
At the same 99.8% specificity score threshold used above, we achieve a positive predictive value of 59.9% and sensitivities with respect to sites and proteins of 59.0% and 65.5%. Interestingly, two proteins account for 50% of all false-positive hits across all of the modeled structures. If we leave these two proteins out of the analysis, the positive predictive value rises to 75.2% and the sensitivity with respect to sites decreases only slightly to 58.3%, while the sensitivity with respect to proteins increases to 66.7% (Fig. 7).
Figure 7.
Performance on homology modeling structures. Two of 29 proteins in a data set of predicted structures produced using MODELLER account for 50% of the observed false positive hits. We compute all sensitivities in these curves with respect to binding sites; values with respect to proteins are slightly higher. (A) The ROC curve is relatively unaffected by the elimination of these two proteins. (B) At the 99.9% specificity score threshold, FEATURE's positive predictive value increases from 59.9% to 75.2% when the two proteins are removed from the data set.
Comparison to MetSite
Since both FEATURE and MetSite were designed to operate on low resolution structures, we compared the two algorithms by scanning all of the modeled protein structures using the MetSite Web server (http://bioinf.cs.ucl.ac.uk/MetSite/MetSite.html). Since MetSite identifies coordinating residues rather than the Cartesian position of the zinc itself, it is difficult to compare the two methods directly. We consider MetSite to have identified a binding site if it detects at least one of its coordinating residues. In the interest of fairness, we use a more stringent definition for FEATURE's true-positive hits than before and require them to be within 3 Å of a coordinating residue. In order to compare FEATURE's false-positive rates to MetSite's, we then map the remaining false-positive hits to the nearest cysteine, histidine, aspartic acid, or glutamic acid residue, as these four amino acids account for 90% of the coordinating residues among the training set's binding sites. Only one of the 349 training sites contains none of these coordinating residues.
At the 99.8% specificity threshold, FEATURE detects 22 of the 39 zinc binding sites among the modeled structures and returns 90 false-positive residues. At the same sensitivity, MetSite finds 191 false-positive residues. At the score cutoff where MetSite finds the same number of false positives as FEATURE, it detects 19 sites among 14 proteins, while FEATURE's 22 sites cover 19 proteins. FEATURE found eight sites not found by MetSite, and MetSite found five not located by FEATURE.
We also considered how the results would change if we filtered MetSite's output to ignore residues other than cysteines, histidines, glutamates, and aspartates. At equal sensitivities, MetSite finds 102 false positives, which brings it in line with FEATURE's 93 false positives. At the score cutoff where the false-positive rates are equal, MetSite finds two fewer sites than does FEATURE, and FEATURE covers four more proteins with its true positives than does MetSite. Again, the list of identified sites differs: FEATURE finds seven unique sites and MetSite finds five. Hence, FEATURE is able to annotate binding sites that MetSite cannot detect, and vice versa.
Analysis of structural genomics targets
We scanned all solved structural genomics targets from the TargetDB Web site (http://targetdb.pdb.org) for zinc binding sites. Since it is difficult to validate predictions made in cases where other functional annotation methods fail, we present several examples where our zinc model supports results obtained via other approaches. The Berkeley Structural Genomics Center crystallized a protein of unknown function from Methanococcus jannaschii (PDB identifier 1s3l) with a number of homologs in archaea and bacteria that are annotated as putative phosphodiesterases (Chen et al. 2004). A scan with the zinc model reveals two clusters of hits with scores well above the 99.8% specificity threshold. The original investigators confirmed phophodiesterase activity and solved additional crystal structures containing either manganese or nickel at the binding site predicted by FEATURE. The metal binding site's close proximity to the active site suggests a role in catalysis (Chen et al. 2004). Since other homologs do bind zinc (e.g., PDB identifier 1aui), it is unclear which metal occupies the binding site in vivo. The biochemical characterization of the protein would have been supported by results using the zinc model, which also may have suggested an additional experiment to assess the protein's level of activity in the presence of zinc.
Another structural genomics target deemed a putative ferritin-like protein was crystallized in the absence of metals (PDB identifier 1vjx), but has high scoring hits to the zinc model. Though the sequence contains no Prosite hits, it matches Pfam's rubrerythrin family, which is a member of the ferritin-like superfamily. At the time of its release, a structural similarity search would have revealed a hit to cytochrome B1 from Escherichia coli (PDB identifier 1bcf), which contains two iron ions at the location predicted by FEATURE. A second structurally similar protein that also matches the rubrerythrin family has one iron and a zinc rather than two irons (PDB identifier 1b71), suggesting that 1vjx may in fact bind zinc.
Finally, we consider a cluster of FEATURE hits to PDB identifier 1zpy, a structural genomics target of unknown function solved in 2005. The sequence contains no hits in the Prosite and Pfam databases, and a BLAST search against the PDB reveals that none of the homologous proteins, whose structures were solved before 1zpy's, contained metals. However, in 2007, an uncharacterized metal binding protein with 29% sequence identity to 1zpy over 44 residues was crystallized in the presence of zinc (PDB identifier 20h3). The sequence identity is relatively low, but superimposing the aligned residues reveals that the zinc ion occupies the predicted binding site.
Discussion
We have built a model for zinc binding sites using the FEATURE algorithm that achieves a high level of sensitivity in apo proteins, many of which undergo conformational changes upon metal binding. The method is general and thus can be applied to other metals. We have previously published a calcium model using a similar approach (Wei and Altman 1998), though this study improves upon the physicochemical features used, adds methodology for enforcing the presence of a reasonable first coordination shell, and applies a significantly more rigorous analysis, particularly in the case of apo proteins. While some methods offer multiple classifiers trained separately on each metal, we are unaware of any previous demonstrations of metal specificity among methods that are robust to variations in side-chain positions. We do not always distinguish between metals such as zinc and iron that have highly similar coordination environments, as their chemical properties are similar enough that they are sometimes capable of occupying each other's binding sites (e.g., Zang et al. 2001). In these cases, one might use FEATURE's results as initial guesses for refinement using a more time-consuming analysis. However, the alkaline earth metals possess properties that make them chemically distinct from zinc, and hence it is theoretically possible to distinguish between these two types of metals. The fact that the zinc model identified approximately half of the calcium sites in the calcium data set suggests that zinc and calcium binding sites have similar chemical and physical characteristics. A more sensitive model obtained by using calcium binding sites as negative training examples helped elucidate the more subtle features that allowed us to discriminate between zinc and calcium with only a small loss in sensitivity.
FEATURE's use of information that extends beyond the identities and orientations of the zinc's ligating residues allows it both to tolerate changes in conformation and sequence and also to detect types of sites that were not present in the training set. As shown in Figure 4, we recognize binding sites in apo proteins even when side chains have moved with respect to the holo protein. In some cases, only one side chain rotates away from the binding site, while in others most or all of the ligating residues undergo significant conformational changes. When one or more of the coordinating atoms are not present in the region expected by the FEATURE model, there is often enough information in the remaining features of the binding site to compensate. Methods based on simple geometric criteria would encounter greater difficulty. Similarly, FEATURE is able to detect a variety of different classes of zinc binding sites regardless of factors such as the number of protein ligands or the separation in sequence space between coordinating residues. At the 99.8% specificity score threshold, we detect 86.0% and 94.8% of sites with three or four protein ligands in the holo test set, respectively. Despite the fact that fewer than a third of the training sites had at least one coordinating residue separated from the others by a distance of at least 30 residues, our sensitivity for such sites in the holo test set is 81.9%. The sensitivity decreases to 66.7% at a separation of at least 100 residues, but this result reflects a remarkable generalization from the training set given that only 4% of training sites exhibited such large separations.
As the number of structures of unknown function deposited into the PDB by structural genomics groups increases, so too does the relevance of structure-based functional annotation methods. As a point of reference, the Midwest Center for Structural Genomics reported that as of September 2005, 42% of solved targets could not be assigned a putative function based on sequence homology with a protein of known function (Watson et al. 2007). FEATURE's high specificity and positive predictive value position it to provide highly confident, experimentally verifiable functional predictions. Of course, function can be defined at many levels; FEATURE is designed to recognize basic binding and active site molecular functions but cannot infer involvement in particular biological pathways. While certainly FEATURE is useful in cases where sequence-based methods are unable to recognize zinc binding sites, it can also increase confidence in these predictions, and it can pinpoint the location of the binding sites in cases where other methods cannot. In particular, although the presence of a zinc binding site does not always yield information regarding the global function of a protein, it may nonetheless increase support for a functional prediction made by other means. In some cases, the coordination environment of a zinc site may suggest its purpose. The presence of a water molecule is highly suggestive of a catalytic site, and coordination by four cysteines is a hallmark of a structural zinc binding site (Auld 2002). Methods such as FEATURE that are based on local similarity may also provide evidence of functional relationships among proteins that are globally dissimilar in sequence and structure.
When faced with the difficult task of identifying functional sites in predicted protein structures, which may be of relatively low quality, it is helpful to apply all available annotation methods. Our comparison of FEATURE and MetSite demonstrates that the two methods are complementary to one another, as each is able to detect binding sites missed by the other. This is not surprising given that they use different sources of information and different methodologies. When both algorithms detect the same binding site, this lends confidence to the prediction. Unlike MetSite and some other methods, FEATURE does not rely on the existence of sequence homologs to provide functional annotation for a protein. It may therefore be able to succeed in cases where a structural genomics target has few sequence homologs.
Our modeling protocol for generating imprecise structural models was quite simple; we used only one template even when others were available, and we used only a BLAST run with default parameters to provide the initial alignment between the template and target proteins. Though we desired imprecise models for the purpose of testing FEATURE, more modern techniques for detecting sequence homology not only may have selected better templates but also may have produced better alignments of the holo sequences to their templates. Both of these factors are critical in producing reliable homology models. It may also be possible to refine the structures of predicted sites by placing zinc at locations with high scoring hits and refining the structure. While previous studies on identifying functional sites in predicted structures have cited varying levels of success (Wei et al. 1999; Arakaki et al. 2004), we point out here that one might scan multiple models for the same protein in the hope that the site will be detectable at least once. Though this method may accumulate false positives, it is also possible to apply more stringent criteria by requiring the site to be identified in a larger fraction of the models.
More complex machine learning algorithms might improve FEATURE's performance, but methods such as neural nets and support vector machines typically lack the interpretability of a Bayesian classifier. The FEATURE model provides biochemical and biophysical information about the environment near zinc binding sites that may be useful in protein engineering and structure prediction efforts that require modeling of metal binding sites. For instance, Marino and Regan (1999) stabilized a zinc binding site that they had engineered into a nonmetal binding protein by introducing mutations that held the zinc's ligating residues in place through a hydrogen-bonding network, thus increasing the site's binding affinity. Similarly, Hunt et al. (1999) demonstrated that mutations affecting hydrophobic residues near a zinc binding site decreased the binding affinity. The nuanced description of the 7.5 Å spherical environment around zinc binding sites provided by FEATURE may be of use in such efforts.
Methods
Nonredundant data set construction
As a starting point from which to build a data set of zinc binding proteins, we identified every biological unit file in the PDB containing at least one zinc atom. To screen out zincs that are nonspecifically bound or that are coordinated by residues from adjacent molecules in a crystal structure, we discard zincs with coordination numbers less than three or with coordinating atoms contributed by fewer than two distinct residues. This requirement applies to the construction of training and test sets but is not part of the FEATURE algorithm itself. We define a coordinating atom as a nitrogen, oxygen, or sulfur from either a residue or a water molecule within 3 Å of the zinc. This allows for some inaccuracies in a structure's coordinates without including extraneous atoms (Harding 2001).
Before selecting a nonredundant data set from among the available zinc binding proteins, we searched the PDB for apo structures for each protein. We aligned all zinc binding proteins to structures in the PDB that do not contain zinc, and retained those with greater than 95% sequence identity. To further ensure that each identified structure is truly an apo form of the holo protein, we discarded proteins with sequence changes among the zinc's coordinating residues or in which coordinates for some or all of the coordinating residues were unavailable. We also discarded proteins whose binding sites are occupied by a different ligand.
Next, we performed all pairwise sequence alignments for all chains with atoms within 5 Å of a zinc binding site and defined the sequence identity between two proteins to be the maximum identity between any two such chains. In order to maximize the number of proteins with apo structures in the final nonredundant data set, we first populated it with apo structures such that no two share greater than 30% sequence identity and then extended it with the remaining holo structures while maintaining the same sequence identity threshold. This yielded a total of 461 proteins, which we partitioned into a training set of 361 proteins and a holo test set of 100 proteins. The test set contains 51 proteins whose apo structures are available (see below), and the remainder were randomly selected.
A calcium training set and holo test set of 207 and 58 proteins, respectively, were created using the same methodology. All data sets are available at http://helix-web.stanford.edu/pubs/zinc.
FEATURE training and scanning
As described previously, FEATURE accepts as input a set of positive training points and a set of negative training points, and produces a Bayesian classifier used to score points in new protein structures for the presence of functional sites (Bagley and Altman 1995, 1996). We use an updated list of features that eliminates some of the redundancy of FEATURE's previous feature set (Table 1). The training sites for the zinc model consist of a single zinc binding site from each of the 361 proteins in the training set. We determined a set of potential zinc coordination environments from those observed in the training set and scanned all training proteins for these environments along a 1 Å cubic grid. Since we do not expect that apo proteins will have well-formed zinc binding sites, we accept a grid point as a potential binding site under a set of relatively lenient constraints. For each point, we make a list of all residues with atoms located within 5 Å. Rather than looking for an exact match to a coordination environment observed in the training set, we require only that residues capable of supplying two thirds of the necessary coordinating atoms be on this list. That is, if a coordination environment consists of four sulfurs contributed by cysteine residues, we accept a grid point if three cysteines have atoms within 5 Å. The negative training set consists of all points at least 10 Å from any grid point identified as a potential binding site.
Table 1.
Description of the FEATURE properties
When scanning a new protein for zinc binding sites, we first embed it in a 1 Å grid, scan for partial matches to the training coordination environments, and score each resulting point using the classifier produced by FEATURE. FEATURE discards all heteroatoms and hence does not use the location of the zinc as a feature in training or in scanning holo proteins. A small number of proteins in the test sets contain metals other than zinc. In these cases, we eliminate grid points within 5 Å of their coordinating residues. Naturally, if a protein in the holo test set contains an unoccupied metal binding site, we will incorrectly consider nearby hits to be false positives. If the site is also unoccupied in the protein's apo form, this error will affect results in the apo test set as well. We also analyze our grid scan results without eliminating binding sites for other metals.
We produced a model designed to distinguish between zinc and calcium binding sites by using the positive training set from the zinc model and a negative training set consisting of one calcium site from each of the 207 calcium training proteins.
Accuracy measures
We employ several standard measurements of accuracy in evaluating results on our test sets. Sensitivity can be defined with respect to either the number of zinc binding sites or the number of proteins. In the former case, it is equal to the number of sites with at least one FEATURE hit divided by the total number of sites. In the latter case, it is equal to the number of proteins with at least one hit in at least one binding site divided by the total number of proteins.
Both specificity and positive predictive value capture the model's tendency to return false-positive results. Specificity is defined as the number of negative sites correctly identified as negative (i.e., without hits), while positive predictive value is the number of true-positive hits divided by the total number of hits.
Homology modeling template selection
For each protein in the zinc holo test set containing a zinc binding site coordinated by residues from a single chain, we scanned the subset of the PDB consisting of proteins without bound zincs for protein chains with 30%–50% sequence identity. The sequence identity is equal to the number of identical residues in a BLAST alignment divided by the length of the holo protein chain, and we did not enforce any constraints on sequence similarity within the zinc binding site itself. We discarded template proteins whose binding sites are occupied by other ligands.
Homology modeling with MODELLER
We used MODELLER (Marti-Renom et al. 2000) to predict the structure of each holo protein for which we identified a suitable template as if the structure were unknown. The BLAST alignments used to identify templates for holo proteins served as the initial alignment for homology modeling using the MODELLER program. A sample python script used to run MODELLER is available at http://helix-web.stanford.edu/pubs/zinc. We produced 10 different models per protein by optimizing loop regions using different random seeds. MODELLER had no access to any information about the holo protein other than its primary amino acid sequence.
Availability
The source code is written in C and C++ and is available at http://helix-web.stanford.edu/pubs/zinc. Users may also submit jobs to a Web interface at http://feature.stanford.edu/metals. The Web site scans a PDB file for zinc binding sites, and provides visualization tools using Jmol (http://jmol.sourceforge.net).
Acknowledgments
We thank Randy Radmer and the Helix Group at Stanford University for helpful discussions regarding this work. This work was supported by NIH grant NIH LM05652 and by a Stanford Dean's Fellowship.
Footnotes
Reprint requests to: Russ B. Altman, Departments of Bioengineering and Genetics, Stanford University, 318 Campus Drive, Clark Center S170, MC 5444 Stanford, CA 94305-5444, USA; e-mail: russ.altman@stanford.edu; fax: (650) 723-8544.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073138508.
References
- Alberts, I., Nadassy, K., Wodak, S. Analysis of zinc binding sites in protein crystal structures. Protein Sci. 1998;7:1700–1716. doi: 10.1002/pro.5560070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloy, P., Querol, E., Aviles, F., Sternberg, M. Automated structure-based prediction of functional sites in proteins: Applications to assessing the validity of inheriting protein function from homology in genome annotation and to protein docking. J. Mol. Biol. 2001;311:395–408. doi: 10.1006/jmbi.2001.4870. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreini, C., Banci, L., Bertini, I., Rosato, A. Counting the zinc-proteins encoded in the human genome. J. Proteome Res. 2006;5:196–201. doi: 10.1021/pr050361j. [DOI] [PubMed] [Google Scholar]
- Arakaki, A.K., Zhang, Y., Skolnick, J. Large-scale assessment of the utility of low-resolution protein structures for biochemical function assignment. Bioinformatics. 2004;20:1087–1096. doi: 10.1093/bioinformatics/bth044. [DOI] [PubMed] [Google Scholar]
- Arnesano, F., Banci, L., Bertini, I., Ciofi-Baffoni, S., Molteni, E., Huffman, D.L., O'Halloran, T.V. Metallochaperones and metal-transporting ATPases: A comparative analysis of sequences and structures. Genome Res. 2002;12:255–271. doi: 10.1101/gr.196802. [DOI] [PubMed] [Google Scholar]
- Auld, D. Zinc coordination sphere in biochemical zinc sites. Biometals. 2002;14:271–313. doi: 10.1023/a:1012976615056. [DOI] [PubMed] [Google Scholar]
- Babor, M., Greenblatt, H., Edelman, M., Sobolev, V. Flexibility of metal binding sites in proteins on a database scale. Proteins. 2005;59:221–230. doi: 10.1002/prot.20431. [DOI] [PubMed] [Google Scholar]
- Bagley, S., Altman, R. Characterizing the microenvironment surrounding protein sites. Protein Sci. 1995;4:622–635. doi: 10.1002/pro.5560040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley, S., Altman, R. Conserved features in the active site of nonhomologous serine proteases. Fold. Des. 1996;1:371–379. doi: 10.1016/S1359-0278(96)00052-1. [DOI] [PubMed] [Google Scholar]
- Banatao, D.R., Altman, R.B., Klein, T.E. Microenvironment analysis and identification of magnesium binding sites in RNA. Nucleic Acids Res. 2003;31:4450–4460. doi: 10.1093/nar/gkg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci, L., Bertini, I., Rosato, A., Varani, G. Mitochondrial cytochromes c: A comparative analysis. J. Biol. Inorg. Chem. 1999;4:824–837. doi: 10.1007/s007750050356. [DOI] [PubMed] [Google Scholar]
- Barker, J., Thornton, J. An algorithm for constraint-based structural template matching: Application to 3D templates with statistical analysis. Bioinformatics. 2003;19:1644–1649. doi: 10.1093/bioinformatics/btg226. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Coin, L., Durbin, R., Finn, R.D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nargkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini, I., Luchinat, C., Provenzani, A., Rosato, A., Vasos, P.R. Browsing gene banks for Fe2S2 ferredoxins and structural modeling of 88 plant-type sequences: An analysis of fold and function. Proteins. 2002;46:110–127. doi: 10.1002/prot.10009. [DOI] [PubMed] [Google Scholar]
- Bixby, K., Nanao, M., Shen, N., Kreusch, A., Bellamy, H., Pfaffinger, P., Choe, S. Zn2+-binding and molecular determinants of tetramerization in voltage-gated K+ channels. Nat. Struct. Biol. 1999;6:38–43. doi: 10.1038/4911. [DOI] [PubMed] [Google Scholar]
- Bork, P., Bairoch, A. Go hunting in sequence databases but watch out for the traps. Trends Genet. 1996;12:425–427. doi: 10.1016/0168-9525(96)60040-7. [DOI] [PubMed] [Google Scholar]
- Brenner, S. Errors in genome annotation. Trends Genet. 1999;15:132–133. doi: 10.1016/s0168-9525(99)01706-0. [DOI] [PubMed] [Google Scholar]
- Brown, R.S., Sander, C., Argos, P. The primary structure of transcription factor TFIIIA has 12 consecutive repeats. FEBS Lett. 1985;186:271–274. doi: 10.1016/0014-5793(85)80723-7. [DOI] [PubMed] [Google Scholar]
- Chandonia, J.M., Brenner, S.E. The impact of structural genomics: Expectations and outcomes. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- Chen, S., Yakunin, A.F., Kuznetsova, E., Busso, D., Pufan, R., Proudfoot, M., Kim, R., Kim, S.H. Structural and functional characterization of a novel phosphodiesterase from Methanococcus jannaschii . J. Biol. Chem. 2004;279:31854–31862. doi: 10.1074/jbc.M401059200. [DOI] [PubMed] [Google Scholar]
- Deng, H., Chen, G., Yang, W., Yang, J. Predicting calcium-binding sites in proteins—a graph theory and geometry approach. Proteins. 2006;64:34–42. doi: 10.1002/prot.20973. [DOI] [PubMed] [Google Scholar]
- Dudev, M., Lim, C. Discovering structural motifs using a structural alphabet: Application to magnesium-binding sites. BMC Bioinformatics. 2007;8:106. doi: 10.1186/1471-2105-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicken, C., Pennella, M., Chen, X., Koshlap, K., VanZile, M., Sacchettini, J., Giedroc, D. A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. J. Mol. Biol. 2003;333:683–695. doi: 10.1016/j.jmb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Fetrow, J., Skolnick, J. Method for prediction of protein function from sequence using the sequence-to-structure-to-function paradigm with application to glutaredoxins/thioredoxins and T1 ribonucleases. J. Mol. Biol. 1998;281:949–968. doi: 10.1006/jmbi.1998.1993. [DOI] [PubMed] [Google Scholar]
- Gaither, L., Eide, D. Eukaryotic zinc transporters and their regulation. Biometals. 2001;14:251–270. doi: 10.1023/a:1012988914300. [DOI] [PubMed] [Google Scholar]
- Gerlt, J., Babbitt, P. Can sequence determine function? Genome Biol. 2000 doi: 10.1186/gb-2000-1-5-reviews0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg, A.M., King, B.O., Roeder, R.G. Xenopus 5S gene transcription factor, TFIIIA: Characterization of a cDNA clone and measurement of RNA levels throughout development. Cell. 1984;39:479–489. doi: 10.1016/0092-8674(84)90455-0. [DOI] [PubMed] [Google Scholar]
- Gregory, D., Martin, A., Cheetham, J., Rees, A. The prediction and characterization of metal binding sites in proteins. Protein Eng. 1993;6:29–35. doi: 10.1093/protein/6.1.29. [DOI] [PubMed] [Google Scholar]
- Hantke, K. Bacterial zinc transporters and regulators. Biometals. 2001;14:239–249. doi: 10.1023/a:1012984713391. [DOI] [PubMed] [Google Scholar]
- Harding, M. Geometry of metal-ligand interactions in proteins. Acta Crystallogr. D Biol. Crystallogr. 2001;57:401–411. doi: 10.1107/s0907444900019168. [DOI] [PubMed] [Google Scholar]
- Hulo, N., Bairoch, A., Bulliard, V., Cerutti, L., De Castro, E., Langendijk-Genevaux, P.S., Pagni, M., Sigrist, C.J. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, J.A., Ahmed, M., Fierke, C.A. Metal binding specificity in carbonic anhydrase is influenced by conserved hydrophobic core residues. Biochemistry. 1999;38:9054–9062. doi: 10.1021/bi9900166. [DOI] [PubMed] [Google Scholar]
- Jambon, M., Imberty, A., Deléage, G., Geourjon, C. A new bioinformatic approach to detect common 3D sites in protein structures. Proteins. 2003;52:137–145. doi: 10.1002/prot.10339. [DOI] [PubMed] [Google Scholar]
- Karlin, S., Zhu, Z. Characterizations of diverse residue clusters in protein three-dimensional structures. Proc. Natl. Acad. Sci. 1996;93:8344–8349. doi: 10.1073/pnas.93.16.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasampalidis, I., Pitas, I., Lyroudia, K. Conservation of metal-coordinating residues. Proteins. 2007;68:123–130. doi: 10.1002/prot.21384. [DOI] [PubMed] [Google Scholar]
- Landau, M., Mayrose, I., Rosenberg, Y., Glaser, F., Martz, E., Pupko, T., Ben-Tal, N. ConSurf 2005: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, R., Watson, J., Thornton, J. ProFunc: A server for predicting protein function from 3D structure. Nucleic Acids Res. 2005a;33:W89–W93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, R., Watson, J., Thornton, J. Protein function prediction using local 3D templates. J. Mol. Biol. 2005b;351:614–626. doi: 10.1016/j.jmb.2005.05.067. [DOI] [PubMed] [Google Scholar]
- Lichtarge, O., Sowa, M. Evolutionary predictions of binding surfaces and interactions. Curr. Opin. Struct. Biol. 2002;12:21–27. doi: 10.1016/s0959-440x(02)00284-1. [DOI] [PubMed] [Google Scholar]
- Lin, C., Lin, K., Yang, C., Chung, I., Huang, C., Yang, Y. Protein metal binding residue prediction based on neural networks. Int. J. Neural Syst. 2005;15:71–84. doi: 10.1142/S0129065705000116. [DOI] [PubMed] [Google Scholar]
- Lin, H., Han, L., Zhang, H., Zheng, C., Xie, B., Cao, Z., Chen, Y. Prediction of the functional class of metal-binding proteins from sequence derived physicochemical properties by support vector machine approach. BMC Bioinformatics. 2006;7(Suppl 5):S13. doi: 10.1186/1471-2105-7-S5-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino, S.F., Regan, L. Secondary ligands enhance affinity at a designed metal-binding site. Chem. Biol. 1999;6:649–655. doi: 10.1016/s1074-5521(99)80116-1. [DOI] [PubMed] [Google Scholar]
- Marti-Renom, M.A., Stuart, A.C., Fiser, A., Sanchez, R., Melo, F., Sali, A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Palmer, D., Garrett, J., Sharma, V., Meganathan, R., Babbitt, P., Gerlt, J. Unexpected divergence of enzyme function and sequence: “N-acylamino acid racemase” is o-succinylbenzoate synthase. Biochemistry. 1999;38:4252–4258. doi: 10.1021/bi990140p. [DOI] [PubMed] [Google Scholar]
- Passerini, A., Punta, M., Ceroni, A., Rost, B., Frasconi, P. Identifying cysteines and histidines in transition-metal-binding sites using support vector machines and neural networks. Proteins. 2006;65:305–316. doi: 10.1002/prot.21135. [DOI] [PubMed] [Google Scholar]
- Passerini, A., Andreini, C., Menchetti, S., Rosato, A., Frasconi, P. Predicting zinc binding at the proteome level. BMC Bioinformatics. 2007;8:39. doi: 10.1186/1471-2105-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, R. Detection of protein three-dimensional side-chain patterns: New examples of convergent evolution. J. Mol. Biol. 1998;279:1211–1227. doi: 10.1006/jmbi.1998.1844. [DOI] [PubMed] [Google Scholar]
- Schymkowitz, J., Rousseau, F., Martins, I., Ferkinghoff-Borg, J., Stricher, F., Serrano, L. Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc. Natl. Acad. Sci. 2005;102:10147–10152. doi: 10.1073/pnas.0501980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi, J., Bryson, K., McGuffin, L., Ward, J., Wernisch, L., Jones, D. Predicting metal-binding site residues in low-resolution structural models. J. Mol. Biol. 2004;342:307–320. doi: 10.1016/j.jmb.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Todd, A.E., Marsden, R.L., Thornton, J.M., Orengo, C.A. Progress of structural genomics initiatives: An analysis of solved target structures. J. Mol. Biol. 2005;348:1235–1260. doi: 10.1016/j.jmb.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Tupler, R., Perini, G., Green, M.R. Expressing the human genome. Nature. 2001;409:832–833. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- Vallee, B., Auld, D. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry. 1990;29:5647–5659. doi: 10.1021/bi00476a001. [DOI] [PubMed] [Google Scholar]
- Watson, J., Sanderson, S., Ezersky, A., Savchenko, A., Edwards, A., Orengo, C., Joachimiak, A., Laskowski, R., Thornton, J. Towards fully automated structure-based function prediction in structural genomics: A case study. J. Mol. Biol. 2007;367:1511–1522. doi: 10.1016/j.jmb.2007.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, L., Altman, R. Recognizing protein binding sites using statistical descriptions of their 3D environments. Pac. Symp. Biocomput. 1998;3:495–506. [PubMed] [Google Scholar]
- Wei, L., Altman, R. Recognizing complex, asymmetric functional sites in protein structures using a Bayesian scoring function. J. Bioinform. Comput. Biol. 2003;1:119–138. doi: 10.1142/s0219720003000150. [DOI] [PubMed] [Google Scholar]
- Wei, L., Huang, E., Altman, R. Are predicted structures good enough to preserve functional sites? Structure. 1999;7:643–650. doi: 10.1016/s0969-2126(99)80085-9. [DOI] [PubMed] [Google Scholar]
- Yamashita, M., Wesson, L., Eisenman, G., Eisenberg, D. Where metal ions bind in proteins. Proc. Natl. Acad. Sci. 1990;87:5648–5652. doi: 10.1073/pnas.87.15.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang, T.M., Hollman, D.A., Crawford, P.A., Crowder, M.W., Makaroff, C.A. Arabidopsis glyoxalase II contains a zinc/iron binuclear metal center that is essential for substrate binding and catalysis. J. Biol. Chem. 2001;276:4788–4795. doi: 10.1074/jbc.M005090200. [DOI] [PubMed] [Google Scholar]
- Zhu, Z., Karlin, S. Clusters of charged residues in protein three-dimensional structures. Proc. Natl. Acad. Sci. 1996;93:8350–8355. doi: 10.1073/pnas.93.16.8350. [DOI] [PMC free article] [PubMed] [Google Scholar]