Abstract
Ipl1p is the budding yeast member of the Aurora family of protein kinases, critical regulators of genomic stability that are required for chromosome segregation, the spindle checkpoint, and cytokinesis. Using time-lapse microscopy, we found that Ipl1p also has a function in mitotic spindle disassembly that is separable from its previously identified roles. Ipl1–GFP localizes to kinetochores from G1 to metaphase, transfers to the spindle after metaphase, and accumulates at the spindle midzone late in anaphase. Ipl1p kinase activity increases at anaphase, and ipl1 mutants can stabilize fragile spindles. As the spindle disassembles, Ipl1p follows the plus ends of the depolymerizing spindle microtubules. Many Ipl1p substrates colocalize with Ipl1p to the spindle midzone, identifying additional proteins that may regulate spindle disassembly. We propose that Ipl1p regulates both the kinetochore and interpolar microtubule plus ends to regulate its various mitotic functions.
Keywords: Ipl1/Aurora protein kinase; spindle; mitosis; microtubule; budding yeast
Introduction
The proper regulation of mitotic spindle dynamics is a fundamental aspect of chromosome segregation and is crucial for the maintenance of genetic information. During S phase, chromosomes replicate and linkage is established between the sister chromatids. The mitotic spindle forms, and microtubules attach to chromosomes via kinetochores, multiprotein complexes that assemble onto centromeric DNA. Once the sister kinetochores attach to microtubules from opposite poles, the linkage between the sister chromatids is dissolved and the chromosomes segregate, resulting in two daughter cells with identical genomes.
Mitotic spindles consist of two types of microtubules: those that bind to kinetochores (kinetochore microtubules) and those that interdigitate with each other (interpolar microtubules) (for review see Desai and Mitchison, 1997). The microtubule minus ends are bound to the microtubule organizing center, whereas the dynamic plus ends either capture kinetochores or interdigitate. In budding yeast, the mitotic spindle is formed after separation of the duplicated spindle pole bodies (SPBs)* (for review see Winey and O'Toole, 2001). During metaphase, cytoplasmic microtubules ensure that the nucleus migrates to the bud neck (nuclear migration) and the spindle aligns parallel to the mother-bud axis (spindle orientation) (for review see Segal and Bloom, 2001). Anaphase then occurs in two phases. In anaphase A, the sister chromatids move toward the SPBs, and in anaphase B a rapid phase of spindle elongation is followed by a second slow phase that drives the SPBs apart (Yeh et al., 1995; Straight et al., 1997). When the spindle has reached its maximal length of ∼10 μm, it disassembles exclusively from the plus ends at the midzone, the site of overlap between the interpolar microtubules (Maddox et al., 2000). Proper regulation of microtubule dynamics is critical for chromosome segregation and is performed in part by motor and nonmotor microtubule-associated proteins.
The fidelity of spindle assembly is monitored by the spindle checkpoint that prevents cells from separating their sister chromatids until kinetochore attachment is complete and kinetochores are under tension (for review see Millband et al., 2002). Spindle checkpoint arrest is achieved through inhibition of the anaphase-promoting complex (APC), a multiprotein ubiquitin ligase that catalyses proteolysis of the mitotic inhibitor proteins Pds1 and Clb2 (for review see Peters, 2002). When the APC targets Pds1p for degradation, the Esp1 protease is liberated to cleave the cohesin Mcd1/Scc1p, resulting in sister chromatid separation (for review see Nasmyth, 2002).
The Ipl1/Aurora protein kinases are key regulators of chromosome segregation and cytokinesis (for reviews see Shannon and Salmon, 2002; Stern, 2002). In mammals, the kinases can be subdivided into three families: Aurora A, B, and C (for review see Nigg, 2001). Aurora A localizes to centrosomes and is required to maintain the separation of centrosomes and to form a bipolar spindle (Glover et al., 1995). Aurora B exhibits a “chromosomal passenger” localization pattern, where it localizes to the chromosomes and kinetochores, transfers to the spindle, and eventually accumulates at the spindle midzone and midbody (Bischoff et al., 1998; Schumacher et al., 1998; Terada et al., 1998; Petersen et al., 2001; Murata-Hori et al., 2002). Aurora B is in a complex with the chromosomal passenger proteins INCENP (inner centromere protein) and Survivin/Bir1 (Kim et al., 1999; Adams et al., 2000; Kaitna et al., 2000; Speliotes et al., 2000; Morishita et al., 2001; Rajagopalan and Balasubramanian, 2002).
In budding yeast, there is a single essential Aurora protein kinase, Ipl1 (Chan and Botstein, 1993; Francisco et al., 1994). In ipl1 mutant cells, sister chromatids are pulled to the same spindle pole instead of opposite poles (Biggins et al., 1999; Kim et al., 1999). Experiments in vitro and in vivo suggest that this defect is due to an inability of ipl1 mutants to release monooriented kinetochore-microtubule attachments in order to make the correct bioriented attachments (Biggins et al., 1999; Tanaka et al., 2002). ipl1 mutants also fail to activate the spindle checkpoint when kinetochores are not under tension (Biggins and Murray, 2001). Like mammalian homologues, Ipl1p localizes to kinetochores and the mitotic spindle (Biggins et al., 1999; Biggins and Murray, 2001; He et al., 2001; Kang et al., 2001; Tanaka et al., 2002). Several Ipl1p/Aurora substrates have been identified in various organisms. CENP-A, the human histone centromeric H3 variant, is an Aurora B kinase substrate, and nonphosphorylatable CENP-A mutants have cytokinesis defects (Zeitlin et al., 2001). In budding yeast, three kinetochore proteins are substrates either in vivo and/or in vitro and are good candidates for essential Ipl1p targets: the INCENP homologue (Sli15p), the Dam1 protein, and the CBF3 component Ndc10p (Biggins et al., 1999; Kang et al., 2001; Cheeseman et al., 2002; Li et al., 2002).
Here we show that the Ipl1p kinase has a role in spindle disassembly that is independent from its previously identified functions. There is a dynamic relocalization of Ipl1p in anaphase, and Ipl1p tracks the plus ends of depolymerizing microtubules. We propose that Ipl1p regulates microtubule plus ends to promote chromosome segregation and spindle disassembly.
Results
Ipl1 localization is similar to chromosomal passenger proteins
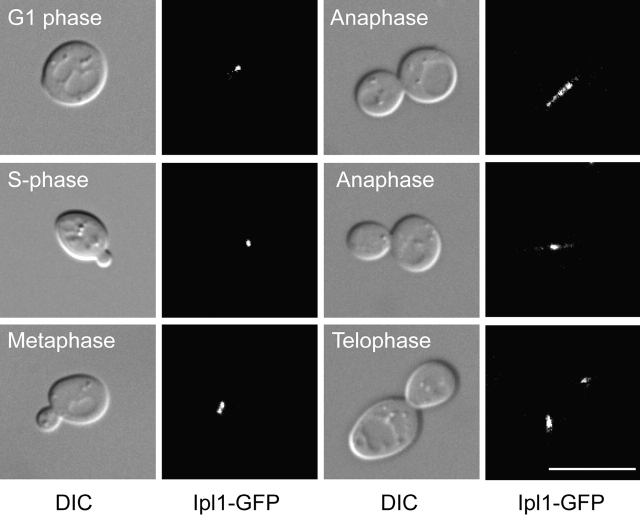
Ipl1p is required for chromosome segregation and the spindle checkpoint when kinetochores are not under tension. To learn more about Ipl1p functions, we examined the localization of an endogenous Ipl1p fusion to GFP at the COOH terminus in living cells using bud size as a marker for cell cycle stage (Fig. 1). In unbudded G1 cells, Ipl1–GFP localizes as a dot or small line. In small budded S phase cells, Ipl1p localizes as a distinct dot that separates into two discrete dots in medium budded metaphase cells and corresponds to precociously separated kinetochores (Goshima and Yanagida, 2000; He et al., 2000, 2001; Tanaka et al., 2000; Biggins and Murray, 2001; Pearson et al., 2001). In large budded cells, Ipl1–GFP exhibits dynamic localization patterns. In some cells, Ipl1–GFP is on the whole spindle but absent from the kinetochores that are clustered at the poles as reported previously (Tanaka et al., 2002). We also found two sites of Ipl1p localization that had not been seen before. Ipl1–GFP is found at the spindle midzone and in small tufts that vary in shape near the spindle poles at telophase. This likely represents Ipl1p bound to the remnants of depolymerized microtubules (Winey et al., 1995). Since the Ipl1–GFP fusion created a temperature-sensitive protein, we also localized endogenous Ipl1p. Immunofluorescence with anti-Ipl1p antibodies on chromosome spreads showed the same localization patterns as the Ipl1–GFP fusion protein (Loidl et al., 1998) (Fig. S1 available at http://www.jcb.org/cgi/content/full/jcb.200209018/DC1). Therefore, Ipl1p localization is similar to Aurora B and other “chromosomal passenger” proteins.
Figure 1.
Ipl1 localization is similar to chromosomal passenger proteins. Live microscopy was performed on cells containing Ipl1–GFP (SBY556). DIC pictures are shown to the left of each corresponding fluorescence picture. In G1, S phase, and metaphase cells, Ipl1p localizes in a discrete dot corresponding to kinetochores. In anaphase and telophase cells, Ipl1p localizes along the whole spindle (top right), at the spindle midzone (middle right), or near the spindle poles (bottom right). Bar, 10 μm.
Ipl1p localizes to kinetochores under tension
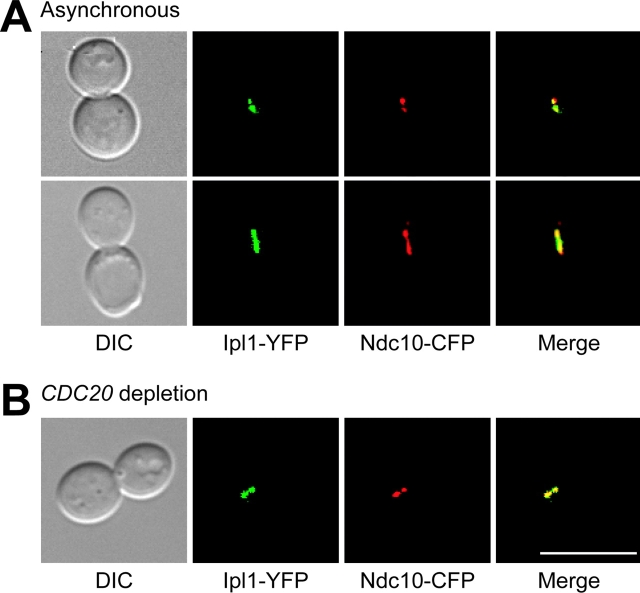
We found previously that Ipl1p localizes to kinetochores that are not under tension and that it is required for the spindle checkpoint at this time (Biggins and Murray, 2001). Once tension is established, Ipl1p likely needs to be inactivated to allow cell cycle progression. Although it was reported that Ipl1p no longer colocalizes with the kinetochore protein Ndc10p at metaphase (Tanaka et al., 2002), we always found Ipl1p localized in discrete dots. Therefore, we repeated their Ipl1–YFP and Ndc10–CFP colocalization experiment. In asynchronously growing cells, Ipl1–YFP and Ndc10–CFP always colocalized as characteristic kinetochore dots in metaphase cells (Fig. 2 A, top). In addition, they also colocalized as a line that looks like a short spindle in some cells (Fig. 2 A, bottom). Since the pole to pole distance is longer than a metaphase spindle, it likely represents cells that are initiating anaphase. We also analyzed Ipl1p localization in a population of metaphase-arrested cells by depleting the Cdc20 protein that activates the APC and found that Ipl1–YFP and Ndc10–CFP always colocalize (Fig. 2 B). Although the majority of cells show two distinct dots that represent the kinetochores, approximately one third of the cells exhibit colocalization in a line similar to the asynchronous population (unpublished data). This may mean that Ipl1p can transfer to the spindle in this arrest or that these cells do not have clustered kinetochores. Therefore, although Ipl1p leaves kinetochores after metaphase, it is still bound to kinetochores that are under tension.
Figure 2.
Ipl1p localizes to metaphase kinetochores that are under tension. (A) Microscopy was performed on cells containing Ipl1–YFP (green) and Ndc10–CFP (red) (SBY1246). DIC pictures are shown on the far left. The merged image (yellow, far right) shows that Ipl1p and Ndc10p colocalize in metaphase cells where kinetochores are precociously separated (top). Some cells also show Ipl1p and Ndc10p costaining on the short spindle (bottom). (B) pGAL-CDC20 cells containing Ipl1–YFP and Ndc10–CFP (SBY1246) were arrested in metaphase by shifting the cells to glucose medium. The merged microscopy images show that Ipl1p and Ndc10p also colocalize in metaphase-arrested cells. Bar, 10 μm.
ipl1–321 mutant cells are defective in spindle disassembly
The dynamic localization of Ipl1p on spindles suggested that it might regulate spindle function. To test this, we performed live cell imaging of wild-type and ipl1–321 mutants containing Tub1–GFP. Cells were synchronized in G1 with α-factor and then released to 35°C to inactivate Ipl1–321p, and time-lapse images were captured every minute. The start of spindle elongation was used as a reference for anaphase initiation and examples of the time-lapse data for a wild-type (video 1 available at http://www.jcb.org/cgi/content/full/jcb.200209018/DC1) and an ipl1–321 mutant cell (video 2 available at http://www.jcb.org/cgi/content/full/jcb.200209018/DC1) are shown in Fig. 3 A. Spindle elongation was quantified by measuring the length of the spindle every minute after the initiation of anaphase B (Fig. 3 B). In wild-type cells, we observed biphasic spindle elongation (Yeh et al., 1995; Straight et al., 1997). Wild-type spindles reach ∼8.4 μm in length ∼14 min after the initiation of anaphase B and then disassemble. In ipl1–321 mutant cells, spindle elongation occurs with kinetics similar to wild-type cells. However, the spindle continues to grow to a length of ∼10.4 μm, delaying spindle disassembly for ∼6 min, a 42% increase in the duration of anaphase B. In two of the ten cells analyzed, the spindle is forced to bend when reaching the cell membrane (Fig. 3 A, A*). Since these phenotypes are not seen in wild-type cells and are similar to mutants in the motor protein Kip3 (Straight et al., 1998), we also analyzed spindle elongation and breakdown kinetics in ipl1–321, kip3Δ, and kip3Δ ipl1–321 mutants and found they exhibit similar delays in spindle breakdown (unpublished data). Therefore, Ipl1p and Kip3p may act in the same pathway to promote spindle disassembly.
Figure 3.
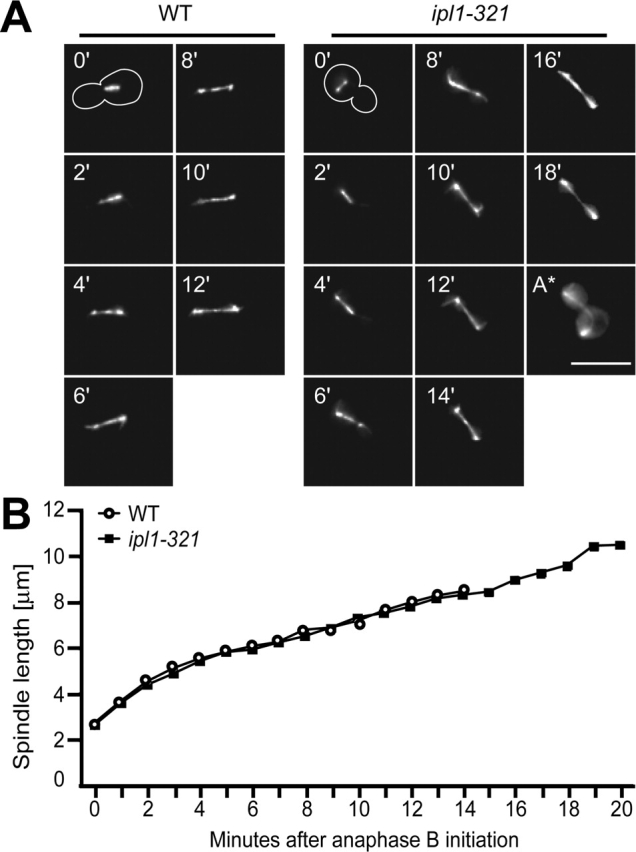
ipl1–321 mutants are defective in spindle disassembly. (A) Live microscopy was performed on wild-type (SBY130, left) and ipl1–321 mutant cells (SBY97, right) containing Tub1–GFP that were released from α-factor at 35°C. Eight z sections at 0.5-μm intervals were acquired every minute. Images of the spindle in a single cell are shown every 2 min after the initiation of anaphase (time 0'). An outline of the cell is shown at time 0'. Spindle disassembly is delayed in ipl1–321 cells, and the spindle orientation changes during the initial phases of anaphase. A hyperelongated spindle in an ipl1–321 mutant cell is shown in A*. See also videos 1 and 2 available at http://www.jcb.org/cgi/content/full/jcb.200209018/DC1. Bar, 10 μm. (B) The spindle length at each time point was measured, and the averages of 10 cells for each strain are graphed. Spindles disassemble in wild-type cells (○) 14 min after anaphase B initiation, whereas ipl1–321 mutant cells (▪) take 20 min.
The ipl1–321 mutant cells also exhibit a spindle orientation defect that we quantified by measuring the angle between the spindle axis and the mother-bud axis every minute starting at metaphase. In wild-type cells, it takes less than 6 min for the spindle to orient itself on the mother-bud axis, whereas it takes more than 11 min in ipl1–321 mutant cells (Fig. 3 A).
The role of Ipl1p in spindle disassembly is an independent function
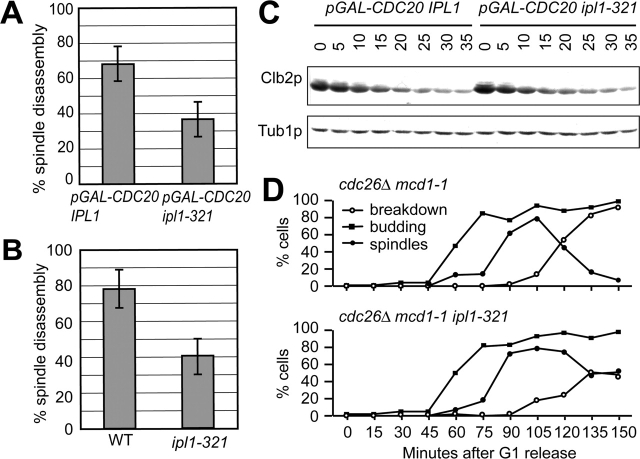
Since Ipl1p is required for chromosome segregation and the spindle checkpoint, we tested whether the spindle disassembly delay was a consequence of defects in these functions. We showed previously that if bipolar spindle assembly occurs before Ipl1p inactivation, chromosome segregation is normal (Biggins and Murray, 2001). Therefore, we arrested cells containing Tub1–GFP in metaphase by depleting the Cdc20 protein, shifted the cells to the restrictive temperature to inactivate Ipl1p, and then released them into the cell cycle. Aliquots were taken every 5 min, and cells with a pole to pole distance corresponding to a late anaphase cell (equal or greater than 9 μm) were analyzed for the presence or absence of a spindle. Since tubulin is always at the SPB, the pole to pole distance can be measured regardless of whether a spindle is present. Spindle disassembly occurred in 68% of Cdc20-depleted cells compared with only 36% of the Cdc20-depleted ipl1–321 double mutant cells (Fig. 4 A). In addition, there was no defect in spindle elongation in either strain. This result suggests that the spindle breakdown defect in ipl1–321 mutants is independent from Ipl1p's role in chromosome segregation.
Figure 4.
Ipl1p's role in spindle disassembly is independent from its role in chromosome segregation. (A) pGAL-CDC20 (SBY952) and pGAL-CDC20 ipl1–321 (SBY943) cells containing Tub1–GFP were shifted to glucose to arrest cells in metaphase and then shifted to 37°C to inactivate Ipl1–321p. Cells were then released into galactose medium at 37°C to restore Cdc20 protein synthesis in the presence of α-factor to arrest cells in the following G1. The percentage of cells with a pole to pole distance ≥9 μm were monitored for the presence or absence of a spindle. Spindle disassembly occurred in 68% of pGAL-CDC20 cells compared with 36% of pGAL-CDC20 ipl1–321 mutant cells released from metaphase. (B) Wild-type (SBY130) and ipl1–321 (SBY97) cells containing Tub1–GFP were released from α-factor (T = 0) into the restrictive temperature (37°C). Time points were taken at 60, 70, and 80 min after release and monitored for the presence or absence of a spindle as in A. In wild-type cells, 78% of the spindles have depolymerized compared with only 40% of ip1l-321 mutant spindles. The bars represent the 95% confidence interval. (C) Cells from A were taken every 5 min, and Clb2p and Tub1p (loading control) protein levels were monitored by immunoblotting. Clb2p levels decline with similar kinetics in both strains, indicating that ipl1–321 mutant cells exit mitosis normally. (D) ipl1–321 mutants have hyperstable microtubules. cdc26Δ mcd1–1 (SBY965) and cdc26Δ mcd1–1 ipl1–321 (SBY2066) cells were arrested in G1 using α-factor then released into the cell cycle at the restrictive temperature (37°C). Cells were monitored for budding index, spindle formation, and spindle breakdown. 93% of cdc26Δ mcd1–1 mutant cells underwent spindle disassembly by 150 min compared with only 48% of cdc26Δ mcd1–1 ipl1–321 cells.
The spindle disassembly defect in the ipl1–321 mutant cell population also occurred when chromosome segregation was defective. We analyzed this by synchronizing wild-type and ipl1–321 mutant cells containing Tub1–GFP in α-factor, releasing them to 37°C, and analyzing spindle disassembly as described above. Whereas 78% of the wild-type cells underwent spindle breakdown, only 40% of ipl1–321 mutant cells had disassembled their spindles (Fig. 4 B). Therefore, the spindle disassembly defect occurs in ipl1–321 mutants when chromosome segregation is normal or defective.
Since mutants defective in the mitotic exit network exhibit a spindle breakdown delay (Stegmeier et al., 2002), we tested whether ipl1–321 mutant cells delayed mitotic exit by monitoring the destruction of Clb2p, the major mitotic B-type cyclin. Cells from the experiment described in Fig. 4 A were collected every 5 min and then immunoblotted with anti-Clb2p antibodies (Fig. 4 C). Clb2p degradation occurred with similar kinetics in wild-type and ipl1–321 mutant cells, indicating Ipl1p is not required for mitotic exit.
Ipl1p also has a function in the spindle checkpoint when kinetochore tension is not generated (Biggins and Murray, 2001) so we tested whether other spindle checkpoint genes are also required for spindle disassembly. We found that the dynamics of spindle elongation and breakdown in wild-type, mad1Δ, and mad2Δ strains containing Tub1–GFP were similar (Fig. S2 available at http://www.jcb.org/cgi/content/full/jcb.200209018/DC1). Therefore, a function in spindle disassembly is not a general property of all checkpoint proteins. Together, these data suggest that Ipl1p's role in spindle disassembly is independent from its roles in chromosome segregation and the spindle checkpoint and identifies a previously unknown function for this protein kinase.
ipl1–321 mutant cells can stabilize fragile spindles
Since ipl1–321 mutants are defective in spindle breakdown, we tested whether they could stabilize fragile spindles that result from loss of sister chromatid cohesion in metaphase. When sister chromatid cohesion is released in the absence of APC function by a mutation in the Mcd1/Scc1 cohesion protein, spindle elongation occurs in the presence of high levels of the Pds1 and Clb2 proteins (Michaelis et al., 1997). This leads to fragile spindles where the spindle elongates but breaks down abnormally fast, creating an “anaphase-like prometaphase” (Severin et al., 2001b). Therefore, we tested whether the addition of an ipl1–321 mutation could stabilize the fragile spindles. We used a cdc26Δ strain, which leads to temperature-sensitive inactivation of the APC, in combination with the cohesin mutation mcd1–1 to create fragile spindles. cdc26Δ mcd1–1 and cdc26Δ mcd1–1 ipl1–321 mutant cells containing Tub1–GFP were arrested in G1 using α-factor, released to the restrictive temperature, and monitored for budding, spindle formation, and spindle breakdown. As reported previously, 93% of cdc26Δ mcd1–1 mutants underwent spindle breakdown within 150 min of release from G1 (Fig. 4 D) (Severin et al., 2001b). However, only 48% of cdc26Δ mcd1–1 ipl1–321 mutant cells had undergone spindle breakdown at this time, indicating that the ipl1–321 mutation stabilizes the fragile spindles.
Ipl1p kinase activity increases when spindles disassemble
To analyze Ipl1p kinase activity when spindles disassemble, we developed a kinase assay using antibodies generated against a recombinant GST-Ipl1 fusion protein. The affinity-purified antibodies specifically recognize a single major band in yeast lysates that migrates just above 45 kD (Fig. S3 available at http://www.jcb.org/cgi/content/full/jcb.200209018/DC1). The antibodies were used to immunoprecipitate wild-type Ipl1p and the Ipl1–321 protein that has reduced kinase activity at high temperatures (Biggins et al., 1999). The majority of Ipl1p present in the yeast lysates (Fig. 5 A, pre) was depleted by the antibody (Fig. 5 A, post). The immunoprecipitates (Fig. 5 A, IP) were then incubated with the histone-fold domain of the kinetochore protein Cse4 in a kinase reaction in vitro. Cse4p was radiolabeled in the presence of wild-type Ipl1p but not the kinase inactive Ipl1–321 protein (Fig. 5 A, right), showing that the assay specifically reflects an Ipl1p-associated kinase activity.
Figure 5.
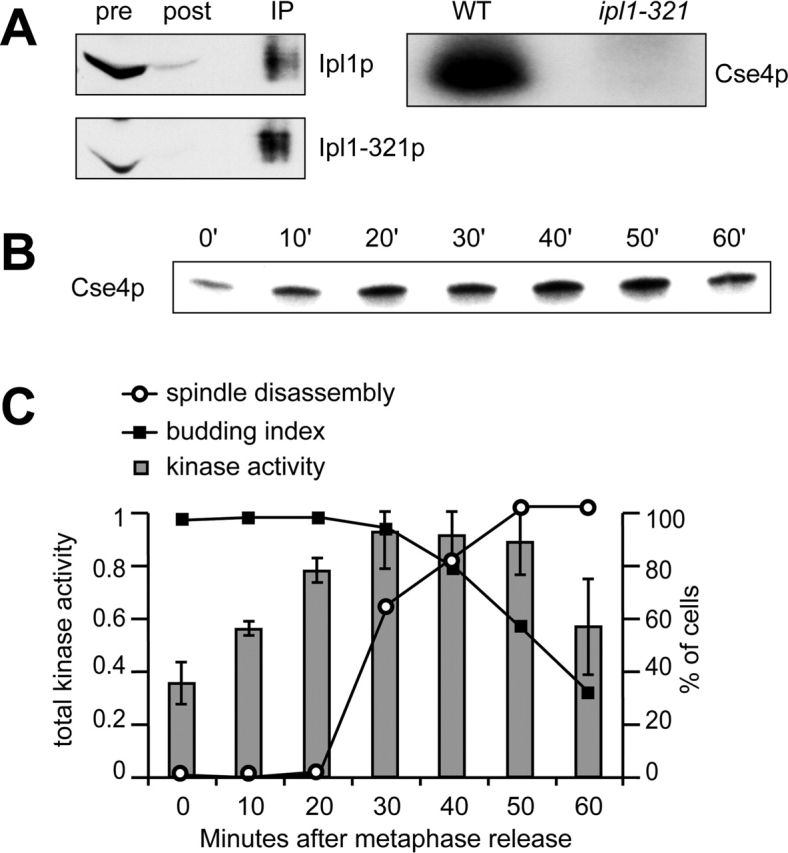
Ipl1p kinase activity increases before spindle disassembly. (A) Ip1lp was immunoprecipitated from a wild-type (SBY3) and a ipl1–321 mutant strain (SBY322) and then incubated with the histone-fold domain of the Cse4 kinetochore protein in a kinase reaction in vitro. The majority of Ipl1p present in the lysates before the immunoprecipitation (pre) was removed (post), and similar amounts of protein were used in the kinase assay (IP). The autoradiogram (right) shows that Cse4p is radiolabeled in the presence of wild-type Ipl1p but not Ipl1–321 mutant protein. (B) pGAL-CDC20 cells expressing Tub1–GFP (SBY952) were synchronized in metaphase by growth in glucose for 3 h. They were then released into galactose medium, and aliquots were taken every 10 min, and kinase assays were performed with the substrate Cse4p in vitro. The autoradiogram shows the phosphate incorporated into Cse4p in one experiment. (C) Microscopy was performed to determine the percent budding (▪) and the percent spindle disassembly (○). The total Ipl1p kinase activity is shown in gray bars (arbitrary units) for three experiments with the standard deviation indicated. Ipl1p kinase activity increases before spindle breakdown.
We analyzed Ipl1p kinase activity as cells exited mitosis by releasing cells arrested in metaphase by Cdc20p depletion and taking time points every 10 min. Ipl1p was immunoprecipitated, and the kinase activity was monitored against the substrate Cse4p (Fig. 5 B). To determine the cell cycle position, we monitored the budding index and spindle disassembly (Fig. 5 C). We quantified the total kinase activity, which represents the protein and its corresponding kinase activity (Fig. 5 C). The Ipl1p kinase activity increases as cells leave metaphase, peaking just before spindle disassembly. Although we do not know whether this increase is due to regulation of the protein levels and/or specific activity, the data is consistent with Ipl1p kinase activity being required for spindle breakdown.
Ipl1p's substrates localize to the spindle midzone, and Ipl1p follows the plus ends of the depolymerizing spindle microtubules
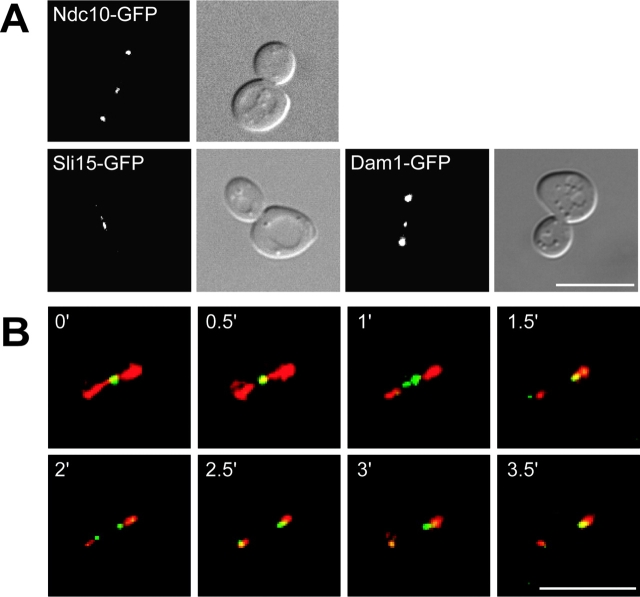
The novel midzone localization pattern for Ipl1p led us to test whether proteins that Ipl1p regulates also localize to the spindle midzone. First, we tested Cse4p which we have shown here is an Ipl1p substrate in vitro. Localization of a Cse4–GFP fusion showed that it does not transfer to the spindle (unpublished data). We next analyzed the localization of COOH-terminal GFP fusions to the Ndc10, Sli15, and Dam1 proteins (Fig. 6 A). Ndc10–GFP localized to the midzone in late anaphase cells in addition to the previously reported spindle and kinetochore localization (Lechner and Carbon, 1991; Goh and Kilmartin, 1993). We found that Sli15–GFP also accumulates at the spindle midzone and exhibits the same localization pattern as Ipl1p throughout the entire cell cycle (Fig. 6 A; unpublished data). Various labs have reported that Dam1p localizes to kinetochores throughout the cell cycle and to the mitotic spindle (Hofmann et al., 1998; He et al., 2001; Jones et al., 2001). Here we show that Dam1–GFP also localizes to the spindle midzone in anaphase cells. Therefore, the majority of known Ipl1p substrates localize to the spindle midzone, providing several potential candidates for Ipl1p regulation of spindle disassembly.
Figure 6.
Ipl1p's substrates localize to the spindle midzone, and Ipl1p follows the plus ends of the depolymerizing spindle. (A) Microscopy was performed on strains containing endogenous Ndc10–GFP (SBY539), Sli15–GFP (SBY875), or Dam1–GFP (SBY1115). The fluorescence images show that all three Ipl1p substrates localize to the spindle midzone. The corresponding DIC pictures are shown (right). (B) Live image analysis was performed on cells expressing Ipl1–GFP and Tub1–CFP (SBY1036). Every 30 s, five z sections at 0.5-μm intervals were acquired while alternating between the two channels (FITC and CFP). The deconvolved video shows tubulin in red, Ipl1p in green, and the overlapping signal in yellow. Before spindle disassembly, Ipl1p localizes to the spindle midzone (0'). When the spindle starts breaking down (1'), the Ipl1p signal splits and then follows the plus ends of the depolymerizing spindle (2') until it reaches the spindle poles (3.5′). Video 3 showing Ipl1p localization is available at http://www.jcb.org/cgi/content/full/jcb.200209018/DC1. Bars, 10 μm.
To learn more about how Ipl1p might regulate spindle disassembly, we analyzed the various Ipl1–GFP anaphase localization patterns more precisely by performing time-lapse microscopy (video 3 available at http://www.jcb.org/cgi/content/full/jcb.200209018/DC1). At anaphase, Ipl1–GFP is distributed along the entire spindle length in a punctate pattern. Ipl1p then accumulates at the spindle midzone, splits into two distinct dots, and then travels back to the poles.
Since Ipl1p localizes to the spindle midzone very late in anaphase and then travels back to the poles, we tested whether it was following the plus ends of the depolymerizing spindle microtubules. Live microscopy was performed on cells coexpressing Tub1–CFP (tubulin) and Ipl1–GFP. Although CFP and GFP have overlapping spectrums, the Ipl1–GFP and Tub1–CFP signals were easily discernible (Fig. 6 B). It was not possible to use the nonoverlapping spectrum of YFP because the Ipl1–YFP signal was not strong enough to perform time-lapse imaging of cells. We started imaging a cell when Ip1lp (Fig. 6 B, green) localized to the midzone of a long spindle (Fig. 6 B, red, 0′). After 1 min, the Ipl1p signal split and there was no longer any tubulin signal in the center of the spindle, indicating that the spindle is starting to break down. As the spindle depolymerized toward the poles, the Ipl1p signal always localized near the plus end of the microtubules (Fig. 6 B, 2′). At the end of spindle disassembly, the remaining tubulin at the pole colocalized with Ipl1p (Fig. 6 B, 3.5′). Therefore, Ipl1p accumulates at the spindle midzone in late anaphase and then follows the plus end of the depolymerizing microtubules back to the poles, suggesting it may specifically regulate the microtubule plus ends.
Discussion
We found that the Ipl1/Aurora protein kinase has a role in spindle microtubule disassembly that is not a consequence of a delay in mitotic exit or a prior defect in chromosome segregation. In addition, it is unlikely to be due to a defect in the spindle checkpoint, since the mad1 and mad2 checkpoint mutants exhibited normal spindle disassembly. Ipl1p localizes to the spindle midzone in anaphase and tracks the plus ends of the depolymerizing spindle microtubules, suggesting it may directly regulate microtubule plus ends. We found that several kinetochore proteins localize to the midzone, suggesting the plus ends of the interpolar microtubules may be regulated in a manner similar to kinetochores. In addition, we found that ipl1 mutants have a spindle orientation defect that may also reflect a role in cytoplasmic microtubule plus end function. Therefore, we propose that Ipl1p is a general microtubule plus end regulator and that its function in spindle disassembly in anaphase is similar to its function in promoting biorientation in prometaphase.
Ipl1p leaves the kinetochores after tension is established
We show here that Ipl1p exhibits a localization pattern similar to chromosomal passenger proteins, making it likely that Ipl1p is an Aurora B homologue. The dynamic localization of Ipl1p reflects its various mitotic functions. During prometaphase, Ipl1p monitors kinetochore tension and promotes microtubule release of monooriented kinetochore attachments, thus ensuring that sister kinetochores establish biorientation before chromosome segregation (Biggins and Murray, 2001; Tanaka et al., 2002). The proposal that Ipl1p might delocalize from kinetochores at metaphase provided an attractive mechanism for inactivating the kinase when tension and biorientation are established (Tanaka et al., 2002). However, we found that both Ipl1–GFP and endogenous Ipl1p localize to kinetochores in metaphase. Our data agrees with the observation that a mammalian Aurora B–GFP fusion protein left kinetochores 0.5 min after the initiation of anaphase, well after tension was established (Murata-Hori et al., 2002). K. Tanaka and T.U. Tanaka recently found that Ipl1p is on metaphase kinetochores by chromatin immunoprecipitation, thus reconciling our data (personal communication). To understand how tension regulates Ipl1p, we are actively trying to elucidate the mechanisms that control Ipl1 protein stability and kinase activity, since these may be alternative modes of regulation at metaphase.
Functions of the Ipl1/Aurora B protein kinase family
We show that the localization of Ipl1p to the mitotic spindle is correlated with spindle disassembly in budding yeast. Since Aurora B localizes to the spindle midzone in all organisms, this may be another conserved function of the Ipl1/Aurora B protein kinase family. Alternatively, this may reflect similarity to the role of Aurora A, since it regulates spindle function and phosphorylates the Eg5 motor protein in frog egg extracts (Giet et al., 1999; Giet and Prigent, 2000). In other organisms, Aurora B is required for cytokinesis, and this may be coupled to defects in spindle microtubule depolymerization that have not been noticed previously. In human cells, cytokinesis is regulated by the phosphorylation of CENP-A by Aurora B (Zeitlin et al., 2001). We report here that the budding yeast histone variant Cse4p is also a good substrate for Ipl1p in vitro. Further analysis of the effects of Cse4p phosphorylation by Ipl1p may reveal more details about spindle disassembly and/or cytokinesis in budding yeast. We show that the dynamics of spindle elongation are not altered in ipl1 mutant cells, unlike mutants in the Aurora–INCENP–Survivin complex in Schizosaccharomyces pombe (Morishita et al., 2001; Rajagopalan and Balasubramanian, 2002). This difference may be due to the number of kinetochore microtubule-binding sites in each organism. In S. pombe, there are multiple binding sites, which results in lagging chromosomes if biorientation is not achieved. In budding yeast where there is a single microtubule binding site, defects in biorientation cannot generate lagging chromosomes. However, when a conditional dicentric chromosome is activated in budding yeast, thus creating a lagging chromosome, spindle elongation is delayed (Yang et al., 1997). Our study in budding yeast had the advantage that defects in biorientation do not interfere with spindle dynamics. To determine whether the spindle disassembly function of Ip1lp is conserved, spindle dynamics will need to be analyzed in situations where chromosome segregation is normal.
Consistent with a role in spindle disassembly, we found that Ipl1p kinase activity increases just before spindle breakdown. Few studies have looked at the regulation of Ipl1p homologues. In Drosophila and rat tissues, Aurora B protein levels and kinase activity peak during mitosis (Bischoff et al., 1998; Terada et al., 1998). However, the time points were not close enough in those studies to determine whether the peak of kinase activity corresponds to spindle breakdown. In fission yeast, Aurora B is not cell cycle regulated (Petersen et al., 2001; Leverson et al., 2002), making it unclear whether there are conserved mechanisms that regulate Ipl1/Aurora B protein levels and kinase activity. Our study also revealed that Ipl1p kinase activity is low when cells are arrested in metaphase with kinetochores under tension. This may reflect an active mechanism that regulates Ipl1p stability and/or activity once tension is established. Future work will be needed to elucidate the mechanisms that lead to changes in Ipl1p kinase activity during mitosis.
How does Ipl1p regulate spindle disassembly?
In support of a role for Ipl1p in direct regulation of microtubules, we found that ipl1 mutants are able to alleviate the spindle fragility of apc mcd1 mutant cells. However, since the mechanism that leads to fragile spindles is not known, it is not clear how Ipl1p stabilizes spindles. Although mutations in Ipl1p affect spindle breakdown, they do not do this by grossly altering the structure of the spindle midzone, since all the midzone proteins we tested still localized in an ipl1 mutant.
Our studies using live microscopy revealed a previously unidentified localization pattern for Ipl1p. At anaphase, Ipl1p is transported along the spindle to the midzone and then tracks the plus ends of the depolymerizing spindle microtubules back to the poles. To our knowledge, the only other protein in budding yeast that exhibits this localization pattern is the Ipl1p substrate Ndc10p and suggests regulated transport of these proteins on microtubules in anaphase, possibly by motor proteins (D. Bouck and K. Bloom, personal communication). The localization to the plus ends may indicate that Ipl1p directly destabilizes microtubules like catastrophe factors, such as the KINI family of motor proteins (Desai et al., 1999). However, although Ipl1p binds microtubules in vitro (Kang et al., 2001), we have not been able to induce microtubule depolymerization with bacterial Ipl1p in vitro (unpublished data). Therefore, Ipl1p may directly promote microtubule depolymerization in a manner that we have not yet detected, or it may instead control a microtubule-binding protein.
There are two nonessential proteins known to be involved in spindle microtubule disassembly in budding yeast: the motor protein Kip3 and the microtubule-associated protein Ase1 (Juang et al., 1997; Straight et al., 1998). Ipl1p and Kip3p may act in the same spindle disassembly pathway because the double mutant exhibits the same spindle breakdown defect as each single mutant. However, we have yet to obtain evidence that Ipl1p regulates Kip3p (unpublished data). There are other potential candidates for Ipl1p regulation that will need to be investigated, such as the midzone protein Stu2 that opposes the Kip3 protein and the Esp1p/Pds1p cell cycle regulation complex that is also found at the midzone and has a function stabilizing spindles during anaphase (Uhlmann et al., 2000; Jensen et al., 2001; Severin et al., 2001a). We also found that three known Ipl1p substrates are at the spindle midzone: Ndc10p, Sli15p, and Dam1p. It is interesting to note that Dam1p was originally identified for its role in regulating spindle dynamics (Hofmann et al., 1998; Jones et al., 1999). Therefore, several potential Ipl1p substrates localize to the spindle midzone, and it will need to be determined whether any of these candidates also promote spindle disassembly.
The spindle midzone: a kinetochore-like structure?
Several kinetochore proteins are now known to localize to the spindle midzone in anaphase, including Stu2p, Slk19p, and the motor protein Cin8 (Hoyt et al., 1992; Zeng et al., 1999; Kosco et al., 2001). Here we show four additional kinetochore proteins localizing to the midzone: the Ipl1/Aurora protein kinase, the INCENP homologue Sli15p, Dam1p, and Ndc10p. Since midzone staining is difficult to detect, it may have been overlooked in several other localization studies and many more kinetochore proteins may be present at the midzone. Most of the spindle midzone proteins have been implicated in the regulation of spindle dynamics in anaphase by either promoting spindle elongation or spindle disassembly. An intriguing possibility is that the microtubule plus ends at the spindle midzone are regulated in anaphase similarly to the kinetochore-microtubule attachments in prometaphase. Future work will determine whether the Ipl1/Aurora protein kinase and other spindle midzone proteins are global regulators of microtubule plus ends.
Materials and methods
Microbial techniques
Media and genetic and microbial techniques were essentially as described (Sherman et al., 1974; Rose et al., 1990). All experiments where cells were released from G1 arrest were performed by adding 1 μg/ml α-factor (stock: 10 mg/ml in DMSO; United Biochemical Research Inc.) at the permissive temperature for 3 h, washing the cells twice in α-factor–free medium, and resuspending them in fresh medium. α-Factor was added back to 1 μg/ml after cells had budded to prevent cells from entering the next cell cycle. Galactose induction was performed by growing cells in 2% raffinose and adding galactose to a final concentration of 4%. All experiments were repeated at least twice with similar results.
Yeast strain construction
Yeast strains are listed in Table I and were constructed by standard genetic techniques. Diploids were isolated on selective medium and subsequently sporulated at 23°C. All strains containing the pGAL-CDC20 construct were obtained through crosses with SLJ577, a gift from S. Jaspersen and M. Winey (University of Colorado, Boulder, Colorado). SBY1036 was created by integration of TUB1–CFP:URA3 (a gift from K. Bloom, University of North Carolina at Chapel Hill, Chapel Hill, NC) with StuI at the URA3 locus. Strains containing CSE4-myc12 were obtained by integrating pSB246 cut with ClaI at the CSE4 locus. Strains containing TUB1–GFP:LEU2 were obtained by integrating plasmid pSB340 cut with AgeI at the LEU2 locus and TUB1–GFP:URA3 by integrating plasmid pMAS27 (a gift from M. Shonn, TUFTS University, Boston MA) cut with StuI at the URA3 locus. SBY554 was obtained by integrating pSB164 (pGAL-IPL1:URA3 [Biggins et al., 1999]) cut with StuI at the URA3 locus. SBY736 was obtained by integrating pSB257 (pGAL-myc12-IPL1:URA3) cut with StuI at the URA3 locus. Deletions in yeast genes and GFP, CFP, and myc epitope tags were made using the PCR-based integration system (Longtine et al., 1998). YFP epitope tags were made using pDH5, a gift from T. Davis (University of Washington, Seattle, WA). Specific primer sequences are available upon request. All deletions and epitope tags were confirmed by PCR.
Table I. Yeast strains used in this study.
| Strain
|
Genotype
|
|---|---|
| SBY3 | MATa ura3-1 leu2-3,112 his3-11 trp1-1 can1-100 ade2-1 bar1Δ |
| SBY97 | MATa ura3-1:TUB1-GFP:URA3 leu2-3,112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 lys2Δ ade2-1 bar1Δ can1-100 ipl1-321 |
| SBY130 | MATa ura3-1:TUB1-GFP:URA3 leu2-3,112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 lys2Δ ade2-1 bar1Δ can1-100 |
| SBY322 | MATa ura3-1 leu2-3,112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 lys2Δade2-1 bar1Δ can1- 100 ipl1-321 |
| SBY539 | MATa ura3-1 leu2-3,112 his3-11 trp1-1 can1-100 ade2-1 bar1Δ NDC10-GFP:KAN |
| SBY554 | MATa ura3-1:pGAL-IPL1:URA3 leu2-3,112 his3-11 trp1-1 can1-100 ade2-1 bar1Δ |
| SBY556 | MATa ura3-1 leu2-3,112 his3-11 trp1-1 ade2-1 bar1Δ can1-100 IPL1-GFP:KAN |
| SBY617 | MATa ura3-1 leu2-3,112 his3-11 trp1-1 ade2-1 bar1Δ can1-100 CSE4-myc12:URA3 |
| SBY736 | MATa ura3-1:pGAL-myc13-IPL1:URA3 leu2-3,112 his3-11 trp1-1 can1-100 ade2-1 bar1Δ |
| SBY875 | MATa ura3-1 leu2-3,112 his3-11 trp1-1 can1-100 ade2-1 bar1Δ SLI15-GFP:HIS3 |
| SBY943 | MATa ura3-1:TUB1-GFP:URA3 leu2-3,112 his3-11 trp1-1 ade2-1 can1-100 cdc20::LEU2 ipl1-321 [pGAL(low)-CDC20-HIS3(CEN)] |
| SBY952 | MATa ura3-1:TUB1-GFP:URA3 leu2-3,112 his3-11 trp1-1 ade2-1 can1-100 cdc20::LEU2 (pGAL[low]- CDC20-HIS3[CEN]) |
| SBY965 | MATa ura3-1 leu2-3,112:TUB1-GFP:LEU2 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:URA3:TRP1 lys2Δ ade2-1 mdc1-1 cdc26::KAN |
| SBY1036 | MATa ura3-1:TUB1-CFP:URA3 leu2-3,112 his3-11 trp1-1 ade2-1 bar1Δ can1-100 IPL1-GFP:KAN |
| SBY1115 | MATα ura3-1 leu2-3,112 his3-11 trp1-1 can1-100 ade2-1 bar1Δ DAM1-GFP:TRP1 |
| SBY1246 | MATα ura3-1 leu2-3,112 his3-11,15 trp1-1 ade2-1 can1-100 cdc20::LEU2 IPL1-YFP:HIS3 NDC10-CFP: KAN (pGAL[low]-CDC20-HIS3[CEN]) |
| SBY1422 | MATa ura3-1:TUB1-GFP:URA3 leu2-3,112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 lys2Δ ade2-1 bar1Δ can1-100 mad2::KAN |
| SBY1423 | MATa ura3-1:TUB1-GFP:URA3 leu2-3,112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:lacO:TRP1 lys2Δ ade2-1 bar1Δ can1-100 mad1::KAN |
| SBY2066 | MATa leu2-3,112 his3-11:LacI12:HIS3 trp1-1:LacO:TRP1 lys2Δ bar1Δ ipl1-321 mcd1-1 cdc26::KAN |
All strains are isogenic with the W303 strain background. Plasmids are indicated in parentheses.
Plasmid constructions
A clone that encoded Cse4p internally tagged with 12 copies of the myc epitope (pSB246) was constructed by PCR amplification of the myc tag from pSB162 (Biggins et al., 1999) using primers SB84 and SB85. The PCR product was digested with SpeI and ligated into the XbaI site of pSB241 (CSE4, URA3 integrating vector). pSB241 was created by digesting a genomic clone containing CSE4 with EcoRI and PstI, and the resulting 1.7-kb fragment was ligated into yiplac211 (Gietz and Sugino, 1988) digested with EcoRI and PstI. To create TUB1–GFP:LEU2, pMAS27 was digested with EagI, and the TUB1–GFP fragment was ligated into the EagI site of pRS305 to create pSB340. To create a pGAL-myc12-IPL1 clone, IPL1 was PCR amplified from genomic DNA, digested with SpeI and SacI, and then ligated into pSB209 (pGAL-myc12, URA3 integrating vector) digested with the same enzymes to create pSB257.
Protein and immunological techniques
Protein extracts were made and immunoblotted as described (Minshull et al., 1996). Anti-Tub1p antibodies were obtained from Accurate Chemical and Scientific and used at a 1:1,000 dilution, anti-Clb2p antibodies (a gift from A. Rudner, Harvard Medical School, Boston, MA) were used as described (Rudner et al., 2000), and anti-Ipl1p antibodies were used at 1:1,000. To generate anti-Ipl1p antibodies, GST-Ipl1p was injected into rabbits at Cocalico Biological Inc. For the fourth and fifth boosts, boiled protein was injected. The antibodies were affinity purified by coupling GST-Ipl1p to SulfoLink Coupling gel (Pierce Chemical Co.).
Microscopy
For live microscopy to analyze GFP fusion proteins, cells were grown in yeast meda, pH 7.0, washed, and resuspended in 1/10 volume minimal medium with casamino acids. For live microscopy at RT, 1.5 μl of cell preparation was put on a 2% agarose pad containing minimal medium with casamino acids. The slide was then sealed with VALAP (1:1:1, vaseline: lanolin: paraffin) and imaged. For live microscopy at 35°C, prepared cells were mounted directly onto a heated stage (Bioptechs). Images were collected through an Olympus 1X17 60× objective with a CH350 CCD camera (Roper Scientific) using the Softwox 2.5 (Applied Precision) software. The same software was used for deconvolution. At least 10 cells were analyzed for all reported experiments.
Chromosome spreads were performed as described (Loidl et al., 1991; Michaelis et al., 1997). Lipsol was obtained from Lip Ltd. 9E10 antibodies that recognize myc tag were used at a 1:500 dilution and obtained from Covance. Anti-Tub1p antibodies (Accurate Chemical and Scientific) were used at a 1:500 dilution. Anti-Ipl1p antibodies were used at a 1:250 dilution. Alexafluor-594 and Alexafluor-488 secondary antibodies were obtained from Molecular Probes and used at a 1:250 dilution.
Cse4 histone fold domain purification
The histone fold domain of CSE4 (encoding aa 121–229) was PCR amplified from Saccharomyces cerevisiae genomic DNA and cloned into a T7 expression vector of the pCRT7/CT TOPO TA cloning kit (Invitrogen). The resulting expression plasmid (pT7Cse4c) was transformed into BL21-CodonPlus (DE3)-RIL cells (Stratagene), and 2 liters were induced with ITPG to 0.2 mM for 2.5 h at 37°C, and Cse4 was purified under denaturing conditions as described (Gelbart et al., 2001). The Cse4 histone fold domain purity was verified by SDS-PAGE and dialyzed against water. A substantial portion of the protein was not soluble in water, but solubility was adequate for kinase assays in vitro.
Ipl1p kinase assays
40-ml cultures of mid-log cells were collected and resuspended in 500 μl lysis buffer (100 mM NaCl, 50 mM Tris, pH 7.5, 50 mM NaF, 50 mM β-glycerophosphate, pH 7.4, 2 mM EDTA, 2 mM EGTA, 0.1% Triton X-100). 2 mM NaVO4, 2 mM PMSF, 10 μg/ml LPC (leupeptin, pepstatin, and chymostatin; Chemicon), 1 mM DTT, 0.1 μg/ml microcystin (Calbiochem) were added fresh. All subsequent steps were performed at 4°C. Cells were lysed with glass beads in a beater (Biospec Products Inc.) for 30 s and then centrifuged for 10 min. 400 μl supernatant was added to 5 μl magnetic protein G beads (Dynal Biotech Inc.) and 4 μl anti-Ipl1p antibodies for 2 h. The beads were washed three times with 400 μl lysis buffer and once with 100 μl kinase buffer without ATP (50 mM Tris, pH 7.4, 1 mM DTT, 25 mM β-glycerophosphate, 5 mM MgCl2) and then resuspended in buffer with 10 μM ATP, 5 μCi [32P]ATP and 5 μg Cse4p for 30 min at 30°C. 2× sample buffer was added, and reactions were separated on SDS-PAGE and subjected to autoradiography using a Phosphorimager Screen (Molecular Dynamics). Kinase assays were quantified using ImageQuant (Molecular Dynamics) software.
Online supplemental material
Videos 1–3 and Figs. S1–S3 are available at http://www.jcb.org/cgi/content/full/jcb.200209018/DC1. Video 1 displays wild-type cells expressing Tub1-GFP from metaphase to anaphase. Video 2 shows ipl1-321 cells expressing Tub1–GFP that are delayed in spindle disassembly. Video 3 shows Ipl1–GFP localization during late anaphase. Fig. S1 shows endogenous Ipl1 localization on chromosome spreads. Fig. S2 demonstrates that the spindle checkpoint mutants mad1Δ and mad2Δ do not have spindle disassembly defects. Fig. S3 shows the Ipl1p antibody specificity.
Supplemental Material
Acknowledgments
We are grateful to Adrian Quintanilla and David McDonald for help with the microscopy, Steve Henikoff for the Cse4p construct, and Delia M. Pinto-Santini for preliminary experiments. We thank members of the Biggins' laboratory and Linda Breeden, Adrian Ferré-D'Amaré, Christian Frei, and Marion Shonn for critical reading of the manuscript and discussions. We thank the following people for strains, plasmids and advice: Kerry Bloom, Trisha Davis, Sue Jaspersen, Kim Nasmyth, Mark Roth, Marion Shonn, Bodo Stern, and Mark Winey. We are grateful to Kerry Bloom and Tomo Tanaka for sharing data prior to publication. We thank Andreas Conzelmann for support.
This work was supported by grants from the Roche Research Foundation and the Department of Defense of the Army to S. Buvelot, a Damon Runyon Cancer Research Foundation fellowship (DRG-1554) to D. Vermaak, and a Sidney Kimmel Scholar award and National Institutes of Health grant to S. Biggins.
The online version of this article contains supplemental material.
Footnotes
Abbreviations used in this paper: APC, anaphase-promoting complex; INCENP, inner centromere protein; SPB, spindle pole body.
References
- Adams, R.R., S.P. Wheatleya, A.M. Gouldsworthy, S.E. Kandels-Lewis, M. Carmena, C. Smythe, D.L. Gerloff, and W.C. Earnshaw. 2000. INCENP binds the aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 10:1075–1078. [DOI] [PubMed] [Google Scholar]
- Biggins, S., and A.W. Murray. 2001. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 15:3118–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S., F.F. Severin, N. Bhalla, I. Sassoon, A.A. Hyman, and A.W. Murray. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13:532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, J.R., L. Anderson, Y. Zhu, K. Mossie, L. Ng, B. Souza, B. Schryver, P. Flanagan, F. Clairvoyant, C. Ginther, et al. 1998. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 17:3052–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, C.S., and D. Botstein. 1993. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 135:677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., S. Anderson, M. Jwa, E.M. Green, J. Kang, J.R. Yates, C.S. Chan, D.G. Drubin, and G. Barnes. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172. [DOI] [PubMed] [Google Scholar]
- Desai, A., and T.J. Mitchison. 1997. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13:83–117. [DOI] [PubMed] [Google Scholar]
- Desai, A., S. Verma, T.J. Mitchison, and C.E. Walczak. 1999. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 96:69–78. [DOI] [PubMed] [Google Scholar]
- Francisco, L., W. Wang, and C.S. Chan. 1994. Type 1 protein phosphatase acts in opposition to Ipl1 protein kinase in regulating yeast chromosome segregation. Mol. Cell. Biol. 14:4731–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart, M.E., T. Rechsteiner, T.J. Richmond, and T. Tsukiyama. 2001. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 21:2098–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet, R., and C. Prigent. 2000. The Xenopus laevis aurora/Ipl1p-related kinase pEg2 participates in the stability of the bipolar mitotic spindle. Exp. Cell Res. 258:145–151. [DOI] [PubMed] [Google Scholar]
- Giet, R., R. Uzbekov, F. Cubizolles, K. Le Guellec, and C. Prigent. 1999. The Xenopus laevis aurora-related protein kinase pEg2 associates with and phosphorylates the kinesin-related protein XlEg5. J. Biol. Chem. 274:15005–15013. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., and A. Sugino. 1988. New yeast—Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 74:527–534. [DOI] [PubMed] [Google Scholar]
- Glover, D.M., M.H. Leibowitz, D.A. McLean, and H. Parry. 1995. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 81:95–105. [DOI] [PubMed] [Google Scholar]
- Goh, P.Y., and J.V. Kilmartin. 1993. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121:503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and M. Yanagida. 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 100:619–633. [DOI] [PubMed] [Google Scholar]
- He, X., S. Asthana, and P.K. Sorger. 2000. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 101:763–775. [DOI] [PubMed] [Google Scholar]
- He, X., D.R. Rines, C.W. Espelin, and P.K. Sorger. 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 106:195–206. [DOI] [PubMed] [Google Scholar]
- Hofmann, C., I.M. Cheeseman, B.L. Goode, K.L. McDonald, G. Barnes, and D.G. Drubin. 1998. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J. Cell Biol. 143:1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt, M.A., L. He, K.K. Loo, and W.S. Saunders. 1992. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J. Cell Biol. 118:109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, S., M. Segal, D.J. Clarke, and S.I. Reed. 2001. A novel role of the budding yeast separin Esp1 in anaphase spindle elongation. Evidence that proper spindle association of Esp1 is regulated by Pds1. J. Cell Biol. 152:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M.H., J.B. Bachant, A.R. Castillo, T.H. Giddings, Jr., and M. Winey. 1999. Yeast Dam1p is required to maintain spindle integrity during mitosis and interacts with the Mps1p kinase. Mol. Biol. Cell. 10:2377–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M.H., X. He, T.H. Giddings, and M. Winey. 2001. Yeast Dam1p has a role at the kinetochore in assembly of the mitotic spindle. Proc. Natl. Acad. Sci. USA. 98:13675–13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang, Y.L., J. Huang, J.M. Peters, M.E. McLaughlin, C.Y. Tai, and D. Pellman. 1997. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 275:1311–1314. [DOI] [PubMed] [Google Scholar]
- Kaitna, S., M. Mendoza, V. Jantsch-Plunger, and M. Glotzer. 2000. INCENP and an Aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr. Biol. 10:1172–1181. [DOI] [PubMed] [Google Scholar]
- Kang, J., I.M. Cheeseman, G. Kallstrom, S. Velmurugan, G. Barnes, and C.S. Chan. 2001. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Cell Biol. 155:763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.H., J.S. Kang, and C.S. Chan. 1999. Sli15 associates with the Ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 145:1381–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosco, K.A., C.G. Pearson, P.S. Maddox, P.J. Wang, I.R. Adams, E.D. Salmon, K. Bloom, and T.C. Huffaker. 2001. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol. Biol. Cell. 12:2870–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner, J., and J. Carbon. 1991. A 240 Kd multisubunit complex, CBF3, is a major component of the budding yeast centromere. Cell. 64:717–725. [DOI] [PubMed] [Google Scholar]
- Leverson, J.D., H.K. Huang, S.L. Forsburg, and T. Hunter. 2002. The Schizosaccharomyces pombe aurora-related kinase Ark1 interacts with the inner centromere protein Pic1 and mediates chromosome segregation and cytokinesis. Mol. Biol. Cell. 13:1132–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., J. Bachant, A.A. Alcasabas, Y. Wang, J. Qin, and S.J. Elledge. 2002. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 16:183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl, J., K. Nairz, and F. Klein. 1991. Meiotic chromosome synapsis in a haploid yeast. Chromosoma. 100:221–228. [DOI] [PubMed] [Google Scholar]
- Loidl, J., F. Klein, and J. Engebrecht. 1998. Genetic and morphological approaches for the analysis of meiotic chromosomes in yeast. Methods Cell Biol. 53:257–285. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie, III, D.J. Demarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Maddox, P.S., K.S. Bloom, and E.D. Salmon. 2000. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2:36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 91:35–45. [DOI] [PubMed] [Google Scholar]
- Millband, D.N., L. Campbell, and K.G. Hardwick. 2002. The awesome power of multiple model systems: interpreting the complex nature of spindle checkpoint signaling. Trends Cell Biol. 12:205–209. [DOI] [PubMed] [Google Scholar]
- Minshull, J., A. Straight, A. Rudner, A. Dernburg, A. Belmont, and A.W. Murray. 1996. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr. Biol. 6:1609–1620. [DOI] [PubMed] [Google Scholar]
- Morishita, J., T. Matsusaka, G. Goshima, T. Nakamura, H. Tatebe, and M. Yanagida. 2001. Bir1/Cut17 moving from chromosome to spindle upon the loss of cohesion is required for condensation, spindle elongation and repair. Genes Cells. 6:743–763. [DOI] [PubMed] [Google Scholar]
- Murata-Hori, M., M. Tatsuka, and Y.L. Wang. 2002. Probing the dynamics and functions of Aurora B kinase in living cells during mitosis and cytokinesis. Mol. Biol. Cell. 13:1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth, K. 2002. Segregating sister genomes: the molecular biology of chromosome separation. Science. 297:559–565. [DOI] [PubMed] [Google Scholar]
- Nigg, E.A. 2001. Cell division mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2:21–32. [DOI] [PubMed] [Google Scholar]
- Pearson, C.G., P.S. Maddox, E.D. Salmon, and K. Bloom. 2001. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 152:1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, J.M. 2002. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell. 9:931–943. [DOI] [PubMed] [Google Scholar]
- Petersen, J., J. Paris, M. Willer, M. Philippe, and I.M. Hagan. 2001. The S. pombe aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J. Cell Sci. 114:4371–4384. [DOI] [PubMed] [Google Scholar]
- Rajagopalan, S., and M.K. Balasubramanian. 2002. Schizosaccharomyces pombe Bir1p, a nuclear protein that localizes to kinetochores and the spindle midzone, is essential for chromosome condensation and spindle elongation during mitosis. Genetics. 160:445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M.D., F. Winston, and P. Hieter. 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 198 pp.
- Rudner, A.D., K.G. Hardwick, and A.W. Murray. 2000. Cdc28 activates exit from mitosis in budding yeast. J. Cell Biol. 149:1361–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, J.M., A. Golden, and P.J. Donovan. 1998. AIR-2: an Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J. Cell Biol. 143:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal, M., and K. Bloom. 2001. Control of spindle polarity and orientation in Saccharomyces cerevisiae. Trends Cell Biol. 11:160–166. [DOI] [PubMed] [Google Scholar]
- Severin, F., B. Habermann, T. Huffaker, and T. Hyman. 2001. a. Stu2 promotes mitotic spindle elongation in anaphase. J. Cell Biol. 153:435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin, F., A.A. Hyman, and S. Piatti. 2001. b. Correct spindle elongation at the metaphase/anaphase transition is an APC-dependent event in budding yeast. J. Cell Biol. 155:711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, K.B., and E.D. Salmon. 2002. Chromosome dynamics: new light on aurora B kinase function. Curr. Biol. 12:R458–R460. [DOI] [PubMed] [Google Scholar]
- Sherman, F., G. Fink, and C. Lawrence. 1974. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 174 pp.
- Speliotes, E.K., A. Uren, D. Vaux, and H.R. Horvitz. 2000. The survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol. Cell. 6:211–223. [DOI] [PubMed] [Google Scholar]
- Stegmeier, F., R. Visintin, and A. Amon. 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 108:207–220. [DOI] [PubMed] [Google Scholar]
- Stern, B.M. 2002. Mitosis: aurora gives chromosomes a healthy stretch. Curr. Biol. 12:R316–R318. [DOI] [PubMed] [Google Scholar]
- Straight, A.F., W.F. Marshall, J.W. Sedat, and A.W. Murray. 1997. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 277:574–578. [DOI] [PubMed] [Google Scholar]
- Straight, A.F., J.W. Sedat, and A.W. Murray. 1998. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 143:687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T., J. Fuchs, J. Loidl, and K. Nasmyth. 2000. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2:492–499. [DOI] [PubMed] [Google Scholar]
- Tanaka, T.U., N. Rachidi, C. Janke, G. Pereira, M. Galova, E. Schiebel, M.J. Stark, and K. Nasmyth. 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 108:317–329. [DOI] [PubMed] [Google Scholar]
- Terada, Y., M. Tatsuka, F. Suzuki, Y. Yasuda, S. Fujita, and M. Otsu. 1998. AIM-1: a mammalian midbody-associated protein required for cytokinesis. EMBO J. 17:667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann, F., W. Wernek, M.-A. Poupart, E. Koonin, and K. Nasmyth. 2000. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 103:375–386. [DOI] [PubMed] [Google Scholar]
- Winey, M., and E.T. O'Toole. 2001. The spindle cycle in budding yeast. Nat. Cell Biol. 3:E23–E27. [DOI] [PubMed] [Google Scholar]
- Winey, M., C.L. Mamay, E.T. O'Toole, D.N. Mastronarde, T.H. Giddings, Jr., K.L. McDonald, and J.R. McIntosh. 1995. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 129:1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S.S., E. Yeh, E.D. Salmon, and K. Bloom. 1997. Identification of a mid-anaphase checkpoint in budding yeast. J. Cell Biol. 136:345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, E., R.V. Skibbens, J.W. Cheng, E.D. Salmon, and K. Bloom. 1995. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 130:687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin, S.G., R.D. Shelby, and K.F. Sullivan. 2001. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 155:1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, X., J.A. Kahana, P.A. Silver, M.K. Morphew, J.R. McIntosh, I.T. Fitch, J. Carbon, and W.S. Saunders. 1999. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 146:415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.