Abstract
Basonuclin is a zinc-finger protein found in basal cells of the epidermis. In human keratinocyte cultures, basonuclin is susceptible to serine-phosphorylation and the addition of the phosphatase inhibitor, okadaic acid, promotes accumulation of basonuclin in the cytoplasm. The region of basonuclin containing the nuclear localization signal of basonuclin is necessary for nuclear localization of the protein and Ser-541, located immediately C-terminal to the nuclear localization signal, is the principal phosphorylation site in vitro. A nearly complete basonuclin transiently expressed in cultured keratinocytes localizes predominantly in the nucleus, but substitution of aspartic acid for Ser-541 promotes cytoplasmic localization. The same substitution of Ser-537 has a similar but weaker effect. Substitution of both serine residues by alanine leads to nuclear localization. These results show that nuclear localization of basonuclin depends on serine dephosphorylation, primarily of Ser-541. Different subcellular locations of basonuclin in different keratinocyte subtypes are therefore most likely to be controlled by the state of phosphorylation of Ser-541.
The keratinocyte is the principal cell type of squamous epithelia. Keratinocytes capable of multiplication are located in or close to the basal layer, whereas cells that lose their capacity to multiply migrate toward the surface of the epithelium as they undergo terminal differentiation.
Basonuclin is a zinc finger protein whose cDNA was cloned from cultured human keratinocytes (1). In the epidermis, it is found mainly in basal and immediately suprabasal keratinocytes. In human plantar epidermis and cells of the outer root sheath of the hair follicle, basonuclin was shown to be present in the nuclei (2), consistent with the presence of a putative nuclear localization signal (NLS) near the middle of the protein. The zinc fingers are in the form of three separated pairs, each consisting of CysX2CysX12HisX4His followed by CysX2CysX12HisX6–8His, and the NLS is located between the first and second pairs. There are similarities between the zinc fingers of basonuclin and those of Disco (3), and of PRDII-BFI (4), which is known to be a transcription factor.
Directly or indirectly, protein kinases affect the nuclear localization of proteins. One form of regulation is phosphorylation of a threonine or serine residue located near the NLS. We show here that the NLS of basonuclin is subject to specific serine phosphorylation and that, as a result, the protein becomes cytoplasmic. Of the 115 serine residues in basonuclin, a single serine seems to be the principal determinant of subcellular localization.
MATERIALS AND METHODS
Cultivation of Keratinocytes.
Unless otherwise stated, keratinocytes were grown in FAD medium (5) in the presence of lethally irradiated 3T3 cells (6). All keratinocytes were of human origin. Except in the case of SCC-13, derived from an epidermoid carcinoma (7), the keratinocytes used were of a normal diploid strain (YF29). Cultures were fed every 3 days with medium containing epidermal growth factor at 10 ng/ml. For growth of small colonies in the absence of 3T3 cells, keratinocyte growth medium (Clonetics, San Diego) was used. For depleting cells of methionine or phosphate, DMEM (BRL) was prepared free of methionine and cysteine or of phosphate, and supplemented with 0.5% fetal bovine albumin and necessary additives.
Phosphoamino Acid Analysis.
Immunoprecipitated [32P]basonuclin was subjected to SDS/PAGE separation and transferred to poly(vinylidene difluoride) (Millipore) in a standard Tris⋅glycine-methanol buffer. The basonuclin on the membrane was hydrolyzed in 6 M HCl at 110°C for 1 hr, dried, and dissolved in water (8). The sample was applied to a Kodak thin-layer plate, and the products were separated in buffer at pH 3.5 (pyridine/acetic acid/H2O = 1:10:189) for 60 min at 800 V, or by chromatography (NH4OH/H2O/isobutyric acid = 2.5:75:200, buffered at pH 1.9) for about 4.5 hr (9).
Preparation of Keratinocyte Extracts and Phosphorylation Reaction in Vitro. A confluent keratinocyte culture was scraped, and the cells were collected by brief centrifugation.
To the pellet, 20 mM Tris⋅HCl buffer, pH 7.4, containing 10% glycerol and the protease inhibitors, was added. The suspension was briefly sonicated and centrifuged for 2 min at 10,000 × g. The resulting extract was used as a source of protein kinase. The phosphotransfer reaction was carried out in a reaction mixture for protein kinase C (10) containing 10 μM [γ-32P]ATP at 25°C. The reaction was initiated by adding the keratinocyte extract and terminated by adding a one-third volume of 3× SDS/PAGE loading buffer.
Construction of Plasmids.
For construction of a plasmid-encoding basonuclin, the coding region was excised from plasmid pM752A173 (1) with NarI and HindIII, and the NarI site was modified to an EcoRI site. The EcoRI-HindIII fragment encodes the entire basonuclin sequence except for the N-terminal three amino acids. This fragment was ligated to plasmid pET-28a(+) (Novagen), or pFLAG-CMV-2 (Kodak), or pEGFP-C2 (CLONTECH), to yield an overproducer in bacteria (pHUB2), or an expression plasmid encoding an N-terminal FLAG tag (pHUB4), or GFP tag (pHUB19) fused to basonuclin. To obtain truncated N-terminal fragments from pHUB4, the enzymes EcoRV, AatII, or SphI were used to cleave at positions 202, 445, or 708.
To obtain substitution mutants, the EcoRI-HindIII fragment of basonuclin was ligated to pBlueScript KS+ to yield pHUB3 and the template single-stranded DNA of plasmids was obtained (11). Nucleotide-directed mutagenesis then was carried out according to Kunkel et al. (12) using a Muta-Gene mutagenesis kit (Bio-Rad) to convert to aspartic acid, Ser-537 (TCC → GAC), Ser-540 (TCC → GAC), and Ser-541 (AGT → GAT). Similarly, Lys-535 was converted to asparagine (AAG → AAT), and Lys-536 to glutamic acid (AAA → GAA). To minimize secondary mutation, each plasmid with the specific mutation was constructed by replacing the AatII-SphI fragment of pHUB4 with a corresponding fragment encoding the mutation, and the absence of secondary mutation was confirmed by sequencing the replaced sequence.
To amplify the partial basonuclin gene encoding a 33-aa sequence containing the NLS, PCR was carried out for 30 cycles with two 33-mer primers in the high-fidelity PCR reaction mixture (Boehringer Mannheim), using pHUB4 as a template. The PCR fragment was inserted 3′ of the glutathione S-transferase (GST) sequence of pGST-5X-3 (Pharmacia) so as to yield plasmids encoding GST-SSS. To prepare mutant plasmids, the 33-codon sequence was replaced with the mutant fragments amplified by PCR. No secondary mutation was found in this region.
Transient Expression of Fusion Proteins of Basonuclin and the Mutant Proteins.
Cultures were transfected with appropriate plasmid (2.5 μg per 3-cm-diameter Petri dish or 6 μg per 6-cm-diameter Petri dish) with DOTAP (N[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate) (Boehringer Mannheim) or by the calcium phosphate method (Stratagene). Cultures were harvested 18 to 22 hr after transfection and fixed for immunostaining.
Purification of GST-Fusion Proteins.
Bacterial strain BL21 was transformed with a plasmid encoding each fusion protein and used to overproduce the protein in Luria–Bertani medium. The bacterial cell pellet was suspended in 10 mM phosphate buffer, pH 7.0, containing 30 mM NaCl and 10 mM EDTA, and treated with 0.5 mg/ml lysozyme for 15 to 30 min on ice, in the presence of added protease inhibitors (1 mM phenylmethylsulfonyl fluoride/1 μM leucinthiol/0.5 μg/ml leupeptin/0.7 μg/ml pepstatin/1 μg/ml aprotinin). The mixture was sonicated and centrifuged for 10 min at 14,000 × g. From the supernatant, the fusion protein was purified according to the manufacturer’s protocol (Pharmacia).
Immunoprecipitation.
Keratinocyte cultures were washed twice, immediately lysed by addition of hot 0.1 M Tris/0.5% SDS buffer, pH 7.4, to 12.7 μl/cm2 of dish area, heated at 95°C for 3 min, and chilled on ice. To avoid proteolysis, the same mixture of protease inhibitors was added to the lysate. The mixture was centrifuged at 14,000 × g for 30 min at room temperature. The cleared supernatant was subjected to immunoprecipitation according to ref. 13. The immunoprecipitate was dissolved in loading buffer and applied to a well of 5.5% polyacrylamide gel for SDS/PAGE analysis. For autoradiography of [35S]methionine, the signal was intensified with 2,5-diphenyloxazole/dimethyl sulfoxide (NEN/DuPont). 32P incorporation was analyzed by autoradiography or the Phosphorimager (Molecular Dynamics).
Basonuclin and DNA Staining.
Immunological staining for basonuclin was carried out as described previously on a glass coverslip or on a plastic culture dish overnight at 4°C (2) using the newly raised polyclonal antibody. FLAG-basonuclin was stained with a mAb against FLAG (Kodak) and DNA was stained with Hoechst 33258.
RESULTS
Posttranslational Modification of Basonuclin by Phosphorylation.
Earlier work on the immunocytology of basonuclin was carried out using an antiserum directed against a 131-aa sequence located near the N-terminal end (2). For the present work, we prepared a new antiserum using as antigen a polypeptide containing 991-aa residues of the total of 994. A plasmid encoding this sequence (pHUB2) was constructed in pET-28a(+) and introduced into Escherichia coli. The protein was overproduced by induction, highly purified by SDS/PAGE, and injected into two rabbits. The resulting antiserum recognized basonuclin of keratinocytes by Western blotting, immunoprecipitation, and immunostaining. The antiserum was purified by affinity chromatography before use.
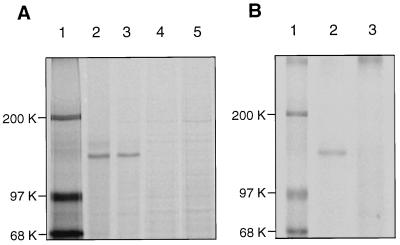
To explore the possibility of posttranslational modification of basonuclin, keratinocyte cultures (about 50% confluent) were starved for methionine and pulse-labeled with [35S]methionine for 2 hr. The cultures were lysed by heating in a Tris/SDS buffer. Parallel cultures were similarly pulse-labeled and then incubated for 3 hr in medium containing unlabeled methionine. The basonuclin was immunoprecipitated and subjected to SDS/PAGE analysis. The immunoprecipitates contained predominantly a single cellular protein with mobility corresponding to about 125 kDa (Fig. 1A, lane 2). Labeled protein did not appear in the immunoprecipitate if the antiserum was preincubated with an excess of the recombinant unlabeled basonuclin (Fig. 1A, lane 4).
Figure 1.
Posttranslational modification of basonuclin in keratinocytes. (A) Reduction of electrophoretic mobility. Two half-confluent cultures were washed twice with 10 mM Hepes buffer, pH 7.5, containing 150 mM sodium chloride (HBS), starved for 30 min in the Met- and Cys-free medium, and labeled for 2 hr with [35S]methionine (1,000 Ci/mmol) at a concentration of 0.125 mCi/ml of culture medium. One of the labeled cultures was washed twice with Tris-buffered saline and immediately dissolved in Tris⋅SDS buffer. The other culture was washed with medium containing unlabeled methionine and cysteine to prevent further incorporation of [35S]methionine, incubated for 3 hr in that medium, washed, and dissolved in Tris/SDS buffer. Immunoprecipitation was carried out with antiserum. The mobility of the modified basonuclin after pulse–chase (lane 3) is slightly reduced in comparison with that of freshly synthesized basonuclin (lane 2). Neutralization of the antiserum with 0.3 mg/ml of the unlabeled recombinant basonuclin (HUB2) eliminated all basonuclin bands (lanes 4 and 5). Lane 1, molecular mass markers. (B) Phosphorylation of basonuclin. A quarter-confluent culture was washed twice with HBS and starved for phosphate for 18 hr. The culture, then about 50% confluent, was labeled for 5 hr with 32Pi (1 Ci/mmol) at a concentration of 0.125 mCi/ml of culture medium. The culture was washed twice with TBS, lysed immediately in Tris⋅SDS buffer, and the basonuclin was immuno-precipitated and resolved by electrophoresis. The basonuclin is clearly labeled with 32P (lane 2). Preliminary neutralization of the antiserum with unlabeled basonuclin eliminated the 32P-labeled basonuclin band (lane 3). Lane 1, molecular mass markers.
Pulse–chase labeling resulted in a slight, but reproducible, reduction of the electrophoretic mobility of the labeled basonuclin (Fig. 1A, lane 3). Further incubation of the labeled basonuclin for up to 9 hr did not result in further reduction of its mobility.
To determine whether the reduction of mobility was associated with phosphorylation, keratinocyte cultures were starved overnight for inorganic phosphate and labeled with 32Pi. When the basonuclin was immunoprecipitated and electrophoretically resolved, it was found to be labeled with 32P (Fig. 1B, lane 2), and the label was absent when immunoprecipitation was preceded by the addition of an excess of unlabeled recombinant basonuclin (Fig. 1B, lane 3).
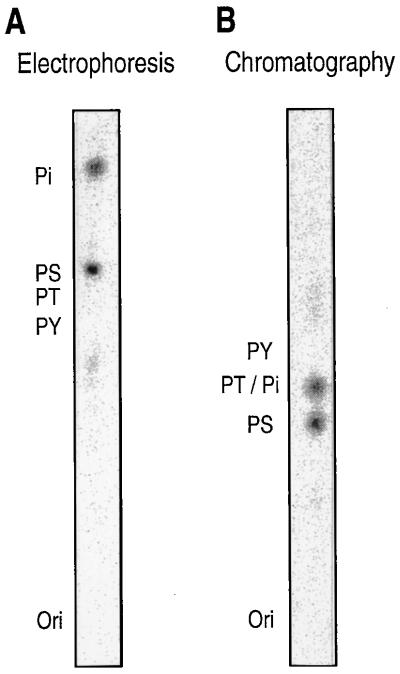
To identify the phosphorylated amino acid residues of basonuclin, 32P-labeled basonuclin synthesized by keratinocytes was immunoprecipitated, acid-hydrolyzed, and analyzed by electrophoresis. The 32P-label comigrated with phosphoserine (Fig. 2A), but not with phosphotyrosine or phosphothreonine. The presence of 32P-labeled serine was confirmed by thin-layer chromatography (Fig. 2B). Basonuclin is therefore phosphorylated only at serine residues.
Figure 2.
Keratinocytes phosphorylate only serine residues of basonuclin. A quarter-confluent culture was starved for phosphate for 18 hr, labeled with 32Pi (9,000 Ci/mmol) at concentration of 0.25 mCi/ml of culture medium for 3 hr, washed, and then lysed. The basonuclin was purified by immunoprecipitation and SDS/PAGE, and then subjected to phosphoamino acid analysis. Added unlabeled phosphoamino acids were detected with ninhydrin, and 32P was located with the PhosphorImager. (A) The positions of electrophoretically resolved phosphoserine, phosphothreonine, phosphotyrosine, and inorganic phosphate are indicated by PS, PT, PY, and Pi. Ori indicates origin. Only serine and inorganic phosphate are labeled with 32P. (B) Chromatography gives the same result.
Phosphorylation Promotes Cytoplasmic Localization of Basonuclin.
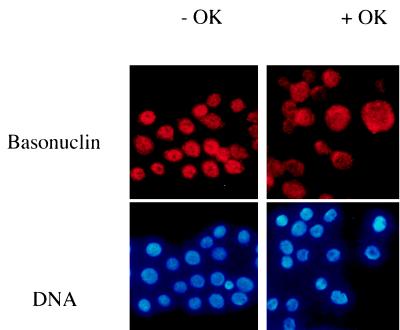
To investigate the relation between the state of phosphorylation of basonuclin and its subcellular localization, we added to keratinocyte cultures a potent phosphatase inhibitor, okadaic acid, which is known to increase phosphorylation of cytoplasmic proteins of keratinocytes (14). Under our conditions, okadaic acid at 1 μM increased the level of 32P in basonuclin by 40–100% within 1 hr (data not shown). The addition of okadaic acid moved basonuclin from nucleus to cytoplasm (Fig. 3). This result suggested that basonuclin accumulation in the nucleus depends on an unphosphorylated state of the protein.
Figure 3.
Effect of okadaic acid on nuclear localization of basonuclin. Keratinocytes growing in KGM medium on a glass coverslip without 3T3 support were treated with 1 μM okadaic acid in 0.1% dimethyl sulfoxide (final concentration) for 1 hr. The control culture contained the same concentration of dimethyl sulfoxide alone. Keratinocytes were stained for basonuclin (red) and for DNA with Hoechst (blue).
The Region Containing the NLS Is Required for Nuclear Localization of Basonuclin.
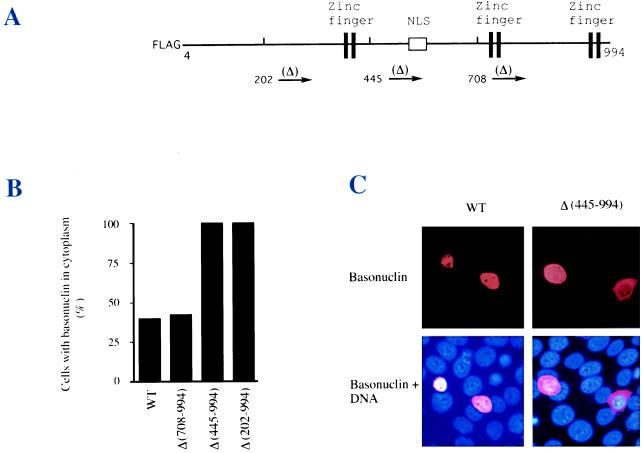
To determine if the putative NLS is functional, we constructed several plasmids, each of which encoded a different deletion mutant of basonuclin fused to a FLAG tag, so that the expression of the plasmid-encoded basonuclin could be distinguished from the endogenous basonuclin. These mutants were expressed under control of the cytomegalovirus promoter (Fig. 4A) in an established keratinocyte cell line (SCC13) whose basonuclin localizes normally to the nucleus. Cells transfected with wild-type basonuclin showed nuclear localization of the protein (Fig. 4C), although 40% of cells also contained detectable cytoplasmic basonuclin (Fig. 4B), probably due to overexpression from the plasmid. Deletion of all basonuclin sequence C-terminal of residue 708 had no effect on the nuclear localization, but deletion of all sequence C-terminal of residues 445 or 202 resulted in cytoplasmic localization in every transfected cell. Cells with strong cytoplasmic staining for the deleted basonuclin, as in Fig. 4C, were never seen in the wild-type control. Therefore, the sequence located between residues 445 and 708, containing the putative NLS, is required for nuclear localization.
Figure 4.
Requirement of the putative NLS for nuclear import. (A) Basonuclin sequence with a FLAG tag and sites of three different deletions, beginning at indicated residue numbers and extending to C-terminal end (residue 994). (B) FLAG-basonuclins each bearing one of the three deletions were expressed for 22 hr after transfection in the SSC-13 line of human keratinocytes (7). The expressed basonuclins were visualized by immunostaining with mAb against the FLAG tag. Length of deletion sequences is indicated in parentheses. Two deletion proteins lacking the NLS, Δ(445–994) and Δ(202–994), were localized in the cytoplasm at a higher concentration than in the nucleus. (C) Examples of nuclear and cytoplasmic basonuclin from experiment described in B. Cell number scored for each transfection was: WT, 82; Δ(708–994), 61; Δ(445–994), 57; and Δ(202–994), 30.
Ser-541 Is the Most Specific Phosphorylation Site.
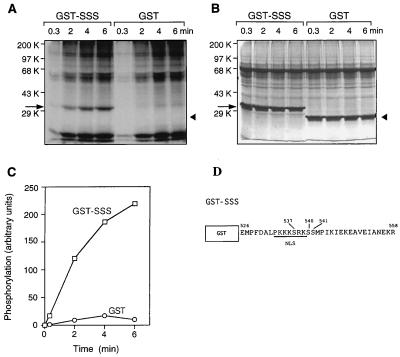
Existence of serine residues within the NLS and immediately C-terminal of it suggested that these residues might be sites of phosphorylation. To identify the phosphorylated serine residues, DNA encoding the same 33-aa residue peptide containing the NLS and its adjacent serine residues was fused to DNA encoding GST (Fig. 5D); the resulting protein (GST-SSS) was overproduced in an E. coli strain, highly purified, and used as the substrate for phosphorylation in vitro. Addition of a keratinocyte extract together with γ-32P-labeled ATP resulted in phosphorylation of the fusion protein in a time-dependent reaction (Fig. 5 A andC), whereas GST itself, which contains eight serine residues, was not appreciable phosphorylated. Therefore, phosphorylation occurs at one or more of the serine residues in the 33-aa sequence containing the NLS.
Figure 5.
Phosphorylation of basonuclin peptide in vitro. Twenty-five micrograms of GST-SSS or GST alone was incubated at 25°C with 85 μg of keratinocyte extract protein in 100 μl of reaction mixture containing components necessary for protein kinase C activity and 1 μM okadaic acid. A 20-μl sample was withdrawn at intervals, the reaction was terminated, and the products were analyzed by SDS/PAGE (10% gel). Arrows indicate location of GST-SSS. Arrowheads indicate position of GST. (A) Time course of 32P accumulation by autoradiography. (B) Proteins stained with Coomassie blue. (C) Plot of time course. Virtually no 32P is incorporated into the GST alone. (D) A scheme of GST-SSS fusion protein.
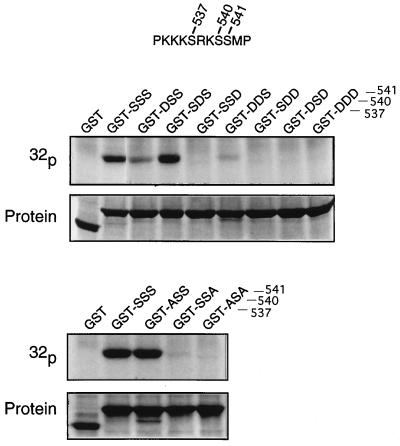
We then determined by mutations which serine residues were phosphorylated (Fig. 6). Substitution of aspartic acid for Ser-540 did not affect phosphorylation, but the same substitution at Ser-537 reduced it, and at Ser-541 eliminated it. Double-substitution of Ser-537 and Ser-540 weakened phosphorylation at Ser-541, but a double-substitution that included Ser-541 prevented all phosphorylation. Similarly, substitution of alanine for Ser-537 did not affect phosphorylation, but the same substitution at serine Ser-541 nearly eliminated it. As in the case of the aspartic acid substitutions, double-substitution with alanine at Ser-537 and Ser-541 eliminated phosphorylation completely. These experiments show that although Ser-537 has slight reactivity, Ser-541 is the principal phosphorylation site.
Figure 6.
Phosphorylation in vitro of a basonuclin fragment containing serine substitutions. Five micrograms of the polypeptide used in Fig. 5, each bearing a substitution for one or two serines, was incubated with 17 μg of keratinocyte extract in the reaction mixture for 15 min at 25°C. Radioactivity was detected by autoradiography and protein by Coomassie blue staining. The three serine residues are numbered. (Upper) Aspartic acid substitutions. (Lower) Alanine substitutions.
Effect of Mutations on the Nuclear Localization of Basonuclin in Intact Cells.
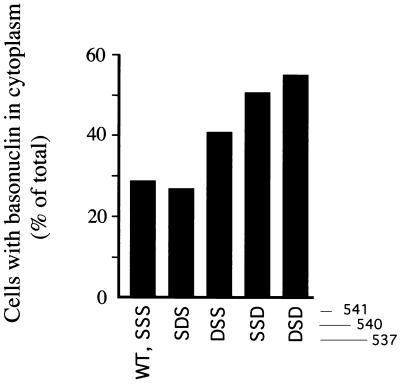
We examined the effect of these serine substitutions on the localization of virtually complete basonuclin labeled with a FLAG tag (Fig. 4A). When the wild-type basonuclin was transfected into keratinocyte cultures, basonuclin bearing the FLAG tag was present in the nucleus. However, 29% of the cells contained a small amount of the protein in the cytoplasm as well (Fig. 7). As in the experiment described in Fig. 4C, we attribute this to overexpression of the plasmid by the strong promoter and saturation of the nuclear import system. Substitution of Ser-540 with aspartic acid did not promote further cytoplasmic localization, but substitution of Ser-537 and particularly Ser-541 led to increasing localization to the cytoplasm; in the double-mutation, 55% of the cells showed cytoplasmic basonuclin. These results are in agreement with earlier findings (15, 16) that acidic substitutions such as glutamic or aspartic acid mimic phosphothreonine in promoting cytoplasmic localization of a protein bearing an NLS. Alanine substitutions at the same positions led to no increase in cytoplasmic localization.
Figure 7.
Effect of aspartic acid substitutions on cytoplasmic localization of complete basonuclin. (A) A keratinocyte culture (YF29) growing in the presence of 3T3 support was transfected with plasmid (pHUB4), encoding FLAG-basonuclin bearing serine substitutions. After 18 hr, the number of cells with cytoplasm stained by anti-FLAG antibody was scored. Total number of cells scored for each transfection was: SSS, 73; SDS, 67; DSS, 61; SSD, 93; DSD, 71. Substitutions at Ser-540 and Ser-541 led to an increase in number of cells with stained cytoplasm.
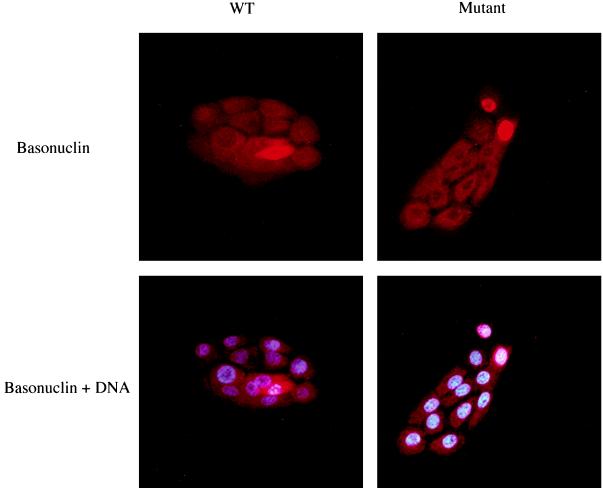
By modifying the culture conditions, it could be shown that alanine substitution promoted nuclear localization of basonuclin. In human keratinocytes cultivated in keratinocyte growth medium at low cell density in the absence of supporting 3T3 cells (a suboptimal culture system for human keratinocytes), basonuclin is localized in the cytoplasm (Fig. 8). When such cultures were transfected with plasmid encoding basonuclin with alanine substitutions for Ser-537 and Ser-541, the protein localized in the nucleus while wild-type plasmid-expressed basonuclin and endogenous basonuclin remained cytoplasmic. From these results we concluded that the determinant of basonuclin localization is the state of phosphorylation of Ser-541 and Ser-537, the latter having a minor role.
Figure 8.
Effect of alanine substitution in promoting nuclear localization of basonuclin. Human keratinocytes grown at low density in keratinocyte growth medium without 3T3 support were transfected with plasmid (pHUB19) encoding GFP basonuclin containing the mutations Ser-537-Ala, Ser-541-Ala. It was found that as the basonuclin overproduced from the plasmid was more brightly stained than the endogenous basonuclin, its localization could be more easily demonstrated by the antiserum to basonuclin than by the GFP tag. After an 18-hr incubation without 3T3 support in FAD medium (suboptimal conditions), the culture was fixed and stained for basonuclin and DNA. Under these conditions, endogenous basonuclin remains cytoplasmic. Wild-type basonuclin expressed from the plasmid is also cytoplasmic, whereas the mutant is strongly concentrated in the nucleus.
DISCUSSION
Regulation of Nuclear Import of Basonuclin.
Most proteins of large size, including transcription factors, protein kinases, structural proteins, and subunits of the proteosome are transported through the nuclear pore by energy-dependent pathways using special machinery (17–19). Two main classes of import machinery have been identified: one is the karyopherin α-β complex, which requires an NLS of the simian virus 40 class (20–23), and the other is the transportin-based system, demonstrated for proteins bearing the M9 signal (24). Because the NLS of basonuclin has an amino acid sequence resembling that of the simian virus 40 T antigen (except for the presence of Ser-537 and two serine residues located immediately downstream of the NLS) we assume that basonuclin will be imported by means of the karyopherin α-β complex. Our experiments show that unphosphorylated Ser-541 of basonuclin is the strongest determinant of nuclear import. The dominance of the NLS region in determining nuclear import is consistent with the fact that a sequence of 28 amino acids, including the NLS and adjacent upstream and downstream sequence, is totally conserved in mouse and human basonuclin, whereas some of the other parts of the molecule are less completely conserved (25).
We have compared the phosphorylation sites adjacent to the NLS for different nuclear proteins. In the case of simian virus 40 T antigen, the site is an upstream threonine (16) and in the case of V-Jun (26) and lamin B2 (27), it is one or two upstream serine residues. For Rum1 (28), ZEX (29), and proteosome subunits (19), it has been shown that there is an adjacent downstream serine, as in basonuclin, but it is not known whether these are subject to phosphorylation. It remains to be determined whether Ser-541 of basonuclin has any counterpart in other proteins.
The Physiological Significance of Serine Phosphorylation.
Subcellular localization of proteins may be subject to short-term changes, as for cell-cycle dependent proteins (26, 30) or may be stable over the long term, as in localization differences between different cell types (31). In the case of basonuclin, there is both short-term and long-term regulation of its localization. For example, we show in Fig. 8 that although, under optimal conditions for multiplication, cultured keratinocytes regularly have basonuclin localized to the nucleus, growth in keratinocyte growth medium in the absence of supporting 3T3 cells results in its cytoplasmic localization. An analogous effect has been reported for a homeodomain protein Exd in Drosophila. This protein is synthesized in endoderm and localized in nucleus when two secreted signaling proteins Dpp and Wg are provided by neighboring visceral mesoderm, but in cytoplasm when the signaling proteins are not available (32).
We will describe elsewhere stable differences in the pattern of localization of basonuclin between different regions of epidermis and between keratinocytes of epidermis and hair follicles. All of these differences, which may reflect different states of cellular activation, are likely to be the result of the specific serine phosphorylation–dephosphorylation described here.
Acknowledgments
We thank Dr. John Rush of the Department of Genetics, Harvard Medical School, for his advice on phosphoamino acid analysis. This investigation was aided by a grant from the National Cancer Institute.
ABBREVIATIONS
- NLS
nuclear localization signal
- GST
glutathione S-transferase
References
- 1.Tseng H, Green H. Proc Natl Acad Sci USA. 1992;89:10311–10315. doi: 10.1073/pnas.89.21.10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tseng H, Green H. J Cell Biol. 1994;126:495–506. doi: 10.1083/jcb.126.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heilig J S, Freeman M, Laverty T, Lee K J, Campos A R, Rubin G M, Steller H. EMBO J. 1991;10:809–815. doi: 10.1002/j.1460-2075.1991.tb08013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keller A D, Maniatis T. Mol Cell Biol. 1992;12:1940–1949. doi: 10.1128/mcb.12.5.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon M, Green H. Cell. 1985;40:677–683. doi: 10.1016/0092-8674(85)90216-8. [DOI] [PubMed] [Google Scholar]
- 6.Rheinwald J G, Green H. Cell. 1975;6:317–330. doi: 10.1016/0092-8674(75)90183-x. [DOI] [PubMed] [Google Scholar]
- 7.Rheinwald J G, Beckett M A. Cancer Res. 1981;41:1657–1663. [PubMed] [Google Scholar]
- 8.Kamps M P, Sefton B M. Anal Biochem. 1989;176:22–27. doi: 10.1016/0003-2697(89)90266-2. [DOI] [PubMed] [Google Scholar]
- 9.Cooper J A, Sefton B M, Hunter T. Methods Enzymol. 1983;99:387–402. doi: 10.1016/0076-6879(83)99075-4. [DOI] [PubMed] [Google Scholar]
- 10.Huang K-P, Huang F L, Nakabayashi H, Yoshida Y. J Biol Chem. 1988;263:14839–14845. [PubMed] [Google Scholar]
- 11.Iuchi S, Lin E C C. J Bacteriol. 1992;174:3972–3980. doi: 10.1128/jb.174.12.3972-3980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 13.Misra R P, Rivera V M, Wang J M, Fan P-D, Greenberg M E. Mol Cell Biol. 1991;11:4545–4554. doi: 10.1128/mcb.11.9.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasahara K, Kartasova T, Ren X-Q, Ikuta T, Chida K, Kuroki T. J Biol Chem. 1993;268:23531–23537. [PubMed] [Google Scholar]
- 15.Li X, Shou W, Kloc M, Reddy B A, Etkin L D. J Cell Biol. 1994;124:7–17. doi: 10.1083/jcb.124.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jans D A, Ackermann M J, Bischoff J R, Beach D H, Peters R. J Cell Biol. 1991;115:1203–1212. doi: 10.1083/jcb.115.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melchior F, Gerace L. Curr Opin Cell Biol. 1995;7:310–318. doi: 10.1016/0955-0674(95)80084-0. [DOI] [PubMed] [Google Scholar]
- 18.Gorlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 19.Nederlof P M, Wang H-R, Baumeister W. Proc Natl Acad Sci USA. 1995;92:12060–12064. doi: 10.1073/pnas.92.26.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adam S A, Gerace L. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- 21.Adam E J H, Adam S A. J Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moroianu J, Hijikata M, Blobel G, Radu A. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore M S, Blobel G. Cell. 1992;69:939–950. doi: 10.1016/0092-8674(92)90613-h. [DOI] [PubMed] [Google Scholar]
- 24.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 25.Matsuzaki, K., Iuchi, S. & Green, H. (1997) Gene, in press. [DOI] [PubMed]
- 26.Tagawa T, Kuroki T, Vogt P K, Chida K. J Cell Biol. 1995;130:255–263. doi: 10.1083/jcb.130.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hennekes H, Peter M, Weber K, Nigg E A. J Cell Biol. 1993;120:1293–1304. doi: 10.1083/jcb.120.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreno S, Nurse P. Nature (London) 1994;367:236–242. doi: 10.1038/367236a0. [DOI] [PubMed] [Google Scholar]
- 29.Schneider-Gadicke A, Beer-Romero P, Brown L G, Mardon G, Luoh S-W, Page D C. Nature (London) 1989;342:708–711. doi: 10.1038/342708a0. [DOI] [PubMed] [Google Scholar]
- 30.Moll T, Tebb G, Surana U, Robitsch H, Nasmyth K. Cell. 1991;66:743–758. doi: 10.1016/0092-8674(91)90118-i. [DOI] [PubMed] [Google Scholar]
- 31.Ghosh S, Baltimore D. Nature (London) 1990;344:678–682. doi: 10.1038/344678a0. [DOI] [PubMed] [Google Scholar]
- 32.Mann R S, Abu-Shaar M. Nature (London) 1996;383:630–633. doi: 10.1038/383630a0. [DOI] [PubMed] [Google Scholar]