Abstract
During mitosis in Saccharomyces cerevisiae, the mitotic spindle moves into the mother–bud neck via dynein-dependent sliding of cytoplasmic microtubules along the cortex of the bud. Here we show that Pac1, the yeast homologue of the human lissencephaly protein LIS1, plays a key role in this process. First, genetic interactions placed Pac1 in the dynein/dynactin pathway. Second, cells lacking Pac1 failed to display microtubule sliding in the bud, resulting in defective mitotic spindle movement and nuclear segregation. Third, Pac1 localized to the plus ends (distal tips) of cytoplasmic microtubules in the bud. This localization did not depend on the dynein heavy chain Dyn1. Moreover, the Pac1 fluorescence intensity at the microtubule end was enhanced in cells lacking dynactin or the cortical attachment molecule Num1. Fourth, dynein heavy chain Dyn1 also localized to the tips of cytoplasmic microtubules in wild-type cells. Dynein localization required Pac1 and, like Pac1, was enhanced in cells lacking the dynactin component Arp1 or the cortical attachment molecule Num1. Our results suggest that Pac1 targets dynein to microtubule tips, which is necessary for sliding of microtubules along the bud cortex. Dynein must remain inactive until microtubule ends interact with the bud cortex, at which time dynein and Pac1 appear to be offloaded from the microtubule to the cortex.
Keywords: Pac1; dynein; Num1; microtubule; nuclear migration
Introduction
To achieve faithful segregation of duplicated chromosomes, cells coordinate the position of the mitotic spindle with the site of cytokinesis. In many types of cells, the site of cytokinesis is determined by the position of the mitotic spindle. Budding yeast Saccharomyces cerevisiae, however, select the site of cell division at the outset of the cell cycle by choosing the site of bud formation. Cytokinesis occurs at the narrow neck between the mother and bud. During the process of budding, the mitotic spindle must therefore move into the neck. The nuclear envelope does not break down, so this process is often termed “nuclear migration.” This movement of the nucleus depends on the interaction of cytoplasmic microtubules with the cell cortex (Stearns, 1997). The mechanism for this movement may be conserved across phyla, because interactions of microtubules with the plasma membrane appear to mediate spindle-positioning processes in higher organisms (Skop and White, 1998; O'Connell and Wang, 2000).
In yeast, nuclear migration and spindle movement occur predominantly in two steps: (1) movement of the nucleus to a position adjacent to the neck, followed by (2) movement of the nucleus into the neck. The first step of nuclear movement involves cytoplasmic microtubules, the kinesin-related protein Kip3, the cortical protein Kar9, and other proteins (Bim1, Bni1, Bud6, Myo2, and actin) that control Kar9 localization or its interaction with microtubules (Cottingham and Hoyt, 1997; DeZwaan et al., 1997; Miller and Rose, 1998; Lee et al., 1999; Miller et al., 1999; Beach et al., 2000; Yin et al., 2000). Early in the cell cycle, a cortical attachment site composed of Kar9 and associated proteins forms at the emerging bud tip. If a growing cytoplasmic microtubule encounters this site, it can be captured. Subsequent shrinkage of the captured microtubule pulls the nucleus toward the nascent bud and orients the preanaphase spindle along the mother–bud axis.
The second step of nuclear movement moves the mitotic spindle into the neck. Cytoplasmic microtubules from the spindle pole body (SPB)* associate laterally with and slide along the bud cortex, pulling the nucleus and the elongating spindle into the neck (Adames and Cooper, 2000). Microtubule sliding depends on the heavy chain of the microtubule-based motor dynein Dyn1, its regulator dynactin complex (which consists of Arp1, Jnm1, and Nip100), and the cortical attachment protein Num1 (Adames and Cooper, 2000; Heil-Chapdelaine et al., 2000). The mechanism of microtubule sliding is poorly understood, but a favored hypothesis is that dynein is anchored in the bud cortex and pulls on the microtubules by walking in the minus end direction toward the SPB (Carminati and Stearns, 1997).
Several known genes of the dynein/dynactin pathway (DYN1, JNM1, NIP100, and NUM1) were isolated in a synthetic lethal screen with the kinesin motor gene cin8; they are called pac (perish in the absence of CIN8) mutants (Geiser et al., 1997). The screen identified four additional genes (PAC1, PAC10, PAC11, and PAC14/BIK1) hypothesized to perform dynein-related functions, based on phenotypic similarity of the mutants with dyn1 mutant.
In this study, we evaluate the function of Pac1 with respect to dynein-mediated nuclear migration. Movies of living individual pac1Δ cells revealed defects in moving the mitotic spindle into the mother–bud neck and microtubule sliding along the bud cortex. We found that Pac1 recruits dynein Dyn1 to the dynamic plus end of microtubules. Analysis of Pac1 and dynein localization in cells lacking other components of the dynein/dynactin pathway revealed novel aspects of the mechanism for how microtubules slide along the bud cortex and move the nucleus.
Results
Pac1 functions in the dynein pathway for nuclear migration
The two processes for nuclear migration in yeast described in the Introduction are genetically redundant. Mutants defective in one process are viable. However, mutations of genes involved in the first process are lethal when combined with mutations of genes in the second process (Cottingham and Hoyt, 1997; DeZwaan et al., 1997; Miller and Rose, 1998; Lee et al., 1999). To identify the process in which Pac1 functions, we crossed a pac1Δ strain with strains carrying deletions of genes of the two processes, followed by sporulation and tetrad dissection (Table I). When pac1Δ was crossed with bim1Δ or kar9Δ, all predicted double mutants (19 of pac1Δ bim1Δ and 17 of pac1Δ kar9Δ) were inviable. The double mutants formed microcolonies consisting of large, swollen, and aberrantly shaped cells that terminated growth at ≤200 cells after 3–4 d incubation at 30°C on rich media. The pac1Δ kip3Δ, pac1Δ bud6Δ, and pac1Δ bni1Δ double mutants were viable and formed colonies that were indistinguishable from those of wild type or parental single mutants. However, these double mutants did exhibit a temperature-sensitive growth defect (see Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1), which was not observed in the parental single mutants, suggesting a weak synthetic interaction of pac1Δ with kip3Δ, bud6Δ, and bni1Δ. No synthetic effect was observed in double mutants of pac1Δ with arp1Δ, dyn1Δ, nip100Δ, jnm1Δ, num1Δ, bik1Δ, or pac11Δ (Table I). These results indicate that Pac1 functions in the dynein-dependent second process.
Table I.
Viability of mutants in combination with pac1 Δ
| Mutant combination | Number of tetrads analyzed | Number of predicted double mutants | Viability of double mutant |
|---|---|---|---|
| pac1Δ bim1Δ | 19 | 19 | Lethal |
| pac1Δ kar9Δ | 18 | 17 | Lethal |
| pac1Δ kip3Δ | 13 | 11 | Viable |
| pac1Δ bud6Δ | 14 | 14 | Viable |
| pac1Δ bni1Δ | 17 | 17 | Viable |
| pac1Δ arp1Δ | 13 | 15 | Viable |
| pac1Δ dyn1Δ | 10 | 5 | Viable |
| pac1Δ nip100Δ | 17 | 17 | Viable |
| pac1Δ jnm1Δ | 16 | 15 | Viable |
| pac1Δ num1Δ | 16 | 16 | Viable |
| pac1Δ bik1Δ | 16 | 13 | Viable |
| pac1Δ pac11Δ | 16 | 16 | Viable |
Pac1 is required for microtubule sliding along the bud cortex
Movement of the nucleus, and hence the mitotic spindle, into the mother–bud neck appears to be mediated by pulling forces produced by the dynein motor acting on cytoplasmic microtubules in the bud. These pulling forces appear to be located at the cortex because cytoplasmic microtubules associate laterally (“plaster”) and slide along the bud cortex (Adames and Cooper, 2000). Free cytoplasmic microtubules not attached to an SPB also slide along the cortex (Adames and Cooper, 2000), further suggesting a cortical location for the pulling force.
To test whether Pac1 is required for microtubule plastering and sliding, we examined cytoplasmic microtubule behavior during movement of the mitotic spindle into the neck in wild type and pac1Δ mutants expressing GFP–tubulin (GFP–Tub1). Movies of living cells were viewed by two independent blinded observers, who evaluated cells in which cytoplasmic microtubules were observed during penetration of the spindle into the neck. In 10 of 30 wild-type cells, microtubules slid along the bud cortex (Fig. 1 A; Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1). This frequency is consistent with previous published data (Adames and Cooper, 2000). In 27 pac1Δ cells, no cases of microtubule sliding along the bud cortex were observed (Fig. 1 B; Videos 2 and 3, available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1). Instead, microtubules in pac1Δ cells swept laterally in the bud, rotating about the SPB. The distal ends of the microtubules occasionally encountered the cortex and appeared attached, but only for a short time (11 ± 6 s; n = 11 events in eight cells). These microtubules then bent, grew, or shrunk, but did not slide, as observed for dynein and dynactin mutants (Adames and Cooper, 2000). Microtubule growth and shrinkage rates were similar in pac1Δ and wild-type mitotic cells. Growth rates were 4.92 μm/min (n = 11) and 4.67 μm/min (n = 13) for pac1Δ and wild-type cells, respectively; and shrinkage rates were 5.59 μm/min (n = 13) and 4.91 μm/min (n = 16). No qualitative differences were observed in the frequency of microtubule catastrophe and rescue for pac1Δ versus wild-type cells. Interestingly, cytoplasmic microtubules laterally plastered along the bud cortex in a few pac1Δ cells. These microtubules did not slide but dissociated from the bud cortex after a short time (10 s; n = 2).
Figure 1.
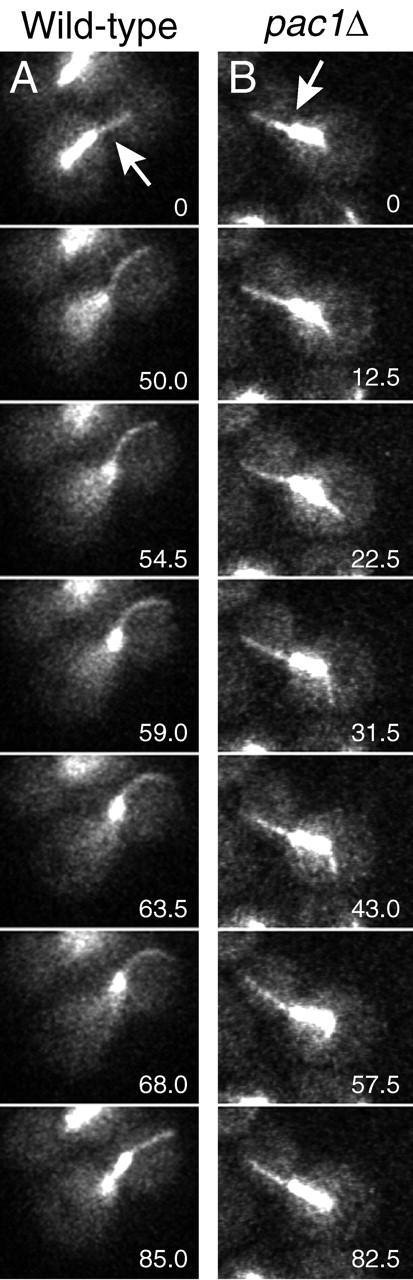
Microtubule and spindle behavior in wild-type and pac1 Δ strains. (A and B) Frames from movies of GFP-labeled microtubules during movement of the mitotic spindle into the mother–bud neck. Arrows indicate the position of the neck. The time elapsed in seconds is indicated. See Videos 1, 2, and 3 (available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1). Strains: wild-type GFP–TUB1, YJC2350; pac1Δ GFP–TUB1, YJC2501.
Spindle elongation appeared to contribute to movement of the spindle into the neck in pac1Δ cells. Upon anaphase onset in pac1Δ cells, well-aligned short spindles near the neck frequently elongated. The elongation was mainly in the direction of the mother. Ultimately, the spindle encountered the cortex of the mother, and further elongation was associated with the other end of the spindle penetrating the neck. At the end of this elongation, the spindle was asymmetrically positioned, with 83 ± 7% (n = 24) of the spindle length located on the mother side of the neck (Videos 2 and 3). We conclude that Pac1 is required for dynein in the bud to pull the spindle into the neck.
PAC1–3GFP and DYN1–3GFP are functional fusion genes
To understand how dynein and Pac1 move the spindle into the neck, we determined their cellular localizations. In previous studies, dynein was found at the SPB and along cytoplasmic microtubules (Shaw et al., 1997), but these studies used truncated and overexpressed tagged versions of dynein heavy chain, so they may not reflect the physiological location of dynein. We designed a tagging vector to integrate three tandem copies of GFP at the 3′ end of the endogenous chromosomal locus of the dynein heavy chain gene DYN1 and PAC1. Multiple copies of GFP were necessary to detect the fusion proteins at endogenous levels. We included a GlyAlaGlyAlaGlyAla linker between the tagged gene and the triple GFP moiety. We performed three assays to evaluate the function of DYN1–3GFP and PAC1–3GFP.
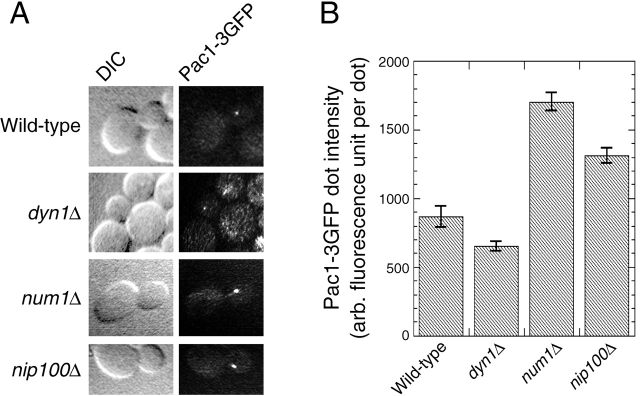
First, we assayed nuclear segregation in DYN1–3GFP and PAC1–3GFP strains (YJC2772 and YJC2770). Loss of DYN1 or PAC1 function causes accumulation of cells with two nuclei in the mother (binucleate cells), more so at lower temperatures (Eshel et al., 1993; Li et al., 1993; Geiser et al., 1997). At 12°C, pac1Δ and dyn1Δ strains in mid-log phase had elevated levels of binucleate cells (Fig. 2). In contrast, strains carrying PAC1–3GFP or DYN1–3GFP as their sole source of Pac1 or dynein, respectively, had a level of binucleate cells similar to that of wild type (Fig. 2). Second, in a more stringent test, PAC1–3GFP and DYN1–3GFP rescued the phenotype of synthetic lethality with bim1Δ (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1). Tetrad dissection produced viable bim1Δ PAC1–3GFP haploids (7 from 7 tetratypes) and bim1Δ DYN1–3GFP haploids (12 from 10 tetratypes and 1 nonparental ditype). PAC1–3GFP also rescued synthetic lethality with kar9Δ in a similar analysis (unpublished data). Third, in liquid rich media (YPD) at 30°C, PAC1–3GFP (YJC2770) and DYN1–3GFP (YJC2772) strains grew with doubling times identical to that of the parental wild-type strain (YJC2296): 106.6 min (n = 2) for PAC1–3GFP; 106.5 min (n = 3) for DYN1–3GFP; and 106.7 min (n = 3) for parental wild type. These results show that the triple GFP tag did not interfere with Pac1 or Dyn1 function.
Figure 2.
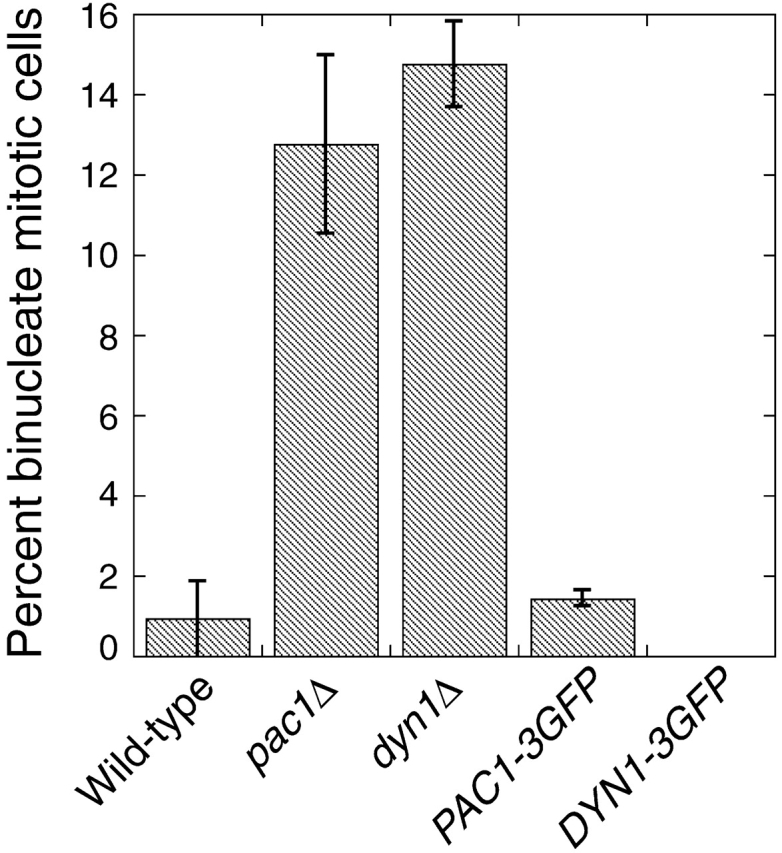
Function of PAC1–3GFP and DYN1–3GFP. Cells grown to mid-log phase at 12°C were fixed, stained for nuclei with DAPI, and imaged. The fraction of budded mitotic cells (Heil-Chapdelaine et al., 2000) with two nuclei in the mother is plotted. Error bars represent standard error (n > 750 cells counted for each strain). Strains: wild type, YJC2296; pac1Δ, YJC1629; dyn1Δ, YJC2007; PAC1–3GFP, YJC2770; DYN1–3GFP, YJC2772.
Localization of Pac1–3GFP
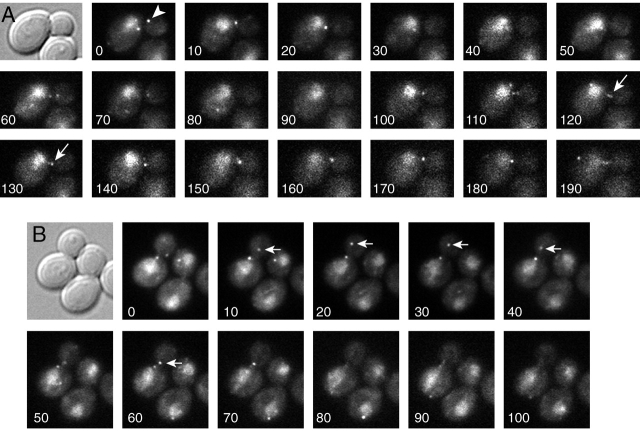
Pac1–3GFP was found in several locations in wild-type cells from asynchronous log-phase cultures. First, Pac1–3GFP localized as dots in the cytoplasm (Fig. 3). Most cells contained one to two dots; the range was zero to four. The dots moved rapidly (3.9 ± 0.7 μm/min, n = 7) and sometimes formed linear streaks (Fig. 3 A, arrows; Videos 4 and 5, available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1). In budded cells, Pac1–3GFP dots moved toward and away from the bud cortex. Once at the cortex, Pac1–3GFP dots were never observed to be stationary for more than 10 s, which was the interval between image acquisitions in the time-lapse movie.
Figure 3.
Localization of Pac1–3GFP. Differential interference contrast (DIC) and Pac1–3GFP wide-field fluorescence images of wild-type cells. (A) Pac1–3GFP is observed in the cytoplasm as dots (arrowhead), which move rapidly and sometimes form linear streaks (arrows). (B) Pac1–3GFP dots move toward and away from the bud (arrows). Pac1–3GFP is also observed in the nucleus, with a diffuse distribution. The time elapsed in seconds is indicated. Strain: PAC1–3GFP, YJC2770. See Videos 4 and 5 (available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1).
Pac1–3GFP dots colocalized with the distal ends of cytoplasmic microtubules (Fig. 4). Imaging of live cells expressing CFP–tubulin and Pac1–3GFP revealed that ∼49% of cytoplasmic microtubules had a Pac1 dot at their distal end, ∼8% had Pac1 along the distal portion of the microtubule, and ∼43% had no Pac1–3GFP (n = 300 microtubules counted). The microtubules and Pac1–3GFP dots were moving during image collection, on a time scale comparable to the interval between images. Thus, these measurements underestimate the extent of colocalization, perhaps by a significant amount.
Figure 4.
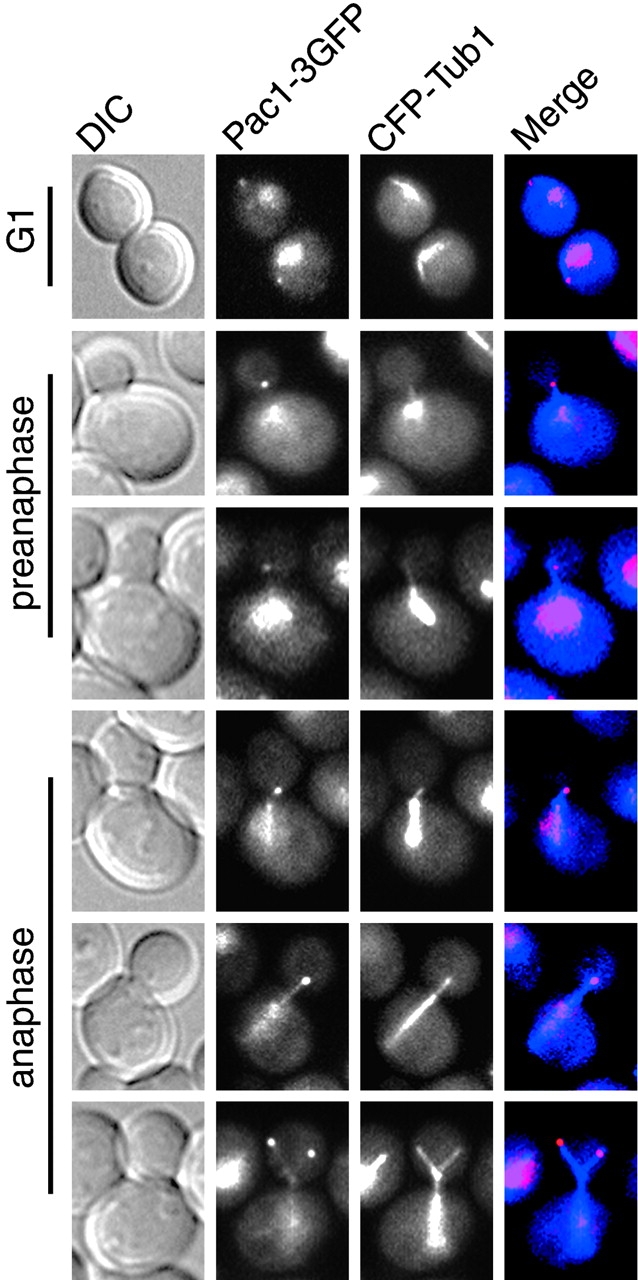
Pac1–3GFP colocalizes with the distal ends of cytoplasmic microtubules. DIC, Pac1–3GFP, and CFP–Tub1 wide-field fluorescence images of wild-type cells. The merged images on the right show Pac1–3GFP in red and CFP–Tub1 in blue. Cytoplasmic dots of Pac1–3GFP colocalize with the distal ends of microtubules at different stages of the cell cycle. Localization of Pac1–3GFP to microtubule ends was observed in the bud before the mitotic spindle moved into the neck (rows 4 and 5 from top) and after the mitotic spindle moved into the neck (row 6). Sometimes Pac1–3GFP was observed along cytoplasmic microtubules (rows 5 and 6). Strain: PAC1–3GFP CFP–TUB1, YJC2814.
Second, Pac1–3GFP was found in the nucleus with a diffuse distribution (Figs. 3 and 4). Nuclear localization was confirmed by costaining with DAPI. Examination of the Pac1 sequence did not reveal any recognizable nuclear localization signal (Fabre and Hurt, 1997), so the mechanism for nuclear targeting remains unclear. Pac1 sometimes appeared at the SPB of unbudded and small-budded cells. This localization is likely due to the presence of short cytoplasmic microtubules, because Pac1 was not observed at the SPB in cases where the distal plus ends of cytoplasmic microtubules were well resolved from the mitotic spindle (Fig. 4).
If Pac1 functions in the bud to assist dynein to move the spindle into the neck, then its localization to distal ends of microtubules may depend on other components of the dynein pathway. To test for such dependence, we examined Pac1–3GFP localization in isogenic mutants carrying deletions of genes in the dynein pathway. The video camera and computer settings for collecting the fluorescence images were the same in all cases, allowing one to compare the intensity of fluorescence between strains. In cells deleted for the cortical attachment molecule Num1, we observed a twofold increase in the intensity of Pac1–3GFP cytoplasmic dots in the bud (P < 0.0001, based on measurements by a blinded observer). Fig. 5 shows the intensities. Videos 6 and 7 (available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1) are representative of wild-type and num1Δ strains, respectively. Pac1 dots in num1Δ cells moved rapidly and sometimes formed streaks across the bud cytoplasm, indicating that they likely correspond to the distal ends of cytoplasmic microtubules. Cytoplasmic microtubules in num1Δ cells are known to have dynamics consistent with these observations (Geiser et al., 1997; Heil-Chapdelaine et al., 2000; Farkasovsky and Kuntzel, 2001). The fluorescence intensity of Pac1–3GFP dots was also increased in the dynactin mutants nip100Δ (P < 0.0001; Fig. 5; Video 8, available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1) and arp1Δ (unpublished data). In contrast, Pac1–3GFP dot intensity was slightly reduced in dyn1Δ cells, but by an insignificant margin compared with wild-type cells (P = 0.015; Fig. 5; Video 9, available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1).
Figure 5.
Localization of Pac1–3GFP in living dyn1 Δ , num1 Δ , and nip100 Δ cells. (A) DIC and a frame from movies of Pac1–3GFP fluorescence in isogenic wild-type and mutant cells. The video camera and microscope settings were the same, allowing one to compare the intensity of fluorescence in the different strains. num1Δ and nip100Δ cells showed increased intensity of Pac1–3GFP dots in the bud. See Videos 6–9 (available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1). (B) Relative fluorescence intensity of motile Pac1–3GFP dots in wild-type and mutant cells. The average corrected fluorescence per dot is plotted; n = 25 dots for wild type, 75 dots for dyn1Δ, 170 dots for num1Δ, 171 dots for nip100Δ. Error bars represent standard error. Strains: PAC1–3GFP, YJC2770; PAC1–3GFP dyn1Δ, YJC2907; PAC1–3GFP num1Δ, YJC2905; PAC1–3GFP nip100Δ, YJC2904.
Localization of Dyn1–3GFP
Dynein Dyn1–3GFP also localized to the distal ends of cytoplasmic microtubules (Fig. 6 B). In wild-type cells, Dyn1–3GFP was observed as dots that moved rapidly in the cytoplasm (Fig. 6 A; Videos 10 and 11, available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1). Dyn1–3GFP dots moved at a rate of 3.8 ± 1.6 μm/min (n = 12) and sometimes formed linear streaks, as seen for Pac1–3GFP. Imaging of live cells expressing CFP–tubulin and Dyn1–3GFP revealed that ∼47% of all observed cytoplasmic microtubules had a Dyn1 dot at their distal end, ∼10% had Dyn1 along the distal portion of the microtubule, and ∼43% had no Dyn1–3GFP (n = 127 microtubules counted). As seen for the case of Pac1–3GFP, the microtubules and Dyn1–3GFP dots were moving on a relatively rapid time scale during image collection. Thus, these measurements represent the lower limit for colocalization of Dyn1–3GFP with microtubule ends. In striking contrast to Pac1–3GFP, some dots of Dyn1–3GFP did not move (>6 min). These stationary dynein dots were found only at the cortex, and only in unbudded cells or the mother of budded cells (Video 11, middle cell).
Figure 6.

Localization of Dyn1–3GFP in living wild-type cells. (A) DIC and movie frames of Dyn1–3GFP fluorescence in a wild-type cell (YJC2772). Each fluorescence image is a two-dimensional projection of a 4-μm Z-stack of confocal images. The time elapsed in seconds is indicated. Dyn1–3GFP is observed as a dot that moves away from and toward the bud (arrows). Dyn1–3GFP sometimes appears as a linear streak (t = 28, 84, and 147 s). Dyn1–3GFP is also observed as stationary cortical dots, but only in the mother (see Videos 10 and 11, available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1). (B) Dyn1–3GFP colocalizes with the distal ends of cytoplasmic microtubules. DIC, Dyn1–3GFP, and CFP–Tub1 wide-field fluorescence images of wild-type cells (YJC2914) at G1 (top), preanaphase (middle), and anaphase (bottom). The merged images show cytoplasmic Dyn1–3GFP dots (red) at the distal ends of microtubules (blue).
We asked whether Dyn1 localization to the distal ends of microtubules depends on other components of the dynein pathway. We examined Dyn1–3GFP localization in isogenic arp1Δ, num1Δ, and pac1Δ mutants. In arp1Δ and num1Δ cells, Dyn1–3GFP was seen as dots in the cytoplasm, which moved rapidly and sometimes formed linear streaks, similar to what was observed in wild-type cells. Furthermore, the fluorescence intensity of the dots was increased relative to wild type, by an amount slightly greater than that seen for Pac1–3GFP dots in these mutants (Fig. 7; compare Video 11 [wild type] with Videos 12 [arp1Δ] and 13 [num1Δ], available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1).
Figure 7.
Localization of Dyn1–3GFP in living arp1 Δ , num1 Δ , and pac1 Δ cells. (A) DIC and Dyn1–3GFP fluorescence images of isogenic wild-type and mutant cells. The video camera and microscope settings were the same for the different strains. arp1Δ and num1Δ cells showed increased fluorescence intensity for Dyn1–3GFP dots in the bud. pac1Δ cells showed the absence of cytoplasmic Dyn1–3GFP motile dots. See Videos 12–15 (available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1). (B) Relative fluorescence intensity of motile Dyn1–3GFP dots in wild-type, arp1Δ, and num1Δ cells. The average corrected fluorescence per dot is plotted; n = 72 dots for wild type, 91 dots for arp1Δ, 84 dots for num1Δ. Error bars represent standard error. Strains: DYN1–3GFP, YJC2772; DYN1–3GFP arp1Δ, YJC2908; DYN1–3GFP num1Δ, YJC2910; DYN1–3GFP pac1Δ, YJC2912.
In contrast, in pac1Δ cells, no localization of Dyn1–3GFP to motile dots in the cytoplasm was observed (Fig. 7 A; Videos 14 and 15, available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1). Immunoblot analysis of Dyn1–3GFP protein in pac1Δ and wild-type cell lysates revealed that the level of Dyn1–3GFP was the same in both strains (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1); thus, the loss of Dyn1–3GFP from the distal ends of microtubules was due to a defect in protein targeting, not protein stability.
Stationary dots of Dyn1–3GFP were observed at the mother cortex in some pac1Δ cells; however, the intensity was quite low (unpublished data). These stationary dots were not observed in num1Δ or arp1Δ cells. The origin and function of stationary dynein dots at the mother cortex remain unclear at this point.
Discussion
In these studies, we found that Pac1 functions in the dynein/dynactin pathway for nuclear migration in budding yeast. In pac1Δ cells, efficient movement of the mitotic spindle into the bud neck is defective due to lack of microtubule sliding along the bud cortex, as seen in dynein- and dynactin-null mutants. We also found that dynein heavy chain, Dyn1, and Pac1 are targeted to the distal ends of microtubules. Pac1 is necessary for targeting dynein, but not vice versa. Neither dynein nor Pac1 require dynactin or the cortical attachment protein Num1 for targeting to microtubule ends. Rather, the amount of dynein and Pac1 at microtubule ends is increased in the absence of dynactin or Num1. These results suggest a model in which microtubule ends with dynein and Pac1 probe the cortex for attachment sites and then deliver the motor to be activated and used in force production for sliding.
Pac1 functions in nuclear migration
PAC1 was isolated in a synthetic lethal screen with cin8, which encodes a kinesin motor involved in spindle elongation (Geiser et al., 1997). This screen identified many components of the dynein/dynactin pathway. Our genetic analysis here supports the conclusion that Pac1 functions in the dynein pathway for nuclear migration. Many of the genetic interactions exhibited by pac1Δ are similar to those observed with mutants in the dynein/dynactin pathway, including strong synthetic interactions with kar9Δ and bim1Δ, two components of the microtubule capture/shrinkage pathway for nuclear migration. A weak synthetic interaction of pac1Δ with bud6Δ, another component of the microtubule capture/shrinkage pathway, was observed, as expected given the relatively mild defects in nuclear migration and Kar9 localization in bud6Δ cells (Miller et al., 1999). Previous studies also found the dyn1Δ bud6Δ double mutant to be viable with a mild growth defect (Miller et al., 1999), as seen here for the pac1Δ bud6Δ mutant. On the other hand, the weak synthetic interactions that we observed for pac1Δ with bni1Δ and kip3Δ (two more components of the microtubule capture/shrinkage pathway) were unexpected, as dyn1Δ bni1Δ and dyn1Δ kip3Δ double mutants were lethal in previous studies (Cottingham and Hoyt, 1997; DeZwaan et al., 1997; Miller and Rose, 1998; Miller et al., 1999). Perhaps a low level of residual dynein function is sufficient for viability in pac1Δ bni1Δ and pac1Δ kip3Δ cells. Alternatively, differences in genetic backgrounds of the strains used for the respective studies may account for the different level of synthetic interaction.
Mechanism of dynein-mediated microtubule sliding
To move the mitotic spindle into the neck, dynein appears to generate force between cytoplasmic microtubules and the bud cortex. As a motor, dynein presumably moves with respect to the microtubule; therefore, we expect that dynein is anchored at the bud cortex as the microtubule slides. We anticipated that dynein and Pac1 might be localized to the bud cortex before cytoplasmic microtubules plaster and slide along the cortex. Instead, we found that Pac1 and dynein localize to the distal ends of microtubules, which appear to grow and shrink in search of attachment sites on the bud cortex. The lack of stationary Pac1–3GFP and Dyn1–3GFP dots along the bud suggests that anchoring of dynein to the bud cortex may be transient or at a low level.
We hypothesize that microtubule sliding along the bud cortex occurs in the following steps (Fig. 8; Video 16, available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1). First, dynein and Pac1 are targeted to plus ends of microtubules. Second, plus ends of microtubules are captured by cortical attachment sites, which contain Num1 and probably other components. Third, dynein and Pac1 are offloaded from the end of the microtubule to the cortex and anchored there. Fourth, the motor activity of dynein is activated, causing it to walk toward the minus end of the microtubule at the SPB. Because dynein is anchored, the microtubule slides, and the spindle is pulled into the bud neck. One intriguing observation in support of this model is that the levels of Pac1 and Dyn1 are higher at the plus ends of microtubules in cells lacking Num1. This observation suggests that a productive interaction of the microtubule end with the bud cortex is required to offload dynein and Pac1 from the microtubule end.
Figure 8.

Proposed mechanism for microtubule sliding along the bud cortex. (Step 1) Dynein Dyn1 (red) and Pac1 (yellow) associated with the distal plus end of a cytoplasmic microtubule probe the bud cortex for attachment sites, which contain Num1 (blue) and probably other components. (Step 2) Upon attachment to a cortical site, dynein and Pac1 are offloaded from the end of microtubule and anchored to the cortex. (Step 3) The motor activity of anchored dynein is activated, causing it to walk toward the SPB. The microtubule slides, and the spindle is pulled into the bud neck. See Video 16 (available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1) and text for further discussion.
Pac1 is required to target dynein to microtubule ends. In other systems, Pac1/LIS1 homologues have been found to bind directly to dynein heavy chain (Sasaki et al., 2000; Hoffmann et al., 2001; Tai et al., 2002), so Pac1 may simultaneously bind the microtubule and dynein Dyn1 at microtubule ends. Pac1 can bind the microtubule directly or indirectly via a microtubule binding protein. We tested whether Pac1 localization depends on Bim1, the yeast EB1 homologue that also localizes to plus ends of microtubules. We observed no qualitative differences in the dynamic and intensity of Pac1 dots in bim1Δ versus wild-type cells (unpublished data). Dynein targeted to microtubule ends can also bind directly to the microtubule, which may enhance targeting. Note that active dynein would move away from the plus end, toward the SPB, so something must be inhibiting dynein or counteracting its activity, along with targeting it to the plus end. Pac1 may have one or more of these roles.
Our results here with dynactin mutants provide insight into the function of dynactin. Dynein and Pac1 accumulate at the plus ends of microtubules in cells lacking dynactin. Therefore, dynactin is not required for targeting of dynein to plus ends. Instead, dynactin appears to function later, in microtubule capture or offloading, anchoring, or activating dynein at the bud cortex. In studies in other systems, dynactin promotes dynein-based movements along microtubules in vitro by increasing the processivity of the motor (King and Schroer, 2000). The NH2-terminal CAP-Gly domain of the dynactin p150Glued subunit binds microtubules, which may tether the cargo to the microtubule during the mechanochemical cycle of dynein, preventing diffusional loss of the cargo (King and Schroer, 2000). The presence of the conserved CAP-Gly domain in the NH2-terminal region of Nip100, the yeast p150Glued homologue, favors this hypothesis (Kahana et al., 1998). Nip100 has been localized at overexpressed, but not endogenous, levels, so its site of action remains somewhat uncertain.
Comparison of Pac1 with Aspergillus NUDF and vertebrate LIS1
Pac1 belongs to the conserved family of LIS1 lissencephaly proteins consisting of members from various organisms, including Schizosaccharomyces pombe, Aspergillus nidulans, Drosophila melanogaster, Caenorhabditis elegans, and human (see phylogenetic analysis in Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1). LIS1 proteins have a predicted coiled-coil region in the NH2 terminus and seven tandem WD40 repeats in the COOH-terminal two thirds of the molecule (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1). LIS1 proteins are found at the plus ends of cytoplasmic microtubules in several organisms (Han et al., 2001; Coquelle et al., 2002).
The role of LIS1 proteins in dynein targeting appears to differ from organism to organism. In Aspergillus, deletion of the LIS1 homologue NUDF did not affect the localization of a GFP fusion of NUDA, the dynein heavy chain homologue, to the distal ends of cytoplasmic microtubules at hyphal tips (Zhang et al., 2002). In cultured mouse fibroblasts, reduction of LIS1 expression appeared to cause redistribution of dynein heavy chain to regions around the nucleus (Sasaki et al., 2000). It is not clear, however, whether this redistribution was due to mislocalization of dynein to microtubule ends or a result of enrichment of microtubules near the nucleus in these LIS1 heterozygous null cells (Smith et al., 2000). We found here, in budding yeast, that pac1Δ mutants show loss of dynein Dyn1 localization to the distal ends of cytoplasmic microtubules, which probably leads to loss of dynein delivery to cortical attachment sites and an inability to carry out productive microtubule–cortex interactions. These results may reflect differences in the approaches used for the respective studies in different organisms. They may also reflect a loss or gain of particular LIS1–ligand interactions subsequent to the divergence of these organisms, as suggested by the weak sequence similarity between vertebrate and yeast dynein/dynactin components (McMillan and Tatchell, 1994; Geiser et al., 1997; Kahana et al., 1998).
Materials and methods
Strains, genetic manipulation, and transformation
Yeast culture, media, and genetic manipulations were performed by standard methods (Kaiser et al., 1994). Lithium acetate transformation was performed as previously described (Knop et al., 1999). Table II lists the S. cerevisiae strains used in this study. Strain YJC1629 (pac1Δ) was crossed to strains from the deletion consortium (Research Genetics genomic resources; bim1Δ, kar9Δ, arp1Δ, num1Δ, dyn1Δ, nip100Δ, jnm1Δ, bik1Δ, kip3Δ, pac11Δ, bud6Δ, or bni1Δ) to construct heterozygous diploid double mutants. From each cross, two independent heterozygous diploids were sporulated and tetrad dissected. Progeny were examined for germination and colony formation in the microscope. Markers were analyzed by replica plating to selective media.
Table II. Strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| YJC1550 | Mata bim1::HIS3 ade2 ade3 ura3 leu2 trp1 lys2 | Muhua et al., 1998 |
| YJC1629 | Matα leu2 ura3 his3-Δ200 pac1::HIS3 | This study |
| YJC2007 | Matα leu2 ura3 his3-Δ200 dyn1::HIS3 | This study |
| YJC2225 | Mata kar9::KanR ade2 ade3 leu2 ura3 trp1 LYS2 [pAFS92 GFP-TUB1::URA3] | This study |
| YJC2296 | Matα ura3-52 lys2-801 leu2-Δ1 his3-Δ200 trp1-Δ63 | YEF473B (Amberg et al., 1997) |
| YJC2350 | Mata lys2-801 leu2-Δ1 his3-Δ200 trp1-Δ63 ura3-52 [pAFS92 GFP-TUB1::URA3] | This study |
| YJC2501 | Mata lys2-801 leu2-Δ1 his3-Δ200 trp1-Δ63 ura3-52 pac1::HIS3 [pAFS92 GFP-TUB1::URA3] | This study |
| YJC2770 | Matα PAC1-3GFP::TRP1 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 trp1-Δ63 | This study |
| YJC2772 | Matα DYN1-3GFP::TRP1 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 trp1-Δ63 | This study |
| YJC2814 | Matα PAC1-3GFP::TRP1 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 trp1-Δ63 [pAFS125C CFP-TUB1::URA3] | This study |
| YJC2904 | Matα PAC1-3GFP::TRP1 nip100::HIS3 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 trp1-Δ63 | This study |
| YJC2905 | Matα PAC1-3GFP::TRP1 num1::HIS3 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 trp1-Δ63 | This study |
| YJC2907 | Matα PAC1-3GFP::TRP1 dyn1::HIS3 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 trp1-Δ63 | This study |
| YJC2908 | Matα DYN1-3GFP::TRP1 arp1::HIS3 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 trp1-Δ63 | This study |
| YJC2910 | Matα DYN1-3GFP::TRP1 num1::HIS3 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 trp1-Δ63 | This study |
| YJC2912 | Matα DYN1-3GFP::TRP1 pac1::HIS3 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 trp1-Δ63 | This study |
| YJC2914 | Matα DYN1-3GFP::TRP1 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 trp1-Δ63 [pAFS125C CFP-TUB1::URA3] | This study |
Construction of PAC1–3GFP and DYN1–3GFP strains
We constructed tagging vectors designed to integrate three tandem copies of GFP at the 3′ end of the PAC1 or DYN1 locus. We first built a plasmid containing three tandemly fused GFP genes and the TRP1 marker. In brief, an engineered BamHI–BglII fragment containing GFP (with S65G and S72A mutations for brighter expression in yeast) without a stop codon was cloned into BamHI- and BglII-digested pBS1479 (Rigaut et al., 1999), a pBluescript-based cloning vector. The resulting plasmid is pBS-1xGFP. We verified the sequence of GFP, excised it with BamHI and BglII, and recloned into the BglII site of pBS-1xGFP to generate pBS-2xGFP. A third BamHI–BglII fragment containing GFP with an engineered stop codon was amplified, verified by sequencing, and cloned into the BglII site of pBS-2xGFP, yielding pBS-3xGFP. We next cloned a BamHI–KpNI fragment containing the TRP1 marker into BglII- and KpNI-digested pBS-3xGFP, creating pBS-3xGFP–TRP1. To target the 3xGFP–TRP1 cassette into the PAC1 or DYN1 locus, an engineered BglII–BamHI fragment containing 3′ coding sequence of PAC1 (575–1482 bp) or DYN1 (11724–12276 bp) and a GlyAlaGlyAlaGlyAla linker was cloned into BamHI-digested pBS-3xGFP–TRP1. The resulting plasmids contain a fragment of PAC1 or DYN1 sequence fused in frame to the triple GlyAla linker and triple GFP. Wild-type haploid cells (YJC2296) were transformed with these integrating vectors linearized by ClaI in the middle of the PAC1 or DYN1 sequence. Stable Trp+ transformants were selected and then screened for proper targeting by PCR.
We deleted various genes in the PAC1–3GFP or DYN1–3GFP strains by oligonucleotide-mediated disruption (Baudin et al., 1993). Stable transformants were tested for appropriate disruption by PCR from genomic DNA. Two independent transformants were chosen for each disruption for subsequent localization studies.
Complementation analysis
We tested triple GFP–tagged Pac1 and Dyn1 for function by assaying nuclear segregation and synthetic lethality with bim1Δ and kar9Δ. To assay for nuclear segregation, mid-log cells grown in YPD at 12°C were harvested, fixed in 70% ethanol, and stained with DAPI. Images of random fields (>80) of cells were collected on an IX70 Olympus fluorescence microscope with a cooled CCD camera (CCD-300T, Dage-MTI). The fraction of mitotic cells with two nuclei in the mother was plotted for each strain. To test for rescue of synthetic lethality, we crossed YJC2770 (PAC1–3GFP) or YJC2772 (DYN1–3GFP) to YJC1550 (bim1Δ) or YJC2225 (kar9Δ). Tetrad analysis of two independent heterozygous diploids from each cross was performed as above.
Fluorescence microscopy
We used GFP–TUB1 (pAFS92, a gift from A. Straight and A. Murray (University of California San Francisco, San Francisco, CA; Straight et al., 1997) and CFP–TUB1 (pAFS125C, a gift from D. Beach and K. Bloom, University of North Carolina at Chapel Hill, Chapel Hill, NC) to visualize microtubules. For analysis of microtubule sliding events, cells expressing GFP–TUB1 were observed with a 100× objective on a BX60 Olympus fluorescence microscope, and images were collected with NIH Image software at two frames per second at a single focal plane using an intensified video camera with a digital image processor (ISIT68 and DSP2000; Dage-MTI) (Heil-Chapdelaine et al., 2000). Living cells from an asynchronous culture were observed during mitosis. When the mitotic spindle moved into the neck, we scored the presence of microtubule sliding along the bud cortex for cells in which cytoplasmic microtubules of the bud were in focus across their entire length. For localization studies, mid-log cells were grown in YPD or selective synthetic defined media (Bio101), washed with nonfluorescent media, and visualized directly on an agarose pad containing nonfluorescent media (Heil-Chapdelaine et al., 2000). Movies of Pac1–3GFP or Dyn1–3GFP were made using QED software (QED Imaging Inc.) by collecting five 1-μm slices at 10-s intervals with an intensified video camera (Dage ISIT68) or an intensified CCD camera (XR-Mega10; Stanford Photonics, Inc.). The fluorescence of Pac1–3GFP and Dyn1–3GFP cytoplasmic dots was measured (NIH image software) and corrected for the background fluorescence from an adjacent region next to each dot.
Cell lysis and immunoblotting
Yeast cultures were grown to mid-log phase in 5 ml YPD or selective media and harvested. Cell pellets were resuspended in 0.5 ml of ice cold lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1.5% Triton X-100, 1 mM PMSF, plus protease inhibitor cocktail tablet [Roche Applied Science]) and lysed by bead beating at 4°C in round-bottom glass tubes, five times for 1 min each, with 2 min on ice between each beating. Crude lysate was clarified at 500 g for 2–5 min and separated by 4–15% gradient or 4% linear SDS-PAGE. Proteins were electroblotted to nitrocellulose in 20 mM CAPS, pH 11.0, supplemented with 0.05% SDS for 30 h at 4°C. Mouse anti-GFP monoclonal antibody (BD Biosciences; CLONTECH) and HRP-conjugated goat anti–mouse antibody (Jackson ImmunoResearch Laboratories) were used at 1:1,000 and 1:10,000 dilutions, respectively.
Online supplemental material
The online version of this article (available at http://www.jcb.org/cgi/content/full/jcb.200209022/DC1) includes additional figures showing synthetic interactions of pac1Δ with kip3Δ, bud6Δ, and bni1Δ, complementation of synthetic lethality with bim1Δ by PAC1–3GFP and DYN1–3GFP, immunoblot analysis of Dyn1–3GFP protein in wild-type and pac1Δ lysates, phylogenetic analysis of Pac1/LIS1 sequences, domain organization of Pac1, movies of microtubule behavior during movement of the spindle into the mother–bud neck, and movies of Pac1–3GFP and Dyn1–3GFP localization in wild-type or mutant backgrounds.
Supplemental Material
Acknowledgments
We thank Dale Beach and Kerry Bloom for pAFS125C plasmid. We are grateful to Guillaume Castillon and Rick Heil-Chapdelaine for extensive advice and assistance. We thank Michelle Kaiser for assistance with double blind experiments, and Michael Young and Magdalena Bezanilla for reading the manuscript.
Wei-Lih Lee was supported by a Damon Runyon Cancer Research Foundation Fellowship, DRG-1671. This work was supported by National Institutes of Health grant GM 47337.
The online version of this article includes supplemental material.
Footnotes
Abbreviations used in this paper: DIC, differential interference contrast; SPB, spindle pole body.
References
- Adames, N.R., and J.A. Cooper. 2000. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 149:863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg, D.C., J.E. Zahner, J.W. Mulholland, J.R. Pringle, and D. Botstein. 1997. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol. Biol. Cell. 8:729–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin, A., O. Ozierkalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach, D.L., J. Thibodeaux, P. Maddox, E. Yeh, and K. Bloom. 2000. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr. Biol. 10:1497–1506. [DOI] [PubMed] [Google Scholar]
- Carminati, J.L., and T. Stearns. 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138:629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquelle, F.M., M. Caspi, F.P. Cordelieres, J.P. Dompierre, D.L. Dujardin, C. Koifman, P. Martin, C.C. Hoogenraad, A. Akhmanova, N. Galjart, et al. 2002. LIS1, CLIP-170's key to the dynein/dynactin pathway. Mol. Cell. Biol. 22:3089–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham, F.R., and M.A. Hoyt. 1997. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J. Cell Biol. 138:1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan, T.M., E. Ellingson, D. Pellman, and D.M. Roof. 1997. Kinesin-related Kip3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J. Cell Biol. 138:1023–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel, D., L.A. Urrestarazu, S. Vissers, J.C. Jauniaux, J.C. vanVliet-Reedijk, R.J. Planta, and I.R. Gibbons. 1993. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc. Natl. Acad. Sci. USA. 90:11172–11176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre, E., and E. Hurt. 1997. Yeast genetics to dissect the nuclear pore complex and nucleocytoplasmic trafficking. Annu. Rev. Genet. 31:277–313. [DOI] [PubMed] [Google Scholar]
- Farkasovsky, M., and H. Kuntzel. 2001. Cortical Num1p interacts with the dynein intermediate chain Pac11p and cytoplasmic microtubules in budding yeast. J. Cell Biol. 152:251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser, J.R., E.J. Schott, T.J. Kingsbury, N.B. Cole, L.J. Totis, G. Bhattacharyya, L. He, and M.A. Hoyt. 1997. Saccharomyces cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol. Biol. Cell. 8:1035–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, G., B. Liu, J. Zhang, W. Zuo, N.R. Morris, and X. Xiang. 2001. The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr. Biol. 11:719–724. [DOI] [PubMed] [Google Scholar]
- Heil-Chapdelaine, R.A., J.R. Oberle, and J.A. Cooper. 2000. The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J. Cell Biol. 151:1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, B., W. Zuo, A. Liu, and N.R. Morris. 2001. The LIS1-related protein NUDF of Aspergillus nidulans and its interaction partner NUDE bind directly to specific subunits of dynein and dynactin and to α- and γ-tubulin. J. Biol. Chem. 276:38877–38884. [DOI] [PubMed] [Google Scholar]
- Kahana, J.A., G. Schlenstedt, D.M. Evanchuk, J.R. Geiser, M.A. Hoyt, and P.A. Silver. 1998. The yeast dynactin complex is involved in partitioning the mitotic spindle between mother and daughter cells during anaphase B. Mol. Biol. Cell. 9:1741–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Plainview, NY. 234 pp.
- King, S.J., and T.A. Schroer. 2000. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat. Cell Biol. 2:20–24. [DOI] [PubMed] [Google Scholar]
- Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 15:963–972. [DOI] [PubMed] [Google Scholar]
- Lee, L., S.K. Klee, M. Evangelista, C. Boone, and D. Pellman. 1999. Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J. Cell Biol. 144:947–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.Y., E. Yeh, T. Hays, and K. Bloom. 1993. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc. Natl. Acad. Sci. USA. 90:10096–10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan, J.N., and K. Tatchell. 1994. The JNM1 gene in the yeast Saccharomyces cerevisiae is required for nuclear migration and spindle orientation during the mitotic cell cycle. J. Cell Biol. 125:143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, R.K., and M.D. Rose. 1998. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140:377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, R.K., D. Matheos, and M.D. Rose. 1999. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J. Cell Biol. 144:963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhua, L., N.R. Adames, M.D. Murphy, C.R. Shields, and J.A. Cooper. 1998. A cytokinesis checkpoint requiring the yeast homolog of an APC-binding protein. Nature. 393:487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, C.B., and Y. Wang. 2000. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol. Biol. Cell. 11:1765–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032. [DOI] [PubMed] [Google Scholar]
- Sasaki, S., A. Shionoya, M. Ishida, M.J. Gambello, J. Yingling, A. Wynshaw-Boris, and S. Hirotsune. 2000. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 28:681–696. [DOI] [PubMed] [Google Scholar]
- Shaw, S.L., E. Yeh, P. Maddox, E.D. Salmon, and K. Bloom. 1997. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J. Cell Biol. 139:985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop, A.R., and J.G. White. 1998. The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegans embryos. Curr. Biol. 8:1110–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D.S., M. Niethammer, R. Ayala, Y. Zhou, M.J. Gambello, A. Wynshaw-Boris, and L.H. Tsai. 2000. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat. Cell Biol. 2:767–775. [DOI] [PubMed] [Google Scholar]
- Stearns, T. 1997. Motoring to the finish: kinesin and dynein work together to orient the yeast mitotic spindle. J. Cell Biol. 138:957–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight, A.F., W.F. Marshall, J.W. Sedat, and A.W. Murray. 1997. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 277:574–578. [DOI] [PubMed] [Google Scholar]
- Tai, C.Y., D.L. Dujardin, N.E. Faulkner, and R.B. Vallee. 2002. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J. Cell Biol. 156:959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, H.W., D. Pruyne, T.C. Huffaker, and A. Bretscher. 2000. Myosin V orientates the mitotic spindle in yeast. Nature. 406:1013–1015. [DOI] [PubMed] [Google Scholar]
- Zhang, J., G. Han, and X. Xiang. 2002. Cytoplasmic dynein intermediate chain and heavy chain are dependent upon each other for microtubule end localization in Aspergillus nidulans. Mol. Microbiol. 44:381–392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.