Abstract
Phloem loading is the initial step in photoassimilate export and the one that creates the driving force for mass flow. It has been proposed that loading occurs symplastically in species that translocate carbohydrate primarily as raffinose family oligosaccharides (RFOs). In these plants, dense fields of plasmodesmata connect bundle sheath cells to specialized companion cells (intermediary cells) in the minor veins. According to the polymer trap model, advanced as a mechanism of symplastic loading, sucrose from the mesophyll diffuses into intermediary cells and is converted there to RFOs. This process keeps the sucrose concentration low and, because of the larger size of the RFOs, prevents back diffusion. To test this model, the RFO pathway was down-regulated in Verbascum phoeniceum L. by suppressing the synthesis of galactinol synthase (GAS), which catalyzes the first committed step in RFO production. Two GAS genes (VpGAS1 and VpGAS2) were cloned and shown to be expressed in intermediary cells. Simultaneous RNAi suppression of both genes resulted in pronounced inhibition of RFO synthesis. Phloem transport was negatively affected, as evidenced by the accumulation of carbohydrate in the lamina and the reduced capacity of leaves to export sugars during a prolonged dark period. In plants with severe down-regulation, additional symptoms of reduced export were obvious, including impaired growth, leaf chlorosis, and necrosis and curling of leaf margins.
Keywords: plasmodesmata, polymer trap, stachyose, galactinol, RNAi
Up to 80% of the carbon fixed in mature leaves by photosynthesis is exported to heterotrophic sinks to enable their growth and development. The first step in the transport pathway, and one that is highly regulated (1, 2), is the transfer of photoassimilate from mesophyll cells to the sieve elements (SEs) and companion cells (CCs) of minor veins (3–6). This process, known as phloem loading, creates the positive hydrostatic pressure difference between source and sink phloem that drives the mass flow of solution.
Two loading mechanisms have been proposed. In one, photoassimilate enters the apoplast and is subsequently loaded into the phloem by specific transport proteins (3, 7). The second mechanism appears to be entirely symplastic (by plasmodesmata) (4–6). The first hint that loading might take place symplastically came from the discovery, in certain species, of specialized CCs (intermediary cells) in the minor veins, which are linked to bundle sheath cells by extremely high numbers of asymmetrically branched plasmodesmata (5).
In all plants with intermediary cells, the phloem sap contains a substantial amount of raffinose family oligosaccharide (RFO), especially the tri- and tetrasaccharides, raffinose, and stachyose. The consistent association of RFOs with intermediary cells suggests that the synthesis of these sugars is an integral part of the phloem-loading mechanism.
This reasoning is the basis of the polymer trap model of symplastic loading (8, 9). According to the model, sucrose diffuses from bundle sheath cells into intermediary cells through the numerous plasmodesmata that connect them. Sucrose is then converted into RFOs in the intermediary cells. A key, although as yet unproven, component of the model is that RFOs exceed the size exclusion limit of the plasmodesmata between bundle sheath cells and intermediary cells, which prevents their movement back to the bundle sheath and vectorizes the transport process out of the leaf.
If the synthesis of RFOs is an essential component of the phloem-loading mechanism, as the polymer trap model suggests, then down-regulating the pathway that produces these sugars by molecular-genetic techniques should inhibit long-distance transport. Although various molecular techniques have provided compelling evidence in favor of apoplastic loading in a number of model plants (10–13), this approach has not been possible with RFO-transporting species because of the lack of an efficient transformation system.
In this paper, we demonstrate that Verbascum phoeniceum (Scrophulariaceae) can be genetically modified by standard techniques, and it transports RFOs. Galactinol synthase (GAS) was down-regulated because the enzyme catalyzes the first committed step in RFO synthesis, the production of galactinol from myo-inositol and UDP-galactose (14). Galactinol then serves as the galactosyl donor in the synthesis of galactosyl oligosaccharides, such as raffinose and stachyose. This role is the only known one for galactinol, so the effects of inhibiting its synthesis are unlikely to be deleterious in other aspects of metabolism. Results of the experiments indicate that simultaneous RNAi suppression of two GAS genes expressed in the intermediary cells of V. phoeniceum inhibits RFO synthesis and the long-distance transport of photoassimilates.
Results
GAS Genes in V. phoeniceum.
To identify GAS genes potentially involved in phloem loading, a V. phoeniceum mature-leaf cDNA library was created. Of the ≈2.4 × 105 clones screened, four were isolated from the tertiary screen. Based on the presence of both putative start codons and polyadenylation sequences, these clones were interpreted to be full length. The isolated clones sorted into two unique contigs. The two cDNA clones were designated VpGAS1 and VpGAS2. The two full-length cDNAs share 72% identity, the ORFs are 78% identical, and the translated amino acid sequences are 83% identical. The deduced amino acid sequences of VpGAS1 and VpGAS2 are highly homologous to known GAS genes from Arabidopsis thaliana (15), Ajuga reptans (16), tomato (17), rice (18), zucchini (19), and soybean (19). Highest amino acid homology (87%) is with the gene from A. reptans, which is most closely related to V. phoeniceum. Translated sequences for both cDNAs contained the putative serine phosphorylation site (residues 254 and 261 in VpGAS1 and VpGAS2, respectively) found in many GAS sequences (15), as well as a hydrophobic motif, APSAA, at the carboxyl terminal, which has been found in all reported GAS sequences (15–17, 20–23).
Localization of VpGAS1 and VpGAS2 mRNA.
Northern blot analysis indicated that VpGAS1 is highly expressed in mature cauline and rosette source leaves, but is barely perceptible in flowers and sink leaves (Fig. 1A). VpGAS2 also is expressed in mature cauline and rosette leaves, and some transcript is detected in sink leaves (Fig. 1B). Galactinol also is produced in the seeds of most species (14), but expression of GAS in seeds was not studied here.
Fig. 1.
RNA gel blot analysis forVpGAS1 (A) and VpGAS2 (B) in different organs of V. phoeniceum. Ethidium bromide-stained rRNA as loading controls are shown in the lower gels.
VpGAS1 and VpGAS2 share visually identical spatial expression patterns, as determined by whole-mount in situ hybridization (Fig. 2). Both transcripts were detected exclusively in the intermediary cells of minor veins, which are easily recognized by their relatively large size, elongated shape, location at the margins of minor veins, and the fact that they occur in pairs [supporting information (SI) Fig. 7] (24). No other cell types in V. phoeniceum minor veins share these characteristics.
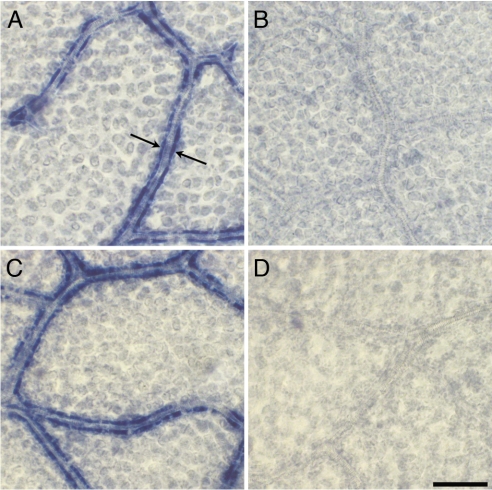
Fig. 2.
In situ hybridization of VpGAS1 and VpGAS2. (A and C) VpGAS1 and VpGAS2 antisense probes, respectively, localize to the intermediary cells. Each arrow in A indicates an intermediary cell. Note the arrangement of intermediary cells in paired files along the lengths of the veins. (B and D) VpGAS1 and VpGAS2 sense probes show only background staining. (Scale bar: 50 μm.)
Phenotype of Transgenic Plants.
By using the RNAi construct pAH-VpGAS2–1, which is designed to suppress the expression of both GAS genes, 70 transgenic plants representing 32 independently transformed lines were generated. In a preliminary screen, nine of these lines were identified as having the most significant reduction in GAS expression by Northern blot analysis (data not shown). The growth of most of these transgenic plants appeared normal. However, plants 8D and 12B displayed distinct abnormalities. In low light, they grew at approximately the same rate as wild-type plants. The young leaves (sink stage) were normal in size and appearance. However, as the leaves reached maturity, they grew more slowly than those of wild-type plants, they developed interveinal chlorosis, and the margins of some leaves curled and died. Finally, entire leaves died, whereas leaves of wild-type plants at the same stage of development were still healthy and green. These symptoms were more pronounced when the plants were transferred to the greenhouse. Under these high-light conditions, they grew more slowly than wild-type plants (Fig. 3). Plant 8D did not flower, whereas plant 12B flowered but did not set seed. Whole plants were regenerated from surface-sterilized leaf tissue, and these plants exhibited the same phenotypes. In electron micrographs, there were no noticeable differences in plasmodesmata structure in either plant 8D or 12B, compared with wild-type plants. The expression levels of both GAS genes in plants 8D and 12B were studied further by quantitative real-time PCR. In plants 8D and 12B, GAS1 expression levels were 5.6% ± 0.4% (±SD) and 3.7% ± 0.9% (±SD) those of wild type, whereas GAS2 expression levels were 2.8% ± 0.4% (±SD) and 4.3% ± 1.1% (±SD) those of wild type, respectively.
Fig. 3.
Wild type (A) and transgenic plant 8D (B) grown in the greenhouse. (Scale bars: 1 cm.)
Accumulation and Export of Carbohydrates.
Galactinol and RFO levels in the leaves of plants 8D and 12B are shown in Fig. 4. Galactinol and RFO levels were greatly reduced, whereas monosaccharide levels were elevated. The composition of sugars transported in the phloem-export stream was determined by labeling leaf blades with 14CO2 for 15 min and analyzing radiolabel distribution in the petiole after a 90-min chase. In the leaf blades of wild-type plants, substantial amounts of radiolabel incorporated into RFOs and galactinol (Fig. 5A). In the petioles of wild-type plants, which contain labeled compounds exported from the lamina, ≈50% of the label was in stachyose, with lesser amounts in raffinose and sucrose (Fig. 5B). Little radiolabeled galactinol was found in the petiole, which is typical of species that translocate RFOs (8, 24–26). In the leaf blades of transgenic plants, the amount of [14C]galactinol was greatly reduced, and 14C-labeled RFOs were barely detectable (Fig. 5A). In proportion to the amount of [14C]sucrose, only small quantities of 14C-labeled RFOs were transported to the petiole in plant 8D, and they were virtually absent in the transport stream of plant 12B (Fig. 5B).
Fig. 4.
Carbohydrates in mature leaves. (A) Mono- and disaccharide concentrations. (B) Galactinol and RFO concentrations. Error bars indicate SE (n ≥ 3).
Fig. 5.
Distribution of radiolabel 105 min after photosynthesis in 14CO2, calculated as a percentage of the neutral fraction. (A) Radiolabel in the lamina. (B) Radiolabel in petioles. m, monosaccharides include glucose, fructose, and galactose; s, sucrose; g, galactinol; r, raffinose; st, stachyose.
To determine whether transgenic plants are able to export accumulated carbohydrate during a prolonged dark period, wild-type plants and the nine lines identified during the original screen were placed in the dark for 20 h. Sucrose and glucose levels in wild-type plants declined to <0.5 μg/mg fresh weight, whereas the levels in transgenic plants remained high, at 1.74 ± 0.33 and 7.57 ± 1.40 (±SE) mg/g fresh weight, respectively. Furthermore, at the end of the 20-h dark period, the mesophyll cells of the transgenic plants stained intensely for starch, whereas the mesophyll cells in leaves of wild-type plants contained no visible starch (Fig. 6).
Fig. 6.
Starch retention in plants kept in the dark for 20 h. Hand sections were stained for starch with iodine. (A) Wild-type tissue does not stain, indicating that all starch has been degraded. (B and C) Transgenic plants 8D and 12B, respectively, with heavy starch staining. (Scale bar: 25 μm.)
Discussion
The essence of the polymer trap model is that the conversion of sucrose to RFOs maintains the sucrose diffusion gradient from mesophyll cells to intermediary cells and prevents back diffusion. Other models of symplastic loading do not incorporate RFO synthesis into the proposed mechanisms (27, 28).
To test the model, we began by demonstrating that GAS gene expression in V. phoeniceum leaves is intermediary cell-specific. GAS has been immunolocalized to intermediary cells in Cucurbita pepo (29), and the expression of one of two GAS genes is intermediary cell-specific in A. reptans (16). Furthermore, the CmGAS1 promoter from melon drives reporter gene expression in minor vein CCs (30). GAS gene expression can be induced in other cell types in the leaves of various species in response to stress (15, 16, 20, 31). However, this finding appears to be an additional function of the biochemical pathway, distinct from the constitutive synthesis of transport RFOs in intermediary cells.
In preliminary experiments, we down-regulated VpGAS1 and VpGAS2 separately by using sequences unique to each mRNA. Although down-regulation was successful, as demonstrated by Northern blots, RFO levels were unaffected, and the plants grew normally (data not shown). This result is not surprising given the redundant expression patterns. However, as reported above, when both genes were coordinately targeted by using a conserved mRNA sequence, synthesis of RFOs was reduced to varying degrees in different transgenic lines. Because RFOs are synthesized in intermediary cells, it was expected that down-regulating the pathway would alter the proportions of sucrose and RFOs in the transport stream. This notion was proven true by exposing leaves to 14CO2 and analyzing the radiolabeled material transported into the petioles.
Although 14C-transport experiments are useful in analyzing the composition of the carbohydrates in the phloem stream, they do not provide information on the absolute amounts transported. However, it is clear that export was reduced in the transgenics because they exhibited well established symptoms of compromised phloem transport. All nine of the transgenics identified in the initial screen were unable to export efficiently, as evidenced by high soluble carbohydrate and starch levels at the end of a prolonged dark period. The most severely affected of the transgenics grew slowly and exhibited chlorosis and curling of leaf margins. Symptoms were especially severe in high light, consistent with an inability of the leaves to match export potential with carbohydrate synthesis. Down-regulation of loading in species that load from the apoplast also results in the symptoms described above (10–13, 32).
It is notable that seven of the nine studied transgenics did not display the most severe symptoms of slow growth and chlorosis. This result is not surprising given that plants that load from the apoplast often continue to grow normally in the face of loading inhibition, and only the most severely affected show visible indications of stress (10, 11, 13, 32). Chlorosis is caused by an oversupply of carbon in leaves (10, 11) and is therefore a secondary symptom of carbohydrate accumulation. Growth will be slowed only if residual loading activity and long-distance transport cannot provide sufficient carbon to meristems.
The residual driving force for transport is not clear in either solanaceous plants with partially down-regulated loading (10, 11, 13, 32) or the transgenic V. phoeniceum plants studied here. It is possible that there is enough of a safety factor built into apoplastic-loading mechanisms in solanaceous species that a reduced amount of activity suffices under most circumstances. In the case of V. phoeniceum, another possibility is that a small amount of apoplastic loading occurs normally or is induced by the down-regulation of symplastic loading. Ordinary CCs, similar in appearance to those in the minor veins of apoplastic-loading plants, are present in the minor veins of Verbascum (SI Fig. 7) (24), especially in large minor veins, and sucrose transporters have been immunolocalized to these cells in the RFO-transporting species Alonsoa meridionalis (33). Yet another possibility is that sucrose diffuses through plasmodesmata into intermediary cells and then into the SEs and is exported without conversion to RFOs. Passive entry of sucrose into the phloem has been documented in willow (34). Although transfer of photoassimilate to the phloem of transgenic V. phoeniceum plants clearly occurs, either actively or passively, it is clear that loading capacity is not sufficient to prevent hyperaccumulation of carbohydrates, and, in the plants with the most complete down-regulation, it is not sufficient to prevent slow growth and other symptoms of compromised export. In conclusion, this study demonstrates that when RFO synthesis is inhibited in V. phoeniceum, classic symptoms of compromised phloem transport ensue. The results indicate that the synthesis of RFOs in intermediary cells is necessary for efficient phloem transport, consistent with the polymer trap model.
Materials and Methods
Plant Material.
For plant transformation and in situ hybridization, V. phoeniceum L. cv. “Flush of White” (Garden Makers, Rowley, MA) seeds were surface-sterilized with 1% (vol/vol) sodium hypochlorite and 0.1% (vol/vol) Tween 20. They were germinated on Murashige and Skoog (MS) medium (35) supplemented with 3% (wt/vol) sucrose and Gamborg's vitamins (36) and solidified with 2.5 g/liter Gelrite (RPI Corporation, Mount Prospect, IL). Soil-grown plants were kept in a growth chamber set to a 16-h light/8-h dark cycle at 25°C, with an average light intensity of 62 μmol of photons per square meter per second or grown in a greenhouse with supplemental lighting.
cDNA Library Construction and Screening.
Mature leaves of V. phoeniceum were used for mRNA isolation by using the PolyATract kit (Promega, Madison, WI) per the manufacturer's instructions. cDNA was synthesized by using oligo(dT) primer and SuperScript RT (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The cDNA library was created by using the λTriplEx Library kit (BD Biosciences Clontech, Palo Alto, CA) and packaged by using Stratagene Gold packaging extract (Stratagene, La Jolla, CA).
A fragment of GAS was amplified by degenerate PCR using V. phoeniceum leaf cDNA as a template and the primers 5′-TTYGCYATGGCYTATTATGT-3′ and 5′-CCRGCRGCACAATAATG-3′ (IDT, Coralville, IA). The primer sequences were based on conserved regions identified by the amino acid alignment of GAS protein sequences found in the GenBank database. The PCR consisted of five cycles of 94°C for 1 min, 48°C for 4 min, and 68°C for 3 min, followed by 30 cycles of 94°C for 1 min, 50°C for 3 min, and 68°C for 3 min. A 400-bp fragment was amplified and cloned into pGEM-T Easy (Promega) according to the manufacturer's instructions.
The ≈2.4 × 105 clones were screened by standard methods (37) using a 32P-labeled probe specific to the fragment of GAS described above. The labeled probe was prepared by standard PCR by using dCT32P (PerkinElmer, Wellesley, MA) and Ex-Taq polymerase (TaKaRa; Pan Vera, Madison, WI). Sequencing of clones was performed by Cornell Bioresource Center (Ithaca, NY). Sequences were analyzed and aligned with Lasergene SeqMan software (DNASTAR, Madison, WI).
Nucleic Acid Extraction and Gel Blot Hybridization.
Total RNA was extracted from frozen tissue by mixing it with a 1:1 mixture of acid phenol/RNA extraction buffer (100 mM LiCl/100 mM Tris, pH 8/10 mM EDTA/1% SDS), followed by 24:1 chloroform/isoamyl alcohol extraction. RNA was precipitated with 4 M LiCl and resuspended in 0.5 ml of diethyl pyrocarbonate-treated water (Sigma–Aldrich, St. Louis, MO). All centrifugation steps were 10,000 × g at 4°C using a Sorvall rotor SS-34 (Newton, CT). The RNA concentration was determined by spectrophotometric measurements by using a Beckman DU-50 spectrophotometer (Beckman Coulter, Fullerton, CA).
Ten micrograms of denatured total RNA was run on a denaturing formaldehyde gel and transferred to Hybond N− nylon membrane (Amersham Biosciences, Piscataway, NJ) by standard methods (37). Membranes were hybridized with 32P-labeled probes, and hybridization was visualized by exposure to BioMax MS autoradiography film (Kodak, New Haven, CT).
Probes specific for each VpGAS gene were synthesized by PCR with dAT32P (PerkinElmer). Primers specific to unique regions of the 3′-UTR of each gene were used (VpGAS1, 5′-TTCATGCAGCTCTATTTTATTATCC-3′ and 5′-GATCAATACTTTGCAGCCAAGC-3′; VpGAS2, 5′GGCTTAATTAATCCCTACAACTTCG-3′ and 5′-TACTTTTAT T A T T CTTATTACATTTC-3′).
In Situ Hybridization.
In situ hybridization was performed as described by Friml et al. (38). Digoxigenin-labeled sense and antisense riboprobes corresponding to the 3′ UTR for each GAS gene were synthesized by using a digoxigenin RNA-labeling kit (Roche Applied Science, Indianapolis, IN) per the manufacturer's instructions. The 3′ UTR for each gene was amplified by PCR using the primer pairs described for Northern blot analysis and subcloned into the pGEM-T Easy vector. Sense probes were synthesized by linearization of plasmid DNA with NcoI (New England Biolabs, Ipswich, MA) and run-off transcription using SP6 polymerase (Roche Applied Science). Antisense probes were synthesized similarly by using SpeI restriction enzyme and T7 polymerase.
Ten-day-old axenically grown seedlings were collected and immediately used for whole-mount in situ hybridization. Probe hybridization occurred overnight at 50°C. Anti-digoxigenin-AP Fab fragments (Roche Applied Science) were used for the secondary detection reaction. Chromogenic development proceeded overnight by using BM Purple AP substrate (Roche Applied Science). The reaction was stopped by rinsing tissue in TE buffer (10 mM Tris-HCl/1.0 mM EDTA, pH 8.0), and samples were then applied to microscope slides with CrystalMount (Biomeda, Foster City, CA) mounting medium.
RNAi Vector Construction.
The silencing construct was made by standard methods (37) using the pHANNIBAL vector (39) and designated pAH-VpGAS2–1. The GAS sequence used to create pAH-VpGAS2–1 was amplified from a 94-bp sequence that shared 93% identity between VpGAS1 and VpGAS2. PCR was used to produce the necessary inverted repeat fragments with primers incorporating restriction sites at the 5′ ends (5′-CTCGAGAAGGGTTTGAGAAA-3′, 5′-GGTACCT C A A AATCTCACG-3′, 5′-GGATCCAAGGGTTTGAGAAA-3′, and 5′-ATCGATTCAAAATCTCACG-3′). Amplified fragments were subcloned into pGEM-T Easy (Promega) and subsequently digested with appropriate enzymes. Plasmid DNA containing VpGAS2 was used as a template for the amplification of fragments for the pAH-VpGAS2–1 construct. The silencing cassette was cloned into the NotI site of the binary vector pART27 (40) for Agrobacterium-mediated transformation into V. phoeniceum.
Plant Transformation.
RNAi constructs were introduced into A. tumefaciens strain GV3101 by standard heat-shock methods (37). T-DNA was transferred into V. phoeniceum by an Agrobacterium-mediated method. A. tumefaciens cultures for transformation were grown by inoculating 20 ml of LB medium containing 100 mg/liter kanamycin, 50 mg/liter rifampicin, and 50 mg/liter gentomycin with a single A. tumefaciens colony and allowing cultures to grow overnight at 28°C with shaking at 250 rpm. Cultures were then centrifuged for 10 min at 4°C at 3,300 × g and resuspended in 20 ml of liquid MS medium. Immature and mature leaves of axenically grown V. phoeniceum were excised, cut into 1-cm squares, bathed in the A. tumefaciens–MS suspension for 10 min, and blotted dry. The explants were placed on cocultivation medium, adapted from Turker et al. (41), containing MS, Gamborg's vitamins, 1 mg/liter napthalene acetic acid, 3 mg/liter 6-benzyl-aminopurine, and 2.5 g/liter Gelrite. After 2 days, the explants were transferred to selection medium containing MS, Gamborg's vitamins, 1 mg/liter napthalene acetic acid, 3 mg/liter 6-benzyl-aminopurine, 100 mg/liter kanamycin, and 400 mg/liter carbenicillin. Cultures were kept in a growth chamber with the same conditions as those for seed germination. Explants were transferred to fresh selection medium every 10 days.
When shoots developed, generally after 14 days or more, they were excised and placed in selection medium without 6-benzyl-aminopurine for root induction. A well developed root system usually formed within 14 days. Plantlets were transferred to a soil mixture of Metromix 360 (Scotts, Marysville, OH) and vermiculite (3:1) and acclimated slowly to growth chamber conditions. Finally, the plantlets were repotted in the same soil mixture in Deepots (cell size D40; Stuewe & Sons, Corvallis, OR), which are elongated pots that accommodate plants with prominent tap roots, and were grown to maturity in the greenhouse. Presence of the transgene was confirmed by PCR amplification of the NPT II gene by using genomic DNA as a template and NPT II-specific primers (5′-GAGGCTATTCGGCTATGACTG-3′ and 5′-ATCGGGAGCGGCGATACCGTA-3′) (42).
Expression Analysis.
Northern blot analysis was performed as described above. To prepare template for quantitative RT-PCR, 0.5–1 μg of total RNA was used to synthesize cDNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Quantitative RT-PCR was performed by using SYBR Green I technology on an PRISM 7000 sequence detection system (Applied Biosystems, Foster City, CA). Primer pairs for the target genes and the endogenous control were designed with Primer Express version 2.0 software (Applied Biosystems, Foster City, CA). The primer sequences were 5′-TAGGCTATTTGGTCTATTTTTGGTG-3′ and 5′-GATCAATACTTTGCAGCCAAGC-3′ for VpGAS1, 5′-TGGGTGTTTAACTATGTTTGTGGGCTA-3′ and 5′-TCAAATTTCAAACCAAACACAACCA-3′ for VpGAS2, and 5′-CGCGGAAGTTTGAGGCAATAA-3′ and 5′-TCGGCCAAGGCTATAGACTCGT-3′ for Verbascum thapsus L. 18S rRNA (accession no. AF207051).
The 20-μl quantitative RT-PCRs contained 1× Power SYBR Green PCR Master Mix (Applied Biosystems), 300 nM of each primer, and 1 μl of template cDNA. The amplification protocol consisted of an initial cycle of 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 55°C for 1 min. All samples were amplified in triplicate, and the average was used for further analysis. Data analysis was performed by using the relative standard curve method.
Analysis of Carbohydrate Content.
Sugars were extracted from source leaves and purified by ion-exchange chromatography as described (34). Sugar profiles were determined by HPLC by using a YMC-Pack Polyamine II column (YMC, Kyoto, Japan). Starch content was assessed by Lugol's iodine (43). One-centimeter-diameter pieces of tissue were removed with a cork borer, cleared in 70% ethanol, and stained for 30 min. Stained tissue was hand-sectioned in water and mounted in water on microscope slides.
14CO2 Labeling.
14CO2 labeling was performed as described (34). Petioles were trimmed of subtending lamina without damaging the vascular tissue and covered with aluminum foil to prevent photosynthesis. Blades were allowed to photosynthesize in the presence of 14CO2 for 15 min, followed by a chase of atmospheric air for 90 min. Blades and petioles were then separately frozen in liquid nitrogen and extracted (34). Sugars were separated by thin-layer chromatography (25). Labeled sugars were identified by autoradiography and removed by scraping the plates, and 14C was quantified by scintillation counting in a Beckman Coulter LS 6500.
Supplementary Material
Acknowledgments
We thank Dr. André Jagendorf for assistance with HPLC analysis, Dr. Susan Henry for use of HPLC equipment, Caren Chang for providing laboratory space during preliminary transformation experiments, Richard Medville for EM services, Cankui Zhang and Edwin Reidel for suggestions on the manuscript, Dr. Elena Kramer for inspiring discussion of in situ hybridization, and Denise Duclos for invaluable assistance with quantitative RT-PCR.
Abbreviations
- CC
companion cell
- GAS
galactinol synthase
- MS
Murashige and Skoog
- RFO
raffinose family oligosaccharide
- SE
sieve element.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF494114 and EF494115).
This article contains supporting information online at www.pnas.org/cgi/content/full/0707368104/DC1.
References
- 1.Vaughn MW, Harrington GN, Bush DR. Proc Natl Acad Sci USA. 2002;99:10876–10880. doi: 10.1073/pnas.172198599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiou TJ, Bush DR. Proc Natl Acad Sci USA. 1998;95:4784–4788. doi: 10.1073/pnas.95.8.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lalonde S, Tegeder M, Throne-Holst M, Frommer WB, Patrick JW. Plant Cell Environ. 2003;26:37–56. [Google Scholar]
- 4.Schulz A. In: Plasmodesmata. Oparka KJ, editor. Oxford: Blackwell; 2005. pp. 135–161. [Google Scholar]
- 5.Turgeon R, Ayre BG. In: Vascular Transport in Plants. Holbrook NM, Zwieniecki MA, editors. Oxford: Elsevier/Academic; 2005. pp. 45–67. [Google Scholar]
- 6.Turgeon R. BioScience. 2006;56:15–24. [Google Scholar]
- 7.Sondergaard TE, Schulz A, Palmgren MG. Plant Physiol. 2004;136:2475–2482. doi: 10.1104/pp.104.048231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turgeon R, Gowan E. Plant Physiol. 1990;94:1244–1249. doi: 10.1104/pp.94.3.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turgeon R. In: Recent Advances in Phloem transport and Assimilate Compartmentation. Bonnemain JL, Delrot S, Dainty J, Lucas WJ, editors. France: Ouest Editions, Nantes; 1991. pp. 18–22. [Google Scholar]
- 10.von Schaewen A, Stitt M, Schmidt R, Sonnewald U, Willmitzer L. EMBO J. 1990;9:3033–3044. doi: 10.1002/j.1460-2075.1990.tb07499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickinson CD, Altabella T, Chrispeels MJ. Plant Physiol. 1991;95:420–425. doi: 10.1104/pp.95.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR. Proc Natl Acad Sci USA. 2000;97:13979–13984. doi: 10.1073/pnas.250473797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riesmeier JW, Willmitzer L, Frommer WB. EMBO J. 1994;13:1–7. doi: 10.1002/j.1460-2075.1994.tb06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterbauer T, Richter A. Seed Sci Res. 2001;11:185–197. [Google Scholar]
- 15.Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. Plant J. 2002;29:417–426. doi: 10.1046/j.0960-7412.2001.01227.x. [DOI] [PubMed] [Google Scholar]
- 16.Sprenger N, Keller F. Plant J. 2000;21:249–258. doi: 10.1046/j.1365-313x.2000.00671.x. [DOI] [PubMed] [Google Scholar]
- 17.Downie B, Gurusinghe S, Dahal P, Thacker RR, Snyder JC, Nonogaki H, Yim K, Fukanaga K, Alvarado V, Bradford KJ. Plant Physiol. 2003;131:1347–1359. doi: 10.1104/pp.016386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi R, Joshee N, Kitagawa Y. Plant Mol Biol. 1994;26:339–352. doi: 10.1007/BF00039544. [DOI] [PubMed] [Google Scholar]
- 19.Kerr PS, Pearlstein RW, Schweiger BJ, Becker-Manley MF, Pierce JW. Int Patent. 1992 PCT/US92/06057. [Google Scholar]
- 20.Liu J-JJ, Krenz DC, Galvez AF, de Lumen BO. Plant Sci. 1998;134:11–20. [Google Scholar]
- 21.Zhao TY, Thacker R, Corum JW, Snyder JC, Meeley RB, Obendorf RL, Downie B. Physiol Plant. 2004;121:634–646. [Google Scholar]
- 22.Zhao TY, Martin D, Meeley RB, Downie B. Physiol Plant. 2004;121:647–655. [Google Scholar]
- 23.Ueda T, Coseoa MP, Harrell TJ, Obendorf RL. Plant Sci. 2005;168:681–690. [Google Scholar]
- 24.Turgeon R, Beebe DU, Gowan E. Planta. 1993;191:446–456. [Google Scholar]
- 25.Turgeon R, Gowan E. Planta. 1992;187:388–394. doi: 10.1007/BF00195663. [DOI] [PubMed] [Google Scholar]
- 26.Ayre BG, Keller F, Turgeon R. Plant Physiol. 2003;131:1518–1528. doi: 10.1104/pp.012054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gamalei Y, Pakhomova MV. Fiziol Rastenii. 1981;28:901–912. [Google Scholar]
- 28.Voitsekhovskaja OV, Koroleva OA, Batashev DR, Knop C, Tomos AD, Gamalei YV, Heldt HW, Lohaus G. Plant Physiol. 2006;140:383–395. doi: 10.1104/pp.105.068312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beebe DU, Turgeon R. Planta. 1992;188:354–361. doi: 10.1007/BF00192802. [DOI] [PubMed] [Google Scholar]
- 30.Haritatos E, Ayre BG, Turgeon R. Plant Physiol. 2000;123:929–937. doi: 10.1104/pp.123.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panikulangara TJ, Eggers-Schumacher G, Wunderlich M, Stransky H, Schoffl F. Plant Physiol. 2004;136:3148–3158. doi: 10.1104/pp.104.042606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bürkle L, Hibberd JM, Quick WP, Kühn C, Hirner B, Frommer WB. Plant Physiol. 1998;118:59–68. doi: 10.1104/pp.118.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knop C, Stadler R, Sauer N, Lohaus G. Plant Physiol. 2004;134:204–214. doi: 10.1104/pp.103.029264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turgeon R, Medville R. Proc Natl Acad Sci USA. 1998;95:12055–12060. doi: 10.1073/pnas.95.20.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murashigi T, Skoog F. Physiol Plant. 1962;136:473–497. [Google Scholar]
- 36.Gamborg OL, Miller RA, Ojima K. Exp Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 38.Friml J, Benkova E, Mayer U, Palme K, Muster G. Plant J. 2003;34:115–124. doi: 10.1046/j.1365-313x.2003.01705.x. [DOI] [PubMed] [Google Scholar]
- 39.Wesley SV, Helliwell CA, Smith NA, Wang M-B, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- 40.Gleave AP. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- 41.Turker AU, Camper ND, Gurel E. Plant. 2001;37:40–43. [Google Scholar]
- 42.Hamill JD, Rounsley S, Spencer A, Todd G, Rhodes MJC. Plant Cell Rep. 1991;10:221–224. doi: 10.1007/BF00232562. [DOI] [PubMed] [Google Scholar]
- 43.Gurr E. Practical Manual of Medical and Biological Staining Techniques. London: Leonard Hill; 1956. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.