Abstract
Anaplasma phagocytophilum is the etiologic agent of human granulocytic anaplasmosis (HGA), one of the major tick-borne zoonoses in the United States. The surface of A. phagocytophilum plays a crucial role in subverting the hostile host cell environment. However, except for the P44/Msp2 outer membrane protein family, the surface components of A. phagocytophilum are largely unknown. To identify the major surface proteins of A. phagocytophilum, a membrane-impermeable, cleavable biotin reagent, sulfosuccinimidyl-2-[biotinamido]ethyl-1,3-dithiopropionate (Sulfo-NHS-SS-Biotin), was used to label intact bacteria. The biotinylated bacterial surface proteins were isolated by streptavidin agarose affinity purification and then separated by electrophoresis, followed by capillary liquid chromatography-nanospray tandem mass spectrometry analysis. Among the major proteins captured by affinity purification were five A. phagocytophilum proteins, Omp85, hypothetical proteins APH_0404 (designated Asp62) and APH_0405 (designated Asp55), P44 family proteins, and Omp-1A. The surface exposure of Asp62 and Asp55 was verified by immunofluorescence microscopy. Recombinant Asp62 and Asp55 proteins were recognized by an HGA patient serum. Anti-Asp62 and anti-Asp55 peptide sera partially neutralized A. phagocytophilum infection of HL-60 cells in vitro. We found that the Asp62 and Asp55 genes were cotranscribed and conserved among members of the family Anaplasmataceae. With the exception of P44-18, all of the proteins were newly revealed major surface-exposed proteins whose study should facilitate understanding the interaction between A. phagocytophilum and the host. These proteins may serve as targets for development of chemotherapy, diagnostics, and vaccines.
Human granulocytic anaplasmosis (HGA) (formerly human granulocytic ehrlichiosis) has been recognized as a zoonotic disease of public health importance and has become one of the most common tick-borne zoonoses in the United States and Europe (1, 13, 47). HGA is an acutely febrile systemic illness accompanied by hematologic and liver enzyme abnormalities. It can cause severe and potentially fatal illness, especially in immunocompromised and elderly people (15). The etiologic agent of HGA, Anaplasma phagocytophilum, is a gram-negative, obligate intracellular bacterium, which initially was known as an organism having tropism for granulocytes (9, 22); it more recently has been shown to infect endothelial cells as well (39).
The surface of A. phagocytophilum provides an important interface for A. phagocytophilum-host interactions, including adherence to and internalization of host cells (45, 58), inhibition of neutrophil apoptosis (6, 18, 20, 60), inhibition of reactive oxygen species production (38), scavenging of exogenous superoxide (M. Herron and J. Goodman, Abstr. Am. Soc. Rickettsiol.-Bartonella Emerg. Pathog. Group 2001 Joint Conf., abstr. 50, 2001) (8), exhibition of antigenic variation to avoid the host immune response (32, 36, 59, 62), mediation of neutralization of infection (27, 58), sensing of the bacterial environment, and exchange of nutrients and metabolites with the host cytoplasm (24). A. phagocytophilum has lost all genes required for the biosynthesis of lipopolysaccharide and most genes required for the biosynthesis of peptidoglycan (30, 48). What's more, there is no pilus or capsule on the surface of organisms in the family Anaplasmataceae (49), suggesting that outer membrane proteins play a crucial role in bacterial interactions with host cells. A. phagocytophilum outer membrane proteins have become the central focus as potential drug targets and as candidates for differential diagnostic antigens and novel vaccines. A series of A. phagocytophilum proteins have been shown to be immunoreactive by Western blotting, such as AnkA, Msp5, GroEL, and approximately 44-, 55-, 72-, 100-, 130-, and 160-kDa proteins (2, 12, 28, 54-56). However, surface exposure of these proteins and their identities have not been well elucidated.
The outer membrane proteins of A. phagocytophilum have not been characterized systematically. The Omp-1/P44/Msp2 superfamily is the most-studied outer membrane protein family of A. phagocytophilum. The A. phagocytophilum genome has three omp-1, one msp2, two msp2 homolog, one msp4, and 113 p44 loci encoding proteins belonging to this superfamily (23). Each A. phagocytophilum P44 consists of a central hypervariable region and conserved flanking sequences (34, 36, 61). Expression of p44 paralog genes occurs via a unique gene conversion mechanism involving the RecF pathway (35, 62). Compared to the well-studied p44 mRNA expression pattern, the P44 paralog proteins are less defined, and only the P44-18 protein has been shown to be surface exposed (27). Recent A. phagocytophilum genome sequencing data have provided a wealth of new genetic information (23). However, there is no experimental evidence demonstrating A. phagocytophilum surface-exposed proteins in addition to P44-18. Furthermore, almost one-half of the predicted open reading frames of A. phagocytophilum encode conserved or novel hypothetical proteins that have never been characterized in any bacterium (23), some of which may be surface proteins. Therefore, it is imperative to use new approaches, including proteomics, to generate a more complete picture of the expression and function of A. phagocytophilum surface proteins.
Cell surface biotinylation has emerged as an important tool for studying cell surface proteins (52). Sulfosuccinimidyl-2-[biotinamido]ethyl-1,3-dithiopropionate (Sulfo-NHS-SS-Biotin) is a thiol-cleavable amine-reactive biotinylation reagent. The N-hydroxysulfosuccinimide (NHS) ester group on this reagent reacts with primary amines on a protein and forms a stable conjugate. It is hydrophilic, making it membrane impermeable and thus appropriate for surface protein labeling. The utility of sulfo-NHS biotin reagents for intracellular bacterial surface labeling has recently been demonstrated by identification of Ehrlichia chaffeensis surface proteins (19).
In this study, to isolate the surface proteins of A. phagocytophilum, bacteria were surface labeled by sulfo-NHS-SS-biotin reagents, and the biotinylated proteins were captured by streptavidin affinity purification. The purified proteins were analyzed using proteomics. The data revealed novel surface proteins of A. phagocytophilum, such as hypothetical proteins APH_0404 (designated Asp62 [62-kDa Anaplasma surface protein]) and APH_0405 (designated Asp55 [55-kDa Anaplasma surface protein]). Not only could recombinant Asp62 and Asp55 be recognized by an HGA patient serum, but anti-Asp62 and anti-Asp55 peptide sera also partially neutralized A. phagocytophilum infection in vitro.
MATERIALS AND METHODS
A. phagocytophilum and cell culture.
The A. phagocytophilum HZ strain (50) was propagated in HL-60 cells in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 5% fetal bovine serum (US Bio-Technologies, Parkerford, PA) and 2 mM l-glutamine (Invitrogen). Cells were incubated in a humidified 5% CO2-95% air atmosphere at 37°C. No antibiotic was used in the culture. The degree of bacterial infection in host cells was assessed by Diff-Quik (Baxter Scientific Products, Obetz, OH) staining of cytocentrifuged preparations. When over 90% of the cells were infected, cells were collected and centrifuged at 500 × g for 10 min. The cell pellets were resuspended in RPMI 1640 medium at a concentration of 2 × 106 cells/ml and homogenized using a 40-ml type B Dounce grinder (Kontes Glass, Vineland, NJ). Each homogenized suspension was subjected to centrifugation at 500 × g for 5 min, and the supernatant was collected and further purified with a 2.7-μm-pore-size, 25-mm, GD/X glass microfiber syringe filter (Whatman, Florham Park, NJ). The filtrate was centrifuged at 10,000 × g for 10 min. The pellets containing the freshly isolated host cell-free A. phagocytophilum were used immediately for biotinylation. The number of purified organisms was estimated as previously described (60).
Bacterial surface biotinylation and affinity purification.
Biotinylation of A. phagocytophilum with Sulfo-NHS-SS-Biotin (Pierce, Rockford, IL) and streptavidin affinity purification of Sulfo-NHS-SS-Biotin-labeled bacterial proteins were performed as described previously (19). Briefly, freshly purified host cell-free bacteria were incubated with Sulfo-NHS-SS-Biotin at a concentration of 1 mg/ml in phosphate-buffered saline (PBS) (pH 8.0) containing 1 mM MgCl2 (PBS2+) at 4°C for 30 min. Free biotin was quenched by washing preparations in PBS containing 500 mM glycine. Biotinylated bacteria were lysed on ice in radioimmunoprecipitation (RIPA) buffer (25 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) containing a 1:100 dilution of protease inhibitor cocktail set II (Calbiochem, San Diego, CA) for 30 min with brief sonication and vortexing. Additional oxidized glutathione (100 μM) was added to the RIPA buffer to protect the disulfide bonds in Sulfo-NHS-SS-Biotin (52). The biotinylated bacterial lysates were cleared by centrifugation at 16,000 × g for 10 min at 4°C. To purify biotinylated proteins, the Sulfo-NHS-SS-Biotin-labeled bacterial lysates were incubated with a streptavidin agarose gel (Pierce) on ice for 2 h. Then the mixture was centrifuged at 500 × g for 1 min, and the supernatant was discarded. The gel slurry was transferred to an Ultrafree-MC centrifugal filter device (Durapore polyvinylidene difluoride; 5.0 μm; Millipore). Unbound proteins were washed away with buffer B-1 (25 mM Tris-HCl [pH 7.6], 0.65 M NaCl, 0.1% NP-40), followed by buffer B-2 (25 mM Tris-HCl [pH 7.6], 1.15 M NaCl, 0.1% NP-40) and 25 mM Tris-HCl buffer (pH 7.6) containing 0.15 M NaCl. The captured bacterial proteins were eluted with 5% 2-mercaptoethanol in PBS. Proteins were precipitated in 10% trichloroacetic acid on ice, followed by washing in cold acetone. The pellets were finally dissolved in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (50 mM Tris-HCl [pH 6.8], 5% 2-mercaptoethanol, 2% SDS, 0.1% bromophenol blue, 10% glycerol) and boiled for 5 min.
Proteomic analysis.
The streptavidin agarose affinity-purified proteins were separated by 10% SDS-PAGE. Seven bands of relatively abundant proteins were submitted to the Mass Spectrometry & Proteomics Facility (Campus Chemical Instrument Center, The Ohio State University). The proteins were identified by capillary liquid chromatography-nanospray tandem mass spectrometry (Nano-LC/MS/MS), and the tandem mass spectrometry data were processed using Mascot Distiller to form a peaklist (.mgf file) and analyzed using the MASCOT tandem mass spectrometry search engine and Turbo SEQUEST algorithm in the BioWorks 3.1 software as described previously (19).
In silico analysis of proteins Asp62 and Asp55.
Amino acid sequences were analyzed using Protean from DNASTAR software (DNASTAR Inc., Madison, WI). Transmembrane β strands and their topology with respect to the outer membrane lipid bilayer were predicted using the web server PRED-TMBB (http://bioinformatics.biol.uoa.gr/PRED-TMBB) (3). A BLAST search for amino acid sequence homology was performed at the web server of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) using the nonredundant database. The gene annotations of A. phagocytophilum HZ and E. chaffeensis Arkansas were obtained from the genome sequencing data (23).
RNA isolation and reverse transcription (RT)-PCR.
Total RNA was extracted from 5 × 106 A. phagocytophilum-infected HL-60 cells using an RNeasy mini RNA extraction kit (QIAGEN, Valencia, CA) according to the manufacturer's instructions. The concentration and purity of the RNA were determined by measuring the A260 and determining the A260/A280 ratio with a GeneQuant II RNA and DNA calculator (Pharmacia Biotech Inc., Piscataway, NJ). Five micrograms of the extracted RNA was treated with 1 U of DNase I (amplification grade; Invitrogen) at 25°C for 10 min. DNase I then was inactivated by addition of 1 μl of 25 mM EDTA and subsequent heating at 65°C for 10 min. The DNase I-treated RNA was added to a 30-μl reaction mixture containing 1× reaction buffer (50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2), 10 mM dithiothreitol, 375 ng random primers (Invitrogen), 60 U of RNaseOUT (Invitrogen), and 0.5 mM of each deoxynucleoside triphosphate (dNTP). After addition of 300 U of SuperScript III reverse transcriptase (Invitrogen), the reaction mixture was incubated for 5 min at 25°C, followed by RT at 50°C for 50 min, and the reaction was terminated by incubation at 70°C for 15 min.
To examine the transcription of the asp62 and asp55 genes and the intergenic region in A. phagocytophilum, 1 μl of cDNA was amplified in a 25-μl reaction mixture containing 1× reaction buffer (20 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2), 0.2 mM of each dNTP, 0.4 μM forward primer, 0.4 μM reverse primer, and 1 U of Taq DNA polymerase (Invitrogen) in a DNA thermal cycler (GeneAmp PCR system 9700; Perkin-Elmer, Foster City, CA). After the mixture was heated at 94°C for 5 min, each PCR cycle consisted of denaturation at 94°C for 60 s, annealing at 55°C for 60 s, and extension at 72°C for 90 s. PCRs were performed for 29 cycles. A final extension was carried out at 72°C for 10 min. PCR products (10 μl) were electrophoresed in a 1.2% agarose gel containing 0.5 μg/ml of ethidium bromide. DNA size markers (1Kb Plus DNA ladder; Invitrogen) were run in parallel. Based on genome sequencing data (23), primers asp62-F1 (nucleotides [nt] 1336 to 1356; 5′CGCAATGATGCTAGGAACGTT3′), asp62-R1 (nt 1532 to 1512; 5′AGCACGCAGCGCATACTCTCC3′), asp55-F1 (nt 67 to 87; 5′GGAGAGCGTGCGTCGGTAACG3′), and asp55-R1 (nt 407 to 387; 5′ATACCAGGCGCACCATGAAAC3′) were designed for this study. The primer pairs used were asp62-F1 and asp62-R1 for asp62, asp55-F1 and asp55-R1 for asp55, and asp62-F1 and asp55-R1 for amplifying the cotranscribed mRNA of asp62 and asp55.
Surface localization of Asp62 and Asp55 by immunofluorescence microscopy.
To design peptides for developing antibodies against extracellular epitopes, two relatively highly antigenic and hydrophilic peptide fragments, located within one of the extracellular loops in the two-dimensional structures predicted by PRED-TMBB as mentioned above, were chosen from the Asp62 and Asp55 amino acid sequences based on Protean analysis (DNASTAR Inc.). A 19-mer peptide, CRYNTRDVYHRDVGYKDHG, corresponding to the sequence from the Asp62 C terminus (amino acids 534 to 552), was synthesized and conjugated to keyhole limpet hemocyanin, and rabbit antibody was developed by Proteintech Group, Inc. (Chicago, IL). A 15-mer peptide, CHEYKSTESSGFVLK (the underlined sequence corresponds to the 14 amino acids from the Asp55 C terminus [amino acids 501 to 514]), was synthesized and conjugated to keyhole limpet hemocyanin, and rabbit antibody was made by Sigma Genosys (St. Louis, MO). According to a BLAST search for short, nearly exactly matching sequences in the NCBI nonredundant database, these two peptide sequences had little or no homology to any other known proteins (E > 25) and thus were unique to the Asp62 and Asp55 proteins.
For immunofluorescence microscopic analysis of A. phagocytophilum Asp62 and Asp55 localization, paraformaldehyde-fixed bacteria were used as described previously (19). Briefly, host cell-free A. phagocytophilum was pelleted and washed in PBS (137 mM NaCl, 2.68 mM KCl, 10 mM Na2HPO4, 1.76 mM KH2PO4; pH 7.4). All subsequent steps were performed at room temperature. Bacteria were fixed in 2% paraformaldehyde for 45 min, followed by quenching in PBS containing 0.1 M glycine. After they were washed in PBS, bacteria were incubated with 1:100-diluted rabbit antisera against Asp62, Asp55 peptide, rabbit preimmune sera, or rabbit anti-irrelevant peptide serum in PG buffer (0.2% gelatin in PBS) for 1 h. After they were washed in PG buffer, the bacteria were then labeled with Alexa Fluor 488 goat anti-rabbit immunoglobulin G (IgG) (Invitrogen) at a dilution of 1:100 in PG buffer for 1 h. The bacteria were washed in PG buffer, resuspended in PBS, and observed using a Nikon Eclipse E400 fluorescence microscope with a xenon-mercury light source (Nikon Instruments, Melville, NY). For protease treatment, bacteria were incubated with pronase E (Sigma) at a concentration of 2 mg/ml in PBS2+ for 5 min at 37°C (60); the pronase was inactivated by adding 10% fetal bovine serum, followed by washing in PBS2+ twice.
Expression of rAsp62 and rAsp55 and Western blot analysis.
A. phagocytophilum genomic DNA was extracted from infected HL-60 cells using a QIAamp DNA blood mini kit (QIAGEN). Primers rAsp62-F (5′CACCATGGCAGGGTATGCGGACGATT3′; NcoI restriction enzyme site underlined) and rAsp62-R (5′GTGAGCTCAAAGCCATCAAGCCAAAG3′; SacI restriction enzyme site underlined) were designed to amplify the Asp62 DNA sequence encoding a 305-amino-acid polypeptide (amino acids 264 to 568). Two microliters of DNA was amplified in a 50-μl reaction mixture containing 1× Phusion HF buffer, 0.2 mM of each dNTP, 0.4 μM forward primer, 0.4 μM reverse primer, and 1 U of Phusion high-fidelity DNA polymerase (New England BioLabs, Espoo, Finland) in the DNA thermal cycler. After initial denaturation at 98°C for 30 s, each PCR cycle consisted of denaturation at 98°C for 10 s, annealing at 65°C for 30 s, and extension at 72°C for 30 s. PCRs were performed for 29 cycles. A final extension was carried out at 72°C for 5 min. The PCR fragment of asp62 was cloned into the pET33b vector (Novagen, Inc., Madison, WI) between NcoI and SacI sites. The full-length gene of Asp55 was cloned into pET33b between the EcoRI and XhoI sites (M. Lin and Y. Rikihisa, unpublished data). The recombinant Asp62 (rAsp62) and recombinant Asp55 (rAsp55) plasmids or the empty pET33b vector was expressed in Escherichia coli BL21(DE3) cells.
The protein samples were subjected to 12% SDS-PAGE and transferred to Trans-Blot nitrocellulose membranes (Bio-Rad, Hercules, CA). The membranes were incubated with serum from an HGA patient (DM981027) or an uninfected human at a dilution of 1:500 or with rabbit anti-Asp62 peptide serum, rabbit anti-Asp55 peptide serum, or rabbit preimmune sera for Asp62 and Asp55 at a dilution of 1:1,000 at 4°C overnight. After four washes in TBST (15 mM NaCl, 5 mM Tris-HCl [pH 7.4], 0.02% Tween 20) (10 min each), the membranes were incubated with 1:3,000-diluted horseradish peroxidase-conjugated goat anti-human IgA, IgM, and IgG or 1:1,000-diluted horseradish peroxidase-conjugated goat anti-rabbit IgG (heavy plus light chains) (KPL, Gaithersburg, MD) at room temperature for 1 h, followed by fours washes in TBST (10 min each). The blots were developed by using an enhanced chemiluminescence kit (Pierce).
Neutralization with Asp62 and Asp55 peptide antisera of A. phagocytophilum infection.
Asp62 and Asp55 rabbit peptide antisera and preimmune rabbit sera were heat inactivated and filtered through a 0.2-μm HT Tuffryn membrane filter (Pall Corporation, Ann Arbor, MI). Approximately 1 × 107 freshly isolated host cell-free A. phagocytophilum cells were preincubated with sera at a 1:50 dilution in triplicate wells in a 48-well plate (BD, Franklin Lakes, NJ) at room temperature for 30 min with gentle shaking. Then HL-60 cells were added at a final concentration of 5 × 105 cells/ml. After gentle shaking for 10 min at room temperature, the plate was incubated in a humidified 5% CO2-95% air atmosphere at 37°C. After 12 h of incubation, the plate was centrifuged at 500 × g for 5 min, and the supernatant was replaced with fresh medium. The cells were cultured for 2 to 3 days, and the infectivity was determined as previously described (31).
RESULTS
Streptavidin affinity purification of biotinylated A. phagocytophilum surface proteins.
To identify biotinylated A. phagocytophilum surface proteins, Sulfo-NHS-SS-Biotin-labeled host cell-free bacteria were solubilized in RIPA buffer. The biotinylated proteins were purified by streptavidin affinity gel chromatography. The disulfide bonds in Sulfo-NHS-SS-Biotin were cleaved with the reducing agent to elute the streptavidin affinity-captured proteins. The eluted proteins were separated by SDS-PAGE. As shown in Fig. 1, with GelCode blue protein staining in the SDS-PAGE gel, there were seven bands of relatively abundant proteins corresponding to molecular masses of approximately 28, 42, 44 to 48, 55, 65, 80, and 105 kDa.
FIG. 1.

Streptavidin affinity purification of Sulfo-NHS-SS-Biotin-labeled A. phagocytophilum surface proteins. In lane 1, Sulfo-NHS-SS-Biotin-labeled A. phagocytophilum surface proteins were separated by 10% SDS-PAGE and stained with GelCode blue. Bands 1 to 7 were subjected to Nano-LC/MS/MS analysis. The marker lane contained Precision Plus prestained protein standards (Bio-Rad).
Nano-LC/MS/MS.
The seven bands of relatively abundant proteins (Fig. 1, bands 1 to 7) were subjected to proteomic analysis. Table 1 summarizes a total of 16 A. phagocytophilum proteins identified by Nano-LC/MS/MS. One-half of these proteins were integral membrane proteins with a cleavable signal peptide and multipass transmembrane β strands as analyzed by PRED-TMBB (the number of β strands is not shown in Table 1). The remaining proteins were either single-pass transmembrane proteins or possible peripheral membrane proteins. Band 1 contained hypothetical protein APH_0441 (accession no. YP_505044). Band 2 contained mainly outer membrane protein Omp85 (YP_505741). Bands 3 and 4 contained mainly hypothetical proteins APH_0404 (YP_505009) (Asp62) and APH_0405 (YP_505010) (Asp55), respectively. Band 5 consisted of several P44 family proteins, including P44-18ES (YP_505752), P44-2 (P44-2a [YP_505715] or P44-2b [YP_505759]; the paralog-specific sequences shared by P44-2a and P44-2b were detected), and P44-59 (AAQ16676). Band 6 contained human β-actin (not shown in Table 1). Band 7 contained mainly Omp-1A, a protein predicted to belong to the Omp-1/P28 outer membrane protein family of Ehrlichia species (23, 42).
TABLE 1.
Surface-exposed proteins of A. phagocytophilum analyzed by Nano-LC/MS/MSa
| Band | Annotation or gene locus tag | Predicted molecular mass (Da) | Nano-LC/ MS/MS sequence coverage by amino acids (%) | Accession no. | Predicted protein function | Pfam family(ies) | Presence of signal peptide sequence (position) |
|---|---|---|---|---|---|---|---|
| 1 | Hypothetical protein APH_0441 | 65,911 | 4 | YP_505044 | Unknown; putative lipoprotein | NAb | Yes (35)c |
| 2 | Outer membrane protein, Omp85 family | 85,709 | 30 | YP_505741 | Cell envelope biogenesis | PF07244, PF01103 | Yes (22)c,d |
| Translation elongation factor G | 76,234 | 32 | YP_505591 | GTPase, power translation | PF00009, PF03144, PF03764, PF00679 | No | |
| Chaperone protein DnaK | 70,019 | 16 | YP_504953 | Chaperone | PF00012 | No | |
| ppiC/parvulin rotamase family protein | 67,535 | 15 | YP_505198 | Accelerate the folding of proteins | NA | No | |
| 3 | Hypothetical protein APH_0404 (Asp62) | 63,987 | 42 | YP_505009 | Unknown | NA | Yes (19)c |
| 60-kDa chaperonin (GroEL) | 57,282 | 35 | YP_504857 | Chaperone | PF00118 | No | |
| Pentapeptide repeat family protein | 61,870 | 5 | YP_505709 | Uncharacterized secreted protein (YP_198411) containing pentapeptide repeats (Wolbachia endosymbiont strain TRS from B. malayi) | PF00805 | No | |
| 4 | Hypothetical protein APH_0405 (Asp55) | 57,595 | 13 | YP_505010 | Unknown | NA | Yes (22)d |
| 5 | P44-18ES, P44 outer membrane protein expression locus with P44-18 | 45,827 | 38 | YP_505752 | Porin | PF01617 | Yes (20)c,d |
| P44-2a and P44-2b | 38,478 and 44,912 | 39 | YP_505715 and YP_505759 | P44 paralog | PF01617 | ||
| P44-59 | NA | 10 | AAQ16676 | P44 paralog | NA | ||
| 7 | Major outer membrane protein Omp-1A | 31,845 | 41 | YP_505858 | Unknown | PF01617 | Yes (19)d |
| Type IV secretion protein VirB8-1 | 27,582 | 16 | YP_505898 | Transportation of macromolecules across bacterial inner and outer membranes | PF04335 | No | |
| Thiol:disulfide oxidoreductase | 27,842 | 8 | YP_504749 | Protein folding and stabilization | PF01323 | Yes (21)d | |
| Cochaperone GrpE | 22,761 | 8 | YP_504670 | Chaperone | PF01025 | No |
The program SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP) was used to predict the presence of N-terminal signal peptides (5). The putative lipoprotein was predicted by LipoP 1.0.
NA, not available.
Predicted by SignalP-NN.
Predicted by SignalP-HMM.
In silico analysis of the Asp62 and Asp55 proteins.
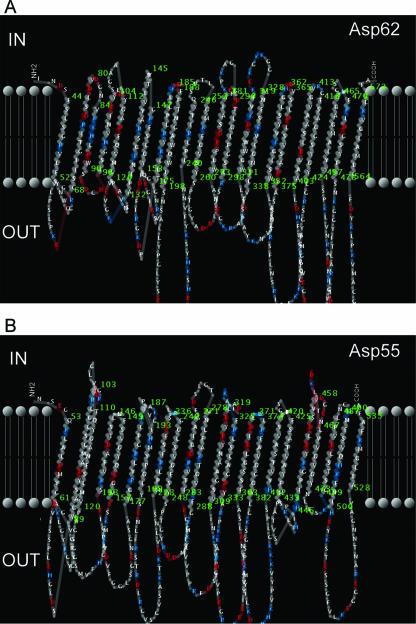
As predicted by the posterior decoding method using the dynamic programming algorithm in PRED-TMBB, there were 22 transmembrane β strands in both the Asp62 (Fig. 2A) and Asp55 (Fig. 2B) proteins; these β strands were connected by 11 long loops on the external side of the bacterium with 10 short turns along the periplasmic space. The discrimination scores of Asp62 and Asp55 for β-barrel proteins were 2.956 and 2.905, respectively. Scores lower than the threshold value of 2.965 were considered significant (3), suggesting that these proteins are β-barrel outer membrane proteins.
FIG. 2.
Two-dimensional structure of A. phagocytophilum Asp62 and Asp55 with respect to the outer membrane lipid bilayer predicted using the Posterior Decoding method available in PRED-TMBB: graphical representations of the predicted topology with respect to the outer membrane lipid bilayers of Asp62 (A) and Asp55 (B).
A physical map of the Asp62 and Asp55 ortholog genes is shown in Fig. 3. Asp62 and Asp55 were paralogs with an E value of 3e−24. The gene encoding one more A. phagocytophilum paralog, hypothetical protein APH_0406, was found downstream of the Asp55 gene, and another gene, aph_0407, was predicted to code for a short 65-amino-acid protein, which was almost identical to a stretch of amino acids within Asp62. Using a BLAST search in the NCBI database, proteins homologous to A. phagocytophilum Asp62 and Asp55 were found in Anaplasma marginale strain St. Maries, E. chaffeensis Arkansas, E. chaffeensis Sapulpa (not shown), Ehrlichia canis Jake, Ehrlichia ruminantium Gardel (not shown), and E. ruminantium Welgevonden. There are three tandem orthologs in A. marginale strain St. Maries, E. chaffeensis Arkansas, and E. canis Jake, whereas in E. ruminantium Welgevonden there are two orthologs. In addition, Wolbachia endosymbiont strain TRS from Brugia malayi has one ortholog, hypothetical protein Wbm0010 with an E value as low as 1e−26 with Asp55; a Wolbachia endosymbiont of Drosophila melanogaster has one ortholog, hypothetical protein WD0745 with an E value of 2e−25; and Rickettsia bellii OSU 85-389 has one ortholog, hypothetical protein RbelO_01000075 with an E value of 8e−11 (not shown in Fig. 3). All of these organisms are obligate intracellular bacteria that belong to the α-proteobacterial order Rickettsiales.
FIG. 3.
Schematic diagram of the organization of genes encoding the APH_0404 (Asp62), APH_0405 (Asp55), APH_0406, and APH_0407 proteins in A. phagocytophilum HZ and the orthologous genes in A. marginale strain St. Maries, E. chaffeensis Arkansas, E. canis Jake, and E. ruminantium Welgevonden. Open reading frames are represented by open arrows that indicate their orientations. Orthologs are indicated by dashed lines at the ends of each open reading frame. The number of amino acid (aa) residues for each open reading frame is shown. The E value cutoff is e−22.
Cotranscription of asp62 and asp55.
The stop codon of asp62 and the start codon of asp55 are separated by only a 65-nt intergenic region (23), leading to the hypothesis that the genes are cotranscribed. To test whether they are cotranscribed experimentally, an RT-PCR analysis of A. phagocytophilum asp62, asp55, and the common transcript of asp62 and asp55 was performed with and without reverse transcriptase as a negative control. As shown in Fig. 4, asp62, asp55 and the common transcript of asp62 and asp55 including the 65-nt intergenic region were transcribed by A. phagocytophilum cultured in HL-60 cells at 37°C. The transcripts for the control without reverse transcriptase were undetectable. These results indicate that the asp62 and asp55 paralog genes are organized in one operon.
FIG. 4.

Cotranscriptional analysis of asp62 and asp55 by RT-PCR. Total RNA was isolated from A. phagocytophilum-infected HL-60 cells. Lane M, marker (1Kb Plus DNA ladder; Invitrogen); lane 1, cotranscript of asp62 and asp55, including the 65-nt intergenic region; lane 3, asp62; lane 5, asp55; lane 2, asp62 and asp55 control reaction without reverse transcriptase; lane 4, asp62 control reaction without reverse transcriptase; lane 6, asp55 control reaction without reverse transcriptase. The amplicon sizes were in agreement with predicted amplicon lengths (i.e., 197 bp for asp62, 340 bp for asp55, and 795 bp for the cotranscript of asp62 and asp55).
Surface localization of Asp62 and Asp55.
To verify the localization of Asp62 and Asp55 on the A. phagocytophilum surface, host cell-free A. phagocytophilum was first paraformaldehyde fixed to prevent antibody permeabilization (19) and then incubated with rabbit antiserum against Asp62 or Asp55 C-terminal peptide. As shown in Fig. 5a and b, both the antisera against Asp62 and Asp55 labeled the surface of individual bacteria of various sizes with a mottled ring-like staining pattern. When bacteria were treated with pronase E, the surface immunofluorescence staining of Asp62 (Fig. 5c) and Asp55 (Fig. 5d) was abolished. Antiserum from a rabbit immunized with an irrelevant peptide (Fig. 5e) or Asp62 or Asp55 preimmune rabbit sera (data not shown) did not recognize A. phagocytophilum. The results confirmed not only the identification of Asp62 and Asp55 by surface biotinylation but also the surface exposure of C-terminal peptides as determined by the in silico analysis.
FIG. 5.
Surface localization of A. phagocytophilum Asp62 and Asp55 as determined by an immunofluorescence assay. Host cell-free A. phagocytophilum bacteria were fixed in paraformaldehyde, incubated with rabbit serum against the Asp62 C-terminal peptide (amino acids 534 to 552) or the Asp55 C-terminal peptide (amino acids 501 to 514), stained with Alexa Fluor 488 goat anti-rabbit IgG, and visualized by fluorescence microscopy. (a) Mottled ring-like bacterial surface staining of Asp62. (b) Mottled ring-like bacterial surface staining of Asp55. (c and d) Bacteria treated with pronase E and then incubated with rabbit anti-Asp62 peptide serum (c) or anti-Asp55 peptide serum (d). (e) Bacteria incubated with rabbit anti-irrelevant peptide serum. Scale bar, 1 μm.
Immunogenicity of Asp62 and Asp55 in an HGA patient and neutralization of A. phagocytophilum infection by anti-Asp62 and anti-Asp55 peptide monospecific sera.
rAsp62 and rAsp55 were expressed in E. coli BL21(DE3) cells and migrated on the SDS-PAGE gel to approximate predicted molecular mass positions (i.e., 35.9 and 63 kDa, respectively) (Fig. 6A). As shown in Fig. 6B, rAsp62 strongly reacted with the rabbit anti-Asp62 peptide serum but not with preimmune rabbit serum. Similarly, rAsp55 strongly reacted with the rabbit anti-Asp55 peptide serum but not with preimmune rabbit serum. High-titer (1:2,560, as determined by immunofluorescence assay) HGA patient serum recognized both the rAsp62 and rAsp55 proteins. In contrast, uninfected human serum did not react with rAsp62 or rAsp55. These data revealed that both Asp62 and Asp55 were immunogenic in the HGA patient.
FIG. 6.
(A) Expression of rAsp62 and rAsp55 in E. coli BL21(DE3) cells. Proteins were separated by 12% SDS-PAGE and stained with GelCode blue. Lane 1, rAsp62 expressed in E. coli BL21(DE3) with a predicted molecular mass of 35.9 kDa; lane 2, rAsp55 expressed in E. coli BL21(DE3) with a predicted molecular mass of 63 kDa; lane 3, pET33b vector expressed in E. coli BL21(DE3). Lane M contained markers (Precision Plus prestained protein standards; Bio-Rad). (B) Western immunoblot analysis of immunogenicity of rAsp62 and rAsp55 in an HGA patient. rAsp62, rAsp55, and the pET33b empty vector expressed in E. coli BL21(DE3) cells were used. The sera used in this study included rabbit preimmune sera for rAsp62 (Pre) and rAsp55 (Pre′), rabbit anti-Asp62 peptide serum (anti-Asp62), rabbit anti-Asp55 peptide serum (anti-Asp55), and HGA patient (HGA) and uninfected human (uninfect) sera.
Asp62 and Asp55 are two of the major A. phagocytophilum immunogenic surface proteins, suggesting that they are neutralizing targets. Therefore, we tested in vitro neutralization of A. phagocytophilum infection of HL-60 cells by adding host cell-free A. phagocytophilum pretreated with Asp62 or Asp55 peptide monospecific antiserum at a 1:50 dilution. Rabbit serum collected prior to immunization with Asp62 or Asp55 peptide was used as the negative control. As shown in Fig. 7, the infectivity of A. phagocytophilum pretreated with anti-Asp62 peptide serum was significantly lower than the infectivity of bacteria pretreated with preimmune serum, as was the infectivity of A. phagocytophilum pretreated with anti-Asp55 peptide serum. These results indicated that A. phagocytophilum anti-Asp62 and anti-Asp55 monospecific sera could partially neutralize A. phagocytophilum infection in vitro.
FIG. 7.
Neutralization of A. phagocytophilum infection of HL-60 cells by Asp62 and Asp55 rabbit peptide antisera. After A. phagocytophilum pretreated with Asp62 or Asp55 rabbit peptide antiserum or preimmune sera was added, HL-60 cells were cultured for 2 to 3 days. The numbers of A. phagocytophilum bacteria were counted after Diff-Quik staining, using 100 cells per well and triplicate wells. The values are the means and standard deviations (n = 3). An asterisk indicates that there is a significant difference between the peptide antiserum and the preimmune serum (P < 0.05). The data are representative of three independent experiments.
DISCUSSION
The present work revealed several novel surface-exposed proteins of A. phagocytophilum using surface biotinylation and proteomics methods. In addition to P44 proteins, the hypothetical proteins APH_0404 and APH_0405 (Asp62 and Asp55) are the other two abundantly expressed A. phagocytophilum surface proteins. In this study, the identification of known or predicted P44 proteins, the Omp-1A outer membrane protein, and conserved outer membrane protein Omp85 attests to the utility of the surface biotinylation experimental approach used.
Both Asp62 and Asp55 have been predicted to be β-barrel outer membrane proteins with a secondary structure consisting of 22 transmembrane β strands by the posterior decoding method on the PRED-TMBB web server, suggesting that Asp62 and Asp55 function as outer membrane transporters (3). The 22-strand β-barrel structure has been revealed by crystal structural data for some bacterial outer membrane siderophore receptors, including FepA (7), FhuA (46), and FecA from E. coli (16) and FpvA (10) and FptA (11) from Pseudomonas aeruginosa, which act as transporters to take up iron. When ligands bind to these normally closed transporters, the transporters exhibit conformational changes that activate them to open, hence their designation “ligand-gated porin”(25). The cotranscription of Asp62 and Asp55 suggests that their mRNA expression responds to the same transcriptional activation signals and is controlled by the same regulatory system. Similarly, the genes of Vibrio parahaemolyticus siderophore receptors (psuA and pvuA) constitute an operon (17). The protein sequences of Asp62 and Asp55 and the gene organization of the Asp62 and Asp55 genes are highly conserved within the family Anaplasmataceae, suggesting that there is evolutionary pressure for conservation within this family. One ortholog, Esp73 (ECH_0525), has recently been revealed to be an E. chaffeensis surface-exposed protein (19). All related orthologs in the family Anaplasmataceae are annotated as hypothetical proteins by genome sequencing data in NCBI, and their functions have not been elucidated. Therefore, it would be interesting to functionally characterize the newly discovered A. phagocytophilum surface proteins Asp62 and Asp55, which is under way in our lab. In addition, the gene for APH_0406 is located 1,715 nt downstream of asp55, suggesting that these two genes may not be cotranscribed. Our lab's recent proteomic data have shown that expression of the APH_0406 protein is undetectable (T. Kikuchi and Y. Rikihisa, unpublished), suggesting that this protein is either not expressed or is expressed at an undetectable level by A. phagocytophilum when it is cultured in HL-60 cells at 37°C. In the future, it would also be interesting to investigate the regulatory mechanisms of expression of these paralogs by A. phagocytophilum at different developmental stages and in different environments, such as different host cell types and different temperatures.
Effective protection against anaplasmosis by the host humoral immune response requires specific recognition of epitopes that are exposed on the surface of Anaplasma and induction of neutralizing antibodies (43, 44). P44 proteins are A. phagocytophilum outer membrane proteins that have been shown to have two neutralizable surface-exposed epitopes that mediate protection by neutralizing antibodies (27, 58). Two P44 monoclonal antibodies, 5C11 and 3E65, which recognize surface-exposed epitopes located in the N-terminal conserved region and the P44-18 central hypervariable region, respectively, almost completely block infection by the A. phagocytophilum population that expresses predominantly P44-18 in HL-60 cells (58). However, development of a vaccine based on P44 proteins is challenging since the surface-exposed central hypervariable regions of P44 molecules undergo antigenic variation in infected horses, mice, or human patients (32, 36, 59). Many members of the p44 family are functional pseudogenes. In other words, although the genes lack the translational start site, the proteins can be expressed as full-length P44 proteins (44 kDa) after RecF-dependent recombination into the p44 expression locus (35). The full-length P44 paralog gene can be either recombined into and expressed in a p44 expression locus or expressed in its own expression site but at a much lower level (58a). The identification of P44-18ES (P44-18 in the expression locus), P44-2 (full length and pseudogene), and P44-59 (pseudogene) revealed the relative abundance of these P44 paralogs, suggesting that they are much more likely to be from different A. phagocytophilum clonal populations. Passive immunization of mice with the two monoclonal antibodies, 5C11 and 3E65, partially protects mice from challenge (27), indicating that neutralizing antibodies against either the P44 paralog-specific region or the P44 conserved region are unable to confer complete in vivo protection from A. phagocytophilum. Consequently, it is necessary to identify other A. phagocytophilum surface proteins as vaccine candidates. The present work revealed two new A. phagocytophilum immunogenic major surface proteins, Asp62 and Asp55, which have surface accessible epitopes and partially mediate the neutralization of A. phagocytophilum infection in vitro. These findings provide an important rationale for design of a new A. phagocytophilum vaccine. While strain variability needs to be determined, Asp62- and Asp55-based vaccines are expected to provide protection against a broader A. phagocytophilum population, since unlike P44 proteins, they do not go through antigenic variation. It is important to determine the potential of these two proteins for mediating protection by the host immune response in vivo.
The predicted molecular mass of the APH_0441 hypothetical protein was 65,911 Da, which was much less than the molecular mass (approximately 105 kDa) deduced from the actual migration distance by 10% SDS-PAGE (Fig. 1). This could be due to posttranslational modification, such as glycosylation, which has been proposed for the gp47 protein of E. chaffeensis (14). Omp85 is a conserved outer membrane protein in gram-negative bacteria (21) that is a central component of the apparatus for outer membrane protein assembly (51, 57). While Omp85 has been shown to be an outer membrane protein of Neisseria gonorrhoeae (37) and E. chaffeensis (19), it has never been shown experimentally to be surface exposed or expressed at the protein or mRNA level by A. phagocytophilum.
The A. phagocytophilum Omp-1A gene is associated with the msp2 locus (distinct from the A. marginale msp2 gene) and is one of the three A. phagocytophilum omp-1 genes belonging to the Omp-1/P44/Msp2 superfamily (23, 33). The homologous protein in A. marginale is Omp11, which has been detected by Western blotting using monospecific peptide antibody (41). Recently, E. chaffeensis Omp-1A, an ehrlichial Omp-1/P28 paralog with the highest homology to A. phagocytophilum Omp-1A, has been shown to be surface exposed by both surface biotinylation and immunofluorescence labeling (19). For the first time, Omp-1A has been directly detected at the protein level and shown to be surface exposed by Anaplasma. Type IV secretion protein VirB8 is a core component of the type IV secretion system apparatus, which recently was proposed to function as the assembly factor for targeting the type IV apparatus to the cell pole (26). In addition to VirB9 (40), VirB8 is an A. phagocytophilum type IV secretion protein that has been shown to be surface exposed. A clustered distribution of VirB8 over the bacterial surface has been demonstrated for Agrobacterium (29), whereas in the recent model for Agrobacterium tumefaciens type IV secretion system VirB8 was referred to as an inner membrane protein (4). It is possible that detection of A. phagocytophilum VirB8 may be due to some damage to the bacterial outer membrane during isolation of cell-free bacteria. However, according to A. phagocytophilum genome sequencing data (23), several type IV secretion proteins, such as VirB1, VirB2, VirB5, and VirB7, are missing. Therefore, the assembly of the A. phagocytophilum type IV secretion apparatus may be different from that of A. tumefaciens.
Some proteins which were considered to be bacterial cytoplasmic, periplasmic, or inner membrane proteins were identified in the present study, such as chaperone protein GroEL, DnaK, translation elongation factor G, and disulfide oxidoreductase. As discussed previously for E. chaffeensis (19), these proteins with well-known functions inside bacteria may be present on the surface and play unexpected roles in the A. phagocytophilum-host interaction too. Similar to the results for E. chaffeensis surface biotinylation (19), one obvious host protein band captured by streptavidin affinity purification was β-actin, which is one of the most abundant cytoskeleton proteins of eukaryotic cells. This may have been due to the binding of host cell actin to bacterial surface proteins during the isolation of host cell-free bacteria or via a functional association. For example, A. marginale assembles an actin filament bundle during intracellular infection (53).
In conclusion, surface biotinylation of A. phagocytophilum was used to identify novel bacterial surface proteins that are promising targets for future study of the interaction between this bacterium and its host, as well as for development of effective vaccines.
Acknowledgments
We thank Kari Green-Church for helpful discussions and technical assistance with the proteomic analysis. We thank Mingqun Lin for help with making the rabbit Asp55 peptide antibody and providing rAsp55 and Kate Hayes for copyediting the manuscript. We are also grateful to G. P. Wormser and H. W. Horowitz of New York Medical College for providing HGA patient serum.
This work was supported by National Institutes of Health grants R01 AI30010 and R01 AI47407.
Footnotes
Published ahead of print on 31 August 2007.
REFERENCES
- 1.Alberti, A., M. F. Addis, O. Sparagano, R. Zobba, B. Chessa, T. Cubeddu, M. L. Parpaglia, M. Ardu, and M. Pittau. 2005. Anaplasma phagocytophilum, Sardinia, Italy. Emerg. Infect. Dis. 11:1322-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alleman, A. R., A. F. Barbet, H. L. Sorenson, N. I. Strik, H. L. Wamsley, S. J. Wong, R. Chandrashaker, F. P. Gaschen, N. Luckshander, and A. Bjoersdorff. 2006. Cloning and expression of the gene encoding the major surface protein 5 (MSP5) of Anaplasma phagocytophilum and potential application for serodiagnosis. Vet. Clin. Pathol. 35:418-425. [DOI] [PubMed] [Google Scholar]
- 3.Bagos, P. G., T. D. Liakopoulos, I. C. Spyropoulos, and S. J. Hamodrakas. 2004. PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res. 32:W400-W404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baron, C. 2006. VirB8: a conserved type IV secretion system assembly factor and drug target. Biochem. Cell Biol. 84:890-899. [DOI] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 6.Borjesson, D. L., S. D. Kobayashi, A. R. Whitney, J. M. Voyich, C. M. Argue, and F. R. Deleo. 2005. Insights into pathogen immune evasion mechanisms: Anaplasma phagocytophilum fails to induce an apoptosis differentiation program in human neutrophils. J. Immunol. 174:6364-6372. [DOI] [PubMed] [Google Scholar]
- 7.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 8.Carlyon, J. A., D. Abdel-Latif, M. Pypaert, P. Lacy, and E. Fikrig. 2004. Anaplasma phagocytophilum utilizes multiple host evasion mechanisms to thwart NADPH oxidase-mediated killing during neutrophil infection. Infect. Immun. 72:4772-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, S. M., J. S. Dumler, J. S. Bakken, and D. H. Walker. 1994. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J. Clin. Microbiol. 32:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobessi, D., H. Celia, N. Folschweiller, I. J. Schalk, M. A. Abdallah, and F. Pattus. 2005. The crystal structure of the pyoverdine outer membrane receptor FpvA from Pseudomonas aeruginosa at 3.6 angstroms resolution. J. Mol. Biol. 347:121-134. [DOI] [PubMed] [Google Scholar]
- 11.Cobessi, D., H. Celia, and F. Pattus. 2005. Crystal structure at high resolution of ferric-pyochelin and its membrane receptor FptA from Pseudomonas aeruginosa. J. Mol. Biol. 352:893-904. [DOI] [PubMed] [Google Scholar]
- 12.de la Fuente, J., R. F. Massung, S. J. Wong, F. K. Chu, H. Lutz, M. Meli, F. D. von Loewenich, A. Grzeszczuk, A. Torina, S. Caracappa, A. J. Mangold, V. Naranjo, S. Stuen, and K. M. Kocan. 2005. Sequence analysis of the msp4 gene of Anaplasma phagocytophilum strains. J. Clin. Microbiol. 43:1309-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demma, L. J., R. C. Holman, J. H. McQuiston, J. W. Krebs, and D. L. Swerdlow. 2005. Epidemiology of human ehrlichiosis and anaplasmosis in the United States, 2001-2002. Am. J. Trop. Med. Hyg. 73:400-409. [PubMed] [Google Scholar]
- 14.Doyle, C. K., K. A. Nethery, V. L. Popov, and J. W. McBride. 2006. Differentially expressed and secreted major immunoreactive protein orthologs of Ehrlichia canis and E. chaffeensis elicit early antibody responses to epitopes on glycosylated tandem repeats. Infect. Immun. 74:711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumler, J. S., K. S. Choi, J. C. Garcia-Garcia, N. S. Barat, D. G. Scorpio, J. W. Garyu, D. J. Grab, and J. S. Bakken. 2005. Human granulocytic anaplasmosis and Anaplasma phagocytophilum. Emerg. Infect. Dis. 11:1828-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferguson, A. D., R. Chakraborty, B. S. Smith, L. Esser, D. van der Helm, and J. Deisenhofer. 2002. Structural basis of gating by the outer membrane transporter FecA. Science 295:1715-1719. [DOI] [PubMed] [Google Scholar]
- 17.Funahashi, T., K. Moriya, S. Uemura, S. Miyoshi, S. Shinoda, S. Narimatsu, and S. Yamamoto. 2002. Identification and characterization of pvuA, a gene encoding the ferric vibrioferrin receptor protein in Vibrio parahaemolyticus. J. Bacteriol. 184:936-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge, Y., and Y. Rikihisa. 2006. Anaplasma phagocytophilum delays spontaneous human neutrophil apoptosis by modulation of multiple apoptotic pathways. Cell. Microbiol. 8:1406-1416. [DOI] [PubMed] [Google Scholar]
- 19.Ge, Y., and Y. Rikihisa. 2007. Surface-exposed proteins of Ehrlichia chaffeensis. Infect. Immun. 75:3833-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge, Y., K. Yoshiie, F. Kuribayashi, M. Lin, and Y. Rikihisa. 2005. Anaplasma phagocytophilum inhibits human neutrophil apoptosis via upregulation of bfl-1, maintenance of mitochondrial membrane potential and prevention of caspase 3 activation. Cell. Microbiol. 7:29-38. [DOI] [PubMed] [Google Scholar]
- 21.Gentle, I. E., L. Burri, and T. Lithgow. 2005. Molecular architecture and function of the Omp85 family of proteins. Mol. Microbiol. 58:1216-1225. [DOI] [PubMed] [Google Scholar]
- 22.Goodman, J. L., C. Nelson, B. Vitale, J. E. Madigan, J. S. Dumler, T. J. Kurtti, and U. G. Munderloh. 1996. Direct cultivation of the causative agent of human granulocytic ehrlichiosis. N. Engl. J. Med. 334:209-215. [DOI] [PubMed] [Google Scholar]
- 23.Hotopp, J. C., M. Lin, R. Madupu, J. Crabtree, S. V. Angiuoli, J. Eisen, R. Seshadri, Q. Ren, M. Wu, T. R. Utterback, S. Smith, M. Lewis, H. Khouri, C. Zhang, H. Niu, Q. Lin, N. Ohashi, N. Zhi, W. Nelson, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, J. Sundaram, S. C. Daugherty, T. Davidsen, A. S. Durkin, M. Gwinn, D. H. Haft, J. D. Selengut, S. A. Sullivan, N. Zafar, L. Zhou, F. Benahmed, H. Forberger, R. Halpin, S. Mulligan, J. Robinson, O. White, Y. Rikihisa, and H. Tettelin. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, H., X. Wang, T. Kikuchi, Y. Kumagai, and Y. Rikihisa. 2007. Porin activity of Anaplasma phagocytophilum outer membrane fraction and purified P44. J. Bacteriol. 189:1998-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang, X., M. A. Payne, Z. Cao, S. B. Foster, J. B. Feix, S. M. Newton, and P. E. Klebba. 1997. Ligand-specific opening of a gated-porin channel in the outer membrane of living bacteria. Science 276:1261-1264. [DOI] [PubMed] [Google Scholar]
- 26.Judd, P. K., R. B. Kumar, and A. Das. 2005. Spatial location and requirements for the assembly of the Agrobacterium tumefaciens type IV secretion apparatus. Proc. Natl. Acad. Sci. USA 102:11498-11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, H. Y., and Y. Rikihisa. 1998. Characterization of monoclonal antibodies to the 44-kilodalton major outer membrane protein of the human granulocytic ehrlichiosis agent. J. Clin. Microbiol. 36:3278-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolbert, C. P., E. S. Bruinsma, A. S. Abdulkarim, E. K. Hofmeister, R. B. Tompkins, S. R. Telford III, P. D. Mitchell, J. Adams-Stich, and D. H. Persing. 1997. Characterization of an immunoreactive protein from the agent of human granulocytic ehrlichiosis. J. Clin. Microbiol. 35:1172-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar, R. B., Y. H. Xie, and A. Das. 2000. Subcellular localization of the Agrobacterium tumefaciens T-DNA transport pore proteins: VirB8 is essential for the assembly of the transport pore. Mol. Microbiol. 36:608-617. [DOI] [PubMed] [Google Scholar]
- 30.Lin, M., and Y. Rikihisa. 2003. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect. Immun. 71:5324-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, M., M. X. Zhu, and Y. Rikihisa. 2002. Rapid activation of protein tyrosine kinase and phospholipase C-γ2 and increase in cytosolic free calcium are required by Ehrlichia chaffeensis for internalization and growth in THP-1 cells. Infect. Immun. 70:889-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, Q., and Y. Rikihisa. 2005. Establishment of cloned Anaplasma phagocytophilum and analysis of p44 gene conversion within an infected horse and infected SCID mice. Infect. Immun. 73:5106-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, Q., Y. Rikihisa, S. Felek, X. Wang, R. F. Massung, and Z. Woldehiwet. 2004. Anaplasma phagocytophilum has a functional msp2 gene that is distinct from p44. Infect. Immun. 72:3883-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, Q., Y. Rikihisa, N. Ohashi, and N. Zhi. 2003. Mechanisms of variable p44 expression by Anaplasma phagocytophilum. Infect. Immun. 71:5650-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, Q., C. Zhang, and Y. Rikihisa. 2006. Analysis of involvement of the RecF pathway in p44 recombination in Anaplasma phagocytophilum and in Escherichia coli by using a plasmid carrying the p44 expression and p44 donor loci. Infect. Immun. 74:2052-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, Q., N. Zhi, N. Ohashi, H. W. Horowitz, M. E. Aguero-Rosenfeld, J. Raffalli, G. P. Wormser, and Y. Rikihisa. 2002. Analysis of sequences and loci of p44 homologs expressed by Anaplasma phagocytophila in acutely infected patients. J. Clin. Microbiol. 40:2981-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manning, D. S., D. K. Reschke, and R. C. Judd. 1998. Omp85 proteins of Neisseria gonorrhoeae and Neisseria meningitidis are similar to Haemophilus influenzae D-15-Ag and Pasteurella multocida Oma87. Microb. Pathog. 25:11-21. [DOI] [PubMed] [Google Scholar]
- 38.Mott, J., and Y. Rikihisa. 2000. Human granulocytic ehrlichiosis agent inhibits superoxide anion generation by human neutrophils. Infect. Immun. 68:6697-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munderloh, U. G., M. J. Lynch, M. J. Herron, A. T. Palmer, T. J. Kurtti, R. D. Nelson, and J. L. Goodman. 2004. Infection of endothelial cells with Anaplasma marginale and A. phagocytophilum. Vet. Microbiol. 101:53-64. [DOI] [PubMed] [Google Scholar]
- 40.Niu, H., Y. Rikihisa, M. Yamaguchi, and N. Ohashi. 2006. Differential expression of VirB9 and VirB6 during the life cycle of Anaplasma phagocytophilum in human leucocytes is associated with differential binding and avoidance of lysosome pathway. Cell. Microbiol. 8:523-534. [DOI] [PubMed] [Google Scholar]
- 41.Noh, S. M., K. A. Brayton, D. P. Knowles, J. T. Agnes, M. J. Dark, W. C. Brown, T. V. Baszler, and G. H. Palmer. 2006. Differential expression and sequence conservation of the Anaplasma marginale msp2 gene superfamily outer membrane proteins. Infect. Immun. 74:3471-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohashi, N., Y. Rikihisa, and A. Unver. 2001. Analysis of transcriptionally active gene clusters of major outer membrane protein multigene family in Ehrlichia canis and E. chaffeensis. Infect. Immun. 69:2083-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer, G. H., A. F. Barbet, W. C. Davis, and T. C. McGuire. 1986. Immunization with an isolate-common surface protein protects cattle against anaplasmosis. Science 231:1299-1302. [DOI] [PubMed] [Google Scholar]
- 44.Palmer, G. H., and T. C. McGuire. 1984. Immune serum against Anaplasma marginale initial bodies neutralizes infectivity for cattle. J. Immunol. 133:1010-1015. [PubMed] [Google Scholar]
- 45.Park, J., K. S. Choi, and J. S. Dumler. 2003. Major surface protein 2 of Anaplasma phagocytophilum facilitates adherence to granulocytes. Infect. Immun. 71:4018-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pawelek, P. D., N. Croteau, C. Ng-Thow-Hing, C. M. Khursigara, N. Moiseeva, M. Allaire, and J. W. Coulton. 2006. Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science 312:1399-1402. [DOI] [PubMed] [Google Scholar]
- 47.Petrovec, M., J. W. Sumner, W. L. Nicholson, J. E. Childs, F. Strle, J. Barlic, S. Lotric-Furlan, and T. Avsic Zupanc. 1999. Identity of ehrlichial DNA sequences derived from Ixodes ricinus ticks with those obtained from patients with human granulocytic ehrlichiosis in Slovenia. J. Clin. Microbiol. 37:209-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rikihisa, Y. 2006. Ehrlichia subversion of host innate responses. Curr. Opin. Microbiol. 9:95-101. [DOI] [PubMed] [Google Scholar]
- 49.Rikihisa, Y. 1991. The tribe Ehrlichieae and ehrlichial diseases. Clin. Microbiol. Rev. 4:286-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rikihisa, Y., N. Zhi, G. P. Wormser, B. Wen, H. W. Horowitz, and K. E. Hechemy. 1997. Ultrastructural and antigenic characterization of a granulocytic ehrlichiosis agent directly isolated and stably cultivated from a patient in New York state. J. Infect. Dis. 175:210-213. [DOI] [PubMed] [Google Scholar]
- 51.Robert, V., E. B. Volokhina, F. Senf, M. P. Bos, P. Van Gelder, and J. Tommassen. 2006. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 4:e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheurer, S. B., J. N. Rybak, C. Roesli, R. A. Brunisholz, F. Potthast, R. Schlapbach, D. Neri, and G. Elia. 2005. Identification and relative quantification of membrane proteins by surface biotinylation and two-dimensional peptide mapping. Proteomics 5:2718-2728. [DOI] [PubMed] [Google Scholar]
- 53.Stich, R. W., G. A. Olah, K. A. Brayton, W. C. Brown, M. Fechheimer, K. Green-Church, S. Jittapalapong, K. M. Kocan, T. C. McGuire, F. R. Rurangirwa, and G. H. Palmer. 2004. Identification of a novel Anaplasma marginale appendage-associated protein that localizes with actin filaments during intraerythrocytic infection. Infect. Immun. 72:7257-7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Storey, J. R., L. A. Doros-Richert, C. Gingrich-Baker, K. Munroe, T. N. Mather, R. T. Coughlin, G. A. Beltz, and C. I. Murphy. 1998. Molecular cloning and sequencing of three granulocytic Ehrlichia genes encoding high-molecular-weight immunoreactive proteins. Infect. Immun. 66:1356-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strik, N. I., A. R. Alleman, A. F. Barbet, H. L. Sorenson, H. L. Wamsley, F. P. Gaschen, N. Luckschander, S. Wong, F. Chu, J. E. Foley, A. Bjoersdorff, S. Stuen, and D. P. Knowles. 2007. Characterization of Anaplasma phagocytophilum major surface protein 5 and the extent of its cross-reactivity with A. marginale. Clin. Vaccine Immunol. 14:262-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Unver, A., S. Felek, C. D. Paddock, N. Zhi, H. W. Horowitz, G. P. Wormser, L. C. Cullman, and Y. Rikihisa. 2001. Western blot analysis of sera reactive to human monocytic ehrlichiosis and human granulocytic ehrlichiosis agents. J. Clin. Microbiol. 39:3982-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voulhoux, R., M. P. Bos, J. Geurtsen, M. Mols, and J. Tommassen. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299:262-265. [DOI] [PubMed] [Google Scholar]
- 58.Wang, X., T. Kikuchi, and Y. Rikihisa. 2006. Two monoclonal antibodies with defined epitopes of P44 major surface proteins neutralize Anaplasma phagocytophilum by distinct mechanisms. Infect. Immun. 74:1873-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58a.Wang, X., Z. Cheng, C. Zhang, T. Kikuchi, and Y. Rikihisa. 2007. Anaplasma phagocytophilum p44 mRNA expression is differentially regulated in mammalian and tick host cells: involvement of the DNA binding protein ApxR. J. Bacteriol, in press. [DOI] [PMC free article] [PubMed]
- 59.Wang, X., Y. Rikihisa, T. H. Lai, Y. Kumagai, N. Zhi, and S. M. Reed. 2004. Rapid sequential changeover of expressed p44 genes during the acute phase of Anaplasma phagocytophilum infection in horses. Infect. Immun. 72:6852-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshiie, K., H. Y. Kim, J. Mott, and Y. Rikihisa. 2000. Intracellular infection by the human granulocytic ehrlichiosis agent inhibits human neutrophil apoptosis. Infect. Immun. 68:1125-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhi, N., N. Ohashi, and Y. Rikihisa. 1999. Multiple p44 genes encoding major outer membrane proteins are expressed in the human granulocytic ehrlichiosis agent. J. Biol. Chem. 274:17828-17836. [DOI] [PubMed] [Google Scholar]
- 62.Zhi, N., N. Ohashi, T. Tajima, J. Mott, R. W. Stich, D. Grover, S. R. Telford III, Q. Lin, and Y. Rikihisa. 2002. Transcript heterogeneity of the p44 multigene family in a human granulocytic ehrlichiosis agent transmitted by ticks. Infect. Immun. 70:1175-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]