Abstract
Many fundamental questions on aging are still unanswered or are under intense debate. These questions are frequently not addressable by examining a single gene or a single pathway, but can best be addressed at the systems level. Here we examined the modular structure of the protein–protein interaction (PPI) networks during fruitfly and human brain aging. In both networks, there are two modules associated with the cellular proliferation to differentiation temporal switch that display opposite aging-related changes in expression. During fly aging, another couple of modules are associated with the oxidative–reductive metabolic temporal switch. These network modules and their relationships demonstrate (1) that aging is largely associated with a small number, instead of many network modules, (2) that some modular changes might be reversible and (3) that genes connecting different modules through PPIs are more likely to affect aging/longevity, a conclusion that is experimentally validated by Caenorhabditis elegans lifespan analysis. Network simulations further suggest that aging might preferentially attack key regulatory nodes that are important for the network stability, implicating a potential molecular basis for the stochastic nature of aging.
Keywords: aging, cell metabolisms, differentiation, module, network, regulation, stability, proliferation
Introduction
Aging is a most prominent factor associated with many complex diseases, such as cancer, diabetes, cardiovascular diseases and neurodegenerative disorders. Decades of research on aging have found hundreds of genes and many biological processes that are associated with the aging process; however, many fundamental questions are still unanswered and/or are under intense debate. As summarized by a few recent reviews (Kirkwood, 2005; Sinclair, 2005; Hekimi, 2006), these questions include the following: (1) Are there many or only a few biological processes contributing to aging? What are they? (2) Are aging changes irreversible or reversible? (3) Is it possible that a single gene mutation recapitulates all the aging phenotypes? (4) Above all, why aging is a stochastic event (Herndon et al, 2002; Rea et al, 2005; Somel et al, 2006)? These questions are extremely important because they shape our basic view of the aging process and influence how we think about and develop strategies to interfere with the aging processes. However, these questions cannot be fully addressed by focusing on a single gene or a single pathway. For example, even to answer if a single gene mutation can recapitulate all the aging phenotypes, we have to have a good knowledge of the ‘phenome' of aging. Systems biology has therefore been viewed as a promising way toward a more comprehensive understanding of aging (Hood, 2003; Kirkwood, 2005).
To study the global transcriptional changes during aging, Lu et al (2004) collected 30 post-mortem human brain frontal cortex samples and through microarray analysis found ∼440 differentially expressed genes between the old and young brains. Pletcher et al (2002) have performed such an analysis on isogenic populations of once-mated female fruitflies at different ages using five flies for each mRNA sample. Lund et al (2002) carried out a microarray analysis on Caenorhabditis elegans using three different strains collected at a variety of ages, and found only 164 genes that showed a significant change with age. These studies identified a large number of genes that display age-related changes in various seemingly unrelated biological processes. However, it is possible that, at the network level, these genes aggregate into a few dynamically organized network modules (Han et al, 2004) and their expressions are changed concertedly through the same regulatory circuitry. Finding these dynamically organized network modules will be an important first step toward revealing the regulatory circuitry.
Here we focus on the dynamic modular structure of the protein–protein interaction (PPI) networks during aging as revealed by gene expression profiles. Recently, we developed a new analytic method, which we termed ‘NP analysis' (Xia et al, 2006). It permits integration of both transcriptome and interactome information. Briefly, ‘NP analysis' finds a sub-PPI network that is active during a specific process such as aging. The subnetwork is termed the ‘NP network' as it contains only PPIs between genes that are positively or negatively correlated during aging. The subnetwork identified this way is dependent only on the variations during the aging process, instead of the amplitude of change, and thus can include even regulatory genes at the top of regulatory cascade that are expressed at low levels and only slightly change during aging. Next, the analysis divides genes in the NP network into coregulated gene groups or clusters through expression profile-based hierarchical clustering followed by manual or automated dissection of the major gene clusters. In the context of the network we called these gene clusters network ‘modules'. The automated cluster dissection procedure uses a threshold of 1% transcriptionally negatively correlated PPIs to define the boundary of a module (Figure 1A; Supplementary data; Xia et al, 2006). Then the relationships among the modules are established through gene expression profiles and PPIs (Figure 1A; Supplementary data). Such an integrative analysis increases the homogeneity and stability of the module compositions and ensures a high probability of identifying the transcriptionally anticorrelated modules, which often represent alternative cellular states separated by a temporal switch (Xia et al, 2006). For example, by examining only the expression profiles, the chance of finding anticorrelated modules is 62–66%, whereas by first extracting the NP network from the PPI network, the chance increases to 98–99%; if randomly constructed PPI network of the same degree of distribution as the real PPI network is used, the modules identified will be ∼30% smaller in size than those found using the real PPI network and the average interaction degree will also be much smaller than that of the real modules. More details can be found in Table II of Xia et al (2006). A pair of anticorrelated modules can be the result of a ‘toggle-switch' regulatory control circuitry that is designed to coordinate different cell fates/states (Hasty et al, 2002). Finding the transcriptionally anticorrelated modules that concurrently upregulate and downregulate during aging will not only allow us to identify the genes whose changes correlate or inversely correlate with chronological age, but also other concerted alterations in expression states that assume different directionalities during aging and developmental stages. More importantly, such an analysis also leads to the identification of the PPI interface between the modules that potentially regulate or coordinate expression of the modules (Xia et al, 2006) and might play regulatory roles in aging.
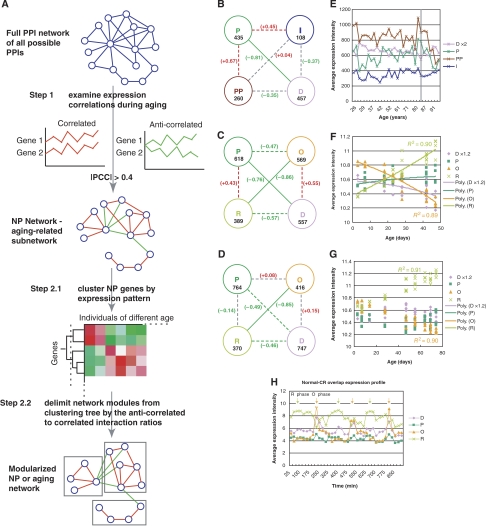
Figure 1.
Network modules during human brain and fruitfly aging. (A) Flow diagram of the NP analysis arriving at the aging-related NP network and modularized NP network. The first step of the NP analysis is to obtain all the PPIs between interactors that are negatively or positively correlated at the transcription level (∣PCC∣>0.4) during aging. Once the subnetwork is found, the genes in the subnetwork are clustered by their expression profiles using hierarchical clustering, then the best separated modules on the clustering tree are found judged by the percentage of negatively correlated interacting gene pairs within a module (<1%). PCC stands for Pearson correlation coefficient. (B) Transcriptional relationships among the modules of the human brain aging NP network. Human D (differentiation), P (proliferation), PP (protein processing) and I (immunity) modules are represented with nodes of lavender, green, brown and dark blue, respectively. Pearson correlation coefficient (PCC) between the two serials of the average expression levels of a pair of the modules over different samples are used to measure the similarity of expression patterns between the modules. The PCC for each module pair is marked at the line connecting the two modules. Solid red and green lines represent strong transcriptional correlations and anticorrelations, respectively (∣PCC∣>=0.7), whereas dotted red and green lines indicate weak correlations and anticorrelations, respectively (0.4<∣PCC∣<0.7). Gray dotted lines represent no obvious transcriptional relationships (∣PCC∣<=0.4). The number of genes of each network module is indicated below the name of the module. (C, D) Transcriptional relationships among the modules of the fruitfly aging under normal (C) and CR (D) conditions. (E) Average expression intensities are plotted against the age of the human subjects for the human brain module genes. The gray vertical line marks age 85. (F, G) Average expression intensities are plotted against fly age for the fly module genes under normal (E) and CR condition (F), respectively. Polynomial fits of the fly module expression level across different populations over age are also displayed, indicating perfect linearity of O and R module expression level with age. Linear regression R2 of the O and R modular changes with age are also displayed. Only the expression levels of the genes overlapping between the corresponding normal diet and CR modules are plotted. Those for the nonoverlapping genes are shown in Supplementary Figure 4. (H) Relationship of O and R modules to metabolic cycle. The average expression levels of the yeast orthologs of the fly R and O genes oscillate with the metabolic cycles and reach the highest levels in the antiphases of reductive and oxidative metabolisms, respectively (marked by the arrows). The P and D modules are also included for comparison. Genes for each module are the overlapping genes between the corresponding modules under normal diet and CR.
Results
Are there many or only a few biological processes contributing to aging?
Aging affects multiple seemingly unrelated biological processes (Finch, 1990). However, by using the ‘NP analysis' on the human brain aging network, we found that the genes aggregate into a small number of network modules. During human brain aging, four modules were found in the NP network. Two of them, the ‘P' and ‘D' modules, are transcriptionally anticorrelated with each other. They are so named because the genes in the modules are enriched in GO annotations related to cell proliferation and differentiation, respectively, and because the former decrease and the latter increase in expression level when cells are induced to switch from proliferation to differentiation state (Figure 1B; Xia et al, 2006). In addition to the P and D modules, there are two other major network modules, the PP and I modules, which are named for ‘protein processing' and ‘immunity,' respectively (named ‘N' and ‘S' in Xia et al, 2006). The PP module is enriched for genes encoding protein translational and degradation activities and the I module is a small module of genes related to immunity (Figure 1B; Xia et al, 2006). Using a single criterion of the percentage of transcriptionally anticorrelated PPIs cannot separate the PP module from the P module because of their high transcriptional correlation. However, the two modules are clearly separable when the clustering results were visualized using the TreeView program. We therefore separated these modules manually.
To determine whether similar modules and their relationships are conserved in other species, we examined the dynamic modular structure of the fruitfly interactome during aging based on the fly yeast two-hybrid (Y2H) interactome (Giot et al, 2003; Formstecher et al, 2005) and fly aging expression profiles (Pletcher et al, 2002) using ‘NP analysis'. As in the human brain NP network, we found in the fly NP network a pair of transcriptionally anticorrelated gene modules corresponding to the proliferation (P module) and differentiation (D module) states at the cellular level (Xia et al, 2006). In addition, we found another pair of transcriptionally anticorrelated gene modules, which we named the reductive metabolism (R) and oxidative metabolism (O) modules as indicated by their associations with cellular processes (see below; Figure 1C, D). The genes and enriched function annotations of each module are listed in Supplementary Tables I and II. Unlike the human PP module, all the four modules in the fly NP network are clearly the only four large clusters visually identifiable using the TreeView program, and additional constraints on module-wise correlations do not generate extra modules.
Through transcriptional relationships, the modules also form a modular network. Under normal condition, module gene expression levels of the P module slightly correlate with R, and D slightly correlates with O across different samples (Figure 1C; Supplementary Figure 1). Upon caloric restriction (CR), the weak expression correlations between P and R and correlation between D and O are lost (Figure 1D; Supplementary Figure 1), due to the apparent lack of a P–D aging pattern during the first half of life in the CR flies (see below).
These modules can also be observed using other interactome data sets (Supplementary data; Supplementary Figure 2; Supplementary Table III), indicating that only a small number of modules are associated with aging, and the number is unlikely to increase significantly with higher genome coverage.
Cross-examination of the orthologous gene expression of fly modules in human and that of the human modules in fly indicates that the lack of O and R modules in human brain and the different composition of fly and human D modules are unlikely due to different coverage of the fly and human interactome and transcriptome (Supplementary data; Supplementary Figure 3; Supplementary Table IV).
Are modular expression changes reversible or irreversible?
Because the genes inside a module have similar expression profiles, we used the average transcription level of the genes inside a module to represent the expression level of a module. We examined the average gene expression levels within a module (module expression levels) relative to age and found that the P and D modules are not only transcriptionally anticorrelated among individuals (Xia et al, 2006), but also undergo opposite changes in expression level with age (Figure 1E). The expression of D decreases and that of P increases with age in the human brain (both significant when only samples of age ⩽85 are included, whereas only D module's decrease is significant if the entire sample set is included; Table I). Such a trend stops and even reverses in the longest-lived people (ages>85), which suggests that their expression levels might also be related to longevity. The reversal is visually clear in Figure 1E, but due to the small sample size, it is only marginally significant (Table I). These changes also suggest that the relationship of P and D module expression with age might be reversible. However, it is not known whether such a reverse is due to the age these individuals achieved or if a difference in the relationship of the P and D modules may have allowed survival to older ages. The expression levels of PP do not have a significant association with age if the longest-lived samples are not included, but are significantly downregulated in these samples (Figure 1E; Table I). The expression of the I module increases steadily in the aging brains, consistent with the previous findings of increased inflammation responses in the aging brain (Lynch, 2004) (Figure 1E; Table I).
Table 1.
Statistical significance of age-related gene expression changes of the human brain and the fly modules
| Module | Slope | P-value | R square | Spearman correlation | P-value | Condition |
|---|---|---|---|---|---|---|
| D | −0.353 | 5.16E−02 | 0.13 | −0.37 | 4.25E−02 | Human brain (entire period) |
| P | 0.693 | 4.19E−01 | 0.02 | 0.10 | 6.06E−01 | |
| PP | −2.327 | 1.71E−02 | 0.19 | −0.33 | 7.83E−02 | |
| I | 0.900 | 1.95E−02 | 0.18 | 0.40 | 2.82E−02 | |
| D | −0.582 | 1.90E−02 | 0.23 | −0.46 | 2.22E−02 | Human brain first period (age⩽85 years) |
| P | 2.661 | 2.04E−02 | 0.22 | 0.39 | 5.77E−02 | |
| PP | −0.429 | 6.82E−01 | 0.01 | −0.12 | 5.83E−01 | |
| I | 1.366 | 1.77E−02 | 0.23 | 0.46 | 2.44E−02 | |
| D | 1.355 | 5.08E−01 | 0.09 | 0.56 | 1.92E−01 | Human brain second period (age⩾85 years) |
| P | −10.220 | 2.09E−01 | 0.29 | −0.83 | 2.12E−02 | |
| PP | −19.511 | 5.64E−02 | 0.55 | −0.77 | 4.08E−02 | |
| I | 4.079 | 6.71E−03 | 0.8 | 0.83 | 2.12E−02 | |
| D | −0.003 | 1.31E−03 | 0.3 | −0.53 | 1.74E−03 | Fly normal condition (7∼47 days) |
| P | 0.003 | 4.81E−03 | 0.24 | 0.47 | 7.17E−03 | |
| O | −0.009 | 4.06E−12 | 0.8 | −0.88 | 1.00E−06 | |
| R | 0.012 | 6.66E−16 | 0.89 | 0.94 | 1.00E−06 | |
| D | 0.001 | 3.70E−01 | 0.07 | 0.21 | 4.71E−01 | Fly CR first half life span (age<40 days) |
| P | −0.004 | 9.21E−02 | 0.22 | −0.31 | 2.86E−01 | |
| O | −0.005 | 6.70E−04 | 0.63 | −0.47 | 9.13E−02 | |
| R | 0.005 | 5.27E−04 | 0.65 | 0.81 | 4.80E−04 | |
| D | −0.004 | 4.54E−05 | 0.52 | −0.73 | 3.50E−05 | Fly CR last half life span (age⩾40 days) |
| P | 0.005 | 1.90E−03 | 0.35 | 0.55 | 4.14E−03 | |
| O | −0.005 | 5.42E−06 | 0.6 | −0.79 | 2.86E−06 | |
| R | 0.006 | 2.24E−04 | 0.45 | 0.64 | 5.23E−04 |
The increase of P and decrease of D gene expression levels can also be observed in the fruitfly (Figure 1F; Table I; Supplementary Figure 4). The expression levels of fly R and O are linear with age (Figure 1F; Table I; Supplementary Figure 4). Although the P and D module expressions display more variation as compared to the O and R modules, both the increase in expression of the P module and the decrease in expression of D module with age are highly significant (P=4.81 × 10−3 and 1.31 × 10−3 for P and D modules, respectively; Table I).
Effects of calorie restriction on aging changes
CR is the single most universal way to slow aging in all species examined (Sinclair, 2005); we therefore asked which, if any, of the aging changes can be reversed or slowed by CR. At the end of life, the expression levels of P, D, R and O are about the same, whether the flies were grown under normal or under CR condition, suggesting that the terminal expression levels of these genes are good indicators of when a life ends. In young flies with normal diet, the expression level of R is lower and that of O is higher, and the difference between R and O is greater when compared to those under CR. This phenomenon is consistent with the metabolic burst in young adults (McCarroll et al, 2004), which is probably enhanced by high food intake. Although the expression levels of fly R and O remain linear with age (Figure 1F, G), under CR the rate of change is slowed by nearly half (Figure 1F, G). The linear change of O and R module expression with age under both normal and CR conditions indicates that the O–R change is an accumulating and likely irreversible event just as is age. The different rates of O–R change under normal diet and CR suggest that these modules reflect biological age rather than chronological age and that the aging processes are associated with concerted changes.
Under diet restriction, the age-related decrease in D expression and increase in P expression only occur at mid-life and through the second half of the lifespan (Figure 1G; Table I; Supplementary Figure 4). In other words, diet restriction delays the onset of P–D aging pattern (decrease of D and increase of P with age) to nearly the end of the normal fly life expectancy (∼40 days) (Figure 1G), which is well before the rapid increase in mortality rate. Once the P–D aging pattern starts, the flies can live about another adult lifetime, as if a span of the P–D aging pattern (from the start to the end of decrease in D and increase in P) demarks a cycle of ‘lifetime' (∼40 days) (Figure 1G). The decrease in D and increase in P with age during the last half lifespan are highly significant (P=4.54 × 10−5 and 1.9 × 10−3, respectively). This again confirms D and P as aging-related modules even under CR.
During the first half lifespan of the diet-restricted flies, the P and D modules almost assume opposite changes. The increase in D with age is not significant (P=0.37) and decrease in P with age is marginal (P=0.09). The lack of aging changes in the P and D modules during this period suggests that, similar to the human brain P and D modules, the fruitfly P and D module changes might be reversible at least within a certain time frame (e.g. first half of lifespan) under certain condition (e.g. CR).
Therefore, there seem to be two different types of changes with age. The changes to the O and R modules are irreversible, whereas in agreement with the human P and D modules' reversibility, the changes to the fly P and D modules seem to be reversible under CR. By ‘reversibility', we refer to the possibility of modules to assume opposite direction of change, not that the aging process is reversed.
What biological processes are the modules related to?
Using Gene Ontology annotations (Ashburner et al, 2000), we found that the D module is enriched in development- and differentiation-related genes, the P is enriched in nuclear transport and cell-cycle genes (Xia et al, 2006; Supplementary Table II). Among other enriched function annotations, mitochondrial and cellular metabolic reactions are enriched in the O and R modules, respectively (Supplementary Table II). The genes in P modules identified from human brain and fly share a significant overlap (P=0.0016, Supplementary Figure 5) and well-conserved functions (Supplementary Table II). In addition, the P modules of both humans and flies are enriched for cell-cycle commitment genes (Supplementary data; Supplementary Figure 6). Although the D modules share no significant overlap between the two species and the enriched functions are also very different, both are enriched in genes required for differentiation and just tailored to the tissue- and species-specific differentiation needs. It is expected that cellular proliferation is conserved between distant species, whereas differentiation is not. Besides being differentiation-related, both the fly and human D modules assume higher expression level when cells are induced to differentiate, and they both have opposite properties toward the P module. We, therefore, termed both modules as the D modules referring to the fly differentiation and human brain differentiation, as the fly and human brain differentiation are explicitly distinctive processes.
As the transcriptionally anticorrelated P and D modules correspond to two alternative cellular states temporally separated by the proliferation to differentiation switch in multiple cell lines (Xia et al, 2006), we hypothesized that the R and O modules could also correspond to a different cellular switch. From the enriched GO annotations in the R and O modules, it is clear that the two modules are related to cellular metabolism. A metabolic cycle has been described in yeast that when synchronized by starvation followed by glucose perfusion, the cells oscillates between oxidative metabolic and reductive metabolic phases. Furthermore, the cell cycle is tightly gated by the metabolic cycle, so that the cell division only occurs during a reductive metabolic phase (Tu et al, 2005). We, therefore, examined whether R and O were temporally separated during the metabolic cycle. Based on the yeast expression profiles during the metabolic cycles (Tu et al, 2005), the average expression levels of the yeast homologs of the R and O module genes strongly oscillate throughout the metabolic cycles and display alternating highest expression levels in antiphases of each metabolic cycle (Figure 1H). The two antiphases correspond to the reductive and oxidative metabolic phases, respectively. Thus, the R and O temporal expression pattern might be introduced by transcription regulators overlapping with those controlling the metabolic cycle. The association of O and R module with the oxidative and reductive metabolic phase is not expected randomly (empirical P<0.01, Supplementary data). In contrast to the R and O modules, the P and D modules display no obvious metabolic cycle-dependent expression (Figure 1G).
Are aging genes unevenly distributed in the aging network?
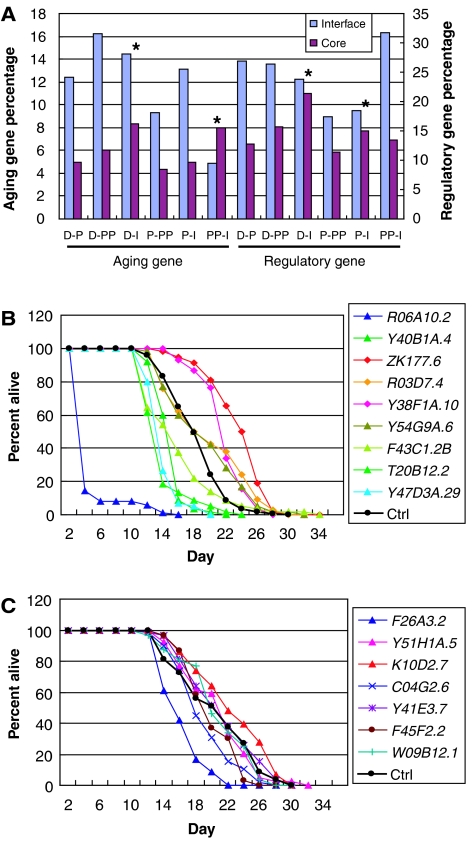
Protein interaction interface usually refers to the structural surface mediating PPIs. Here, we use the term ‘interface' to refer to the module–module interactions mediated by PPIs, so that the proteins on the module interface have PPIs connecting two modules. Our previous study indicates that the module interface between the human brain P and D modules may have regulatory roles coordinating the P and D modules in that the proteins at the module interfaces are enriched in regulatory genes, tumor suppressors and oncogenes and feedback loops, which are defined as cyclic paths (whose start and end nodes are the same) of length between 2 to 10 directed PPI or regulatory interactions (Xia et al, 2006). To further investigate whether the module interfaces may regulate the aging process, we examined in each interface and ‘cores,' or noninterfaces of the human brain aging modules, the proportion of known ‘aging genes,' which are the genes that have been observed to affect cellular or organism aging reported in the literature and/or curated in online databases (Materials and methods). Except for the human brain PP–I interface, all other pair-wise module interfaces consistently have two- to threefold enrichment of known aging genes over that of the cores or the background level in the genome (Figure 2A; Supplementary Table V). There is also a significant enrichment of transcription regulators (obtained as in Xia et al, 2006) at the module interfaces (Figure 2A; Supplementary Table V). When we examined the genes for disease association, we found that as expected, genes associated with cancers are only enriched in the P–D interface but not any other interfaces (data not shown).
Figure 2.
Aging regulatory role of the interface genes. (A) Aging genes and transcription regulators are more enriched on the module interfaces than in the cores. * indicates insignificant differences. Proportion test P-values for the differences can be found in Supplementary Table V. (B) Worm lifespan upon RNAi inactivation of worm orthologs of selected human module interface genes. Wild-type N2 worms were fed bacteria expressing dsRNA that target the genes indicated on the right of the panel. The figure shows the results of one representative experiment carried out at 20°C. See Table II for statistical differences. (C) Worm lifespan upon RNAi inactivation of randomly selected worm genes that have human orthologs.
To examine whether the high percentages of aging and regulatory genes at the module interfaces are a mere consequence of the ‘module interface' definition, we randomly grouped genes in the NP network into modules of the same sizes as the D, P, PP and I modules, and examined the percentages of aging and regulatory genes on their module interfaces. We performed such simulations 100 times, and found only the transcriptionally anticorrelated D–P and D–PP interfaces having significantly higher percentages of aging (P=0.04 for D-P and P<0.01 for D–PP interface) and regulatory genes (P<0.01 for both D–P and D–PP interfaces) than randomly expected. This highlights the importance of finding the transcriptionally anticorrelated modules and their interfaces.
Together these results suggest that the module interfaces, especially those that are transcriptionally anticorrelated, have specific regulatory roles in coordinating the two modules it bridges. Such properties led to a testable hypothesis that the genes on the interface, especially those located in the feedback loops at the anticorrelated module interfaces may constitute the key regulatory components in coordinating different modules during the aging process, which we validated with lifespan assays in C. elegans upon RNAi knockdown of the genes.
We randomly selected eight C. elegans orthologs of human genes that locate in the regulatory feedback loops at the human brain module interfaces and one other gene on the interface but not in a feedback loop (POLA) as testing genes, and seven randomly selected C. elegans orthologs of human genes as control genes. We then analyzed the lifespan of the worms upon RNAi knockdown of both the testing and control genes in parallel with at least two repeated experiments. RNAi knockdown of three of the nine testing genes extended the worm lifespan and that of five other genes shortened the lifespan significantly (log-rank test, P-value<0.01 for both repeated experiments, Figure 2B; Table II), whereas RNAi of only one of the seven control randomly selected human ortholog genes shortened the lifespan and none extended the lifespan of the worm under the same significance level (Figure 2C; Table II). The significant difference (Fisher exact test P=0.006) of the effect on the worm lifespan between the P–D or PP–D interface genes and the random genes proved that these interface genes indeed play an important regulatory role on longevity of the worm. Although without more detailed genetic and biochemical analysis, we cannot rule out that some of the lifespan-shortening effect might be due to developmental defects, the mere difference in the proportion of genes that can extend lifespan upon RNAi is dramatic (33 versus 0%). Moreover, in contrast to the control genes, each of the testing genes always displayed consistent lifespan increase or decrease (same direction of change) among replicates, despite the variations in the extent of change between different experiments (Table II).
Table 2.
Significance of changes in C. elegans lifespan upon RNAi knockdown of testing and control genes
| Group | Gene name | Human orthorlog | P-value 1 | % change in lifespan 1 | P-value 2 | % change in lifespan 2 | Module |
|---|---|---|---|---|---|---|---|
| Testing genes | R06A10.2a | GNAS | 0 | −82.46 | 0 | −84.83 | PP interface |
| Y40B1A.4a | SP3 | 1.20E−12 | −20.07 | 5.42E−12 | −28.35 | P core | |
| F43C1.2Ba | MAPK1 | 0.00366 | −14.23 | 0.0103b | −8.24 | D interface | |
| T20B12.2a | TBP | 9.66E−15 | −26.88 | 3.19E−11 | −15.31 | D interface | |
| Y47D3A.29a | POLA | 0 | −25.68 | 2.55E−15 | −18.69 | D interface | |
| Y54G9A.6 | BUB3 | 0.027 | +5.78 | 0.0497 | +7.09 | P interface | |
| ZK177.6a | CDC20 | 5.11E−15 | +32.43 | 0.000342 | +17.19 | D interface | |
| R03D7.4a | TCEB3 | 0.00282 | +8.73 | 0.000011 | +16.74 | D interface | |
| Y38F1A.10a | PAK3 | 1.72E−08 | +21.22 | 5.32E−13 | +28.91 | D interface | |
| Control genes | F26A3.2a | NCBP2 | 6.40E−07 | −22.79 | 1.11E−08 | −25.27 | |
| Y51H1A.5 | HDAC10 | 0.917 | +1.58 | 0.149 | −3.36 | ||
| K10D2.7 | MOCS2 | 0.0316 | +11.09 | 0.434 | −1.00 | ||
| C04G2.6 | KIAA1008 | 0.0212 | −7.82 | 0.00301 | −13.12 | ||
| Y41E3.7 | ACBD3 | 0.608 | +3.74 | 0.255 | −2.00 | ||
| F45F2.2 | HIST2H2BE | 0.0633 | −2.91 | 0.0861 | −1.08 | ||
| W09B12.1 | ACHE | 0.607 | +0.11 | 0.0423 | −8.10 |
aAfter the gene names denote that RNAi of the genes significantly extend (shorten) C. elegans lifespan (P<0.01 for both duplicate experiments).
bA third time repeat gives rise to a more significant P-value, 2.68E-009.
All the three genes we found to extend worm lifespan upon RNAi knockdown are known to be associated with cell proliferation, growth and/or differentiation at the cellular level. Again all the five genes whose RNAi shortens lifespan are known to be associated with cell proliferation, growth and/or differentiation; there is direct evidence that one of the them affect cellular senescence (MAPK1) (Ota et al, 2006), and the other four are associated with aging or neural degeneration, which is a phenotype of aging. This is consistent with our prediction that the coordination of the proliferation, differentiation and metabolic modules are important for regulating aging (Supplementary data). These experiments also indicate that a high proportion of the regulatory genes are functionally conserved between human and worms.
The enrichment of aging regulatory genes at the module interface suggests that the regulation or the coordination of the modular relationship has strong effect on aging. This implicates that the misregulation of modular relationships is a more direct cause of aging or altering the regulation has stronger impacts, as it may change many genes related to the process at once. Such a scenario will make it possible for a single or a few regulatory genes in combination to recapitulate all the aging phenotypes.
Are aging genes the key nodes to maintain system homeostasis or network stability?
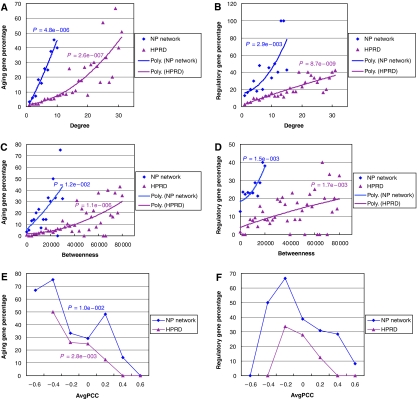
It is known that hubs (nodes making many links) are important to the stability of a network (Albert et al, 2000). We, therefore, examined whether the aging genes are preferentially hubs in the network. In both the HPRD network and the active subnetworks (the NP network), during human brain aging, the percentage of known aging genes increases with the increasing degrees of PPI of the genes (Figure 3A). Transcription regulators, which are also enriched at the module interfaces, also increase in percentage with increasing PPI degree in both the NP network and the HPRD network (Figure 3B).
Figure 3.
Correlations of the percentage of aging genes and regulatory genes to PPI degree, betweeness of the proteins and the AvgPCC of the hubs. (A, B) The percentage of aging genes (A) or transcription regulators (B) among the NP or HPRD proteins of distinct PPI degrees are plotted against the PPI degrees of the proteins. The linear regression significance P-values are indicated above the polynomial fitted trend lines. (C, D) The percentage of aging genes (C) or transcription regulators (D) among the NP or HPRD proteins that are within each betweeness value interval of 2000 is plotted against minimal betweeness value of the interval. The linear regression significance P-values are indicated above the polynomial fitted trend lines. (E, F) The percentage of aging genes (E) or transcription regulators (F) among the NP or HPRD hubs that are within each AvgPCC value interval of 0.2 is plotted against minimal AvgPCC value of the interval. The linear regression significance P-values are indicated above the connected lines in (E).
Node ‘betweeness' is the number of shortest distance paths between any two nodes in the network that passes through a node. As betweeness and PPI degree are highly correlated and both correlate well with the essentiality of genes in the yeast PPI networks (Yu et al, 2007), we also examined its relationship to the likelihood of being an aging gene. Similarly, the percentages of aging genes and transcription regulators increase with increasing node betweeness as well (Figure 3C, D). Furthermore, the percentages of both aging genes and regulatory genes are higher and the increase with degree or betweeness is sharper inside the NP network as compared to the full PPI network or the HPRD network, confirming the NP network as an aging-related subnetwork. The insufficient number of annotated aging genes or transcription regulators in fruitfly precludes any statistically meaningful results from such analysis.
It has been shown that, based on interaction dynamics, hubs in the yeast PPI networks can be categorized into ‘date' and ‘party' hubs. Party hubs are those that interact with their interactors at the same time and space, whereas date hubs interact with their interactors at different time or space. The dynamic behavior of the two types of hubs can be estimated by the average PCC (AvgPCC) between the expression profiles of a hub and its partners. High AvgPCC is characteristic of party hubs, and a relatively low AvgPCC signifies date hubs (Han et al, 2004; Bertin et al, 2007). As aging genes are enriched at the module interface, which by definition would have relatively low AvgPCC compared with those inside the modules, we examined if aging genes are preferentially associated with date hubs. Defining NP and HPRD hubs as the top 10% highly connected genes in the NP and HPRD networks, respectively, we plotted the AvgPCC of the hubs against either the percentage of aging genes or the percentage of transcription regulators within equal intervals of AvgPCC. Both aging genes and transcription regulators are preferentially associated with low AvgPCC or ‘date hubs' (Figure 3E, F). The bias is much stronger for aging genes where there is an inverse relationship between the percentage of aging genes and the AvgPCC of the hubs (Figure 3E), suggesting that aging genes are not only predominately date hubs, but hubs participating in inhibitory interactions. The negative values of AvgPCC (Figure 3E, F) for the majority of aging genes and transcription regulators indicate that both types of genes are more likely to connect anticorrelated modules, consistent with the observation that the interface between the highly correlated P and PP modules have the lowest percentage of aging and transcription regulators among all the module interfaces (Figure 2A). These results suggest that aging is characterized by a change in network stability and especially dynamic stability.
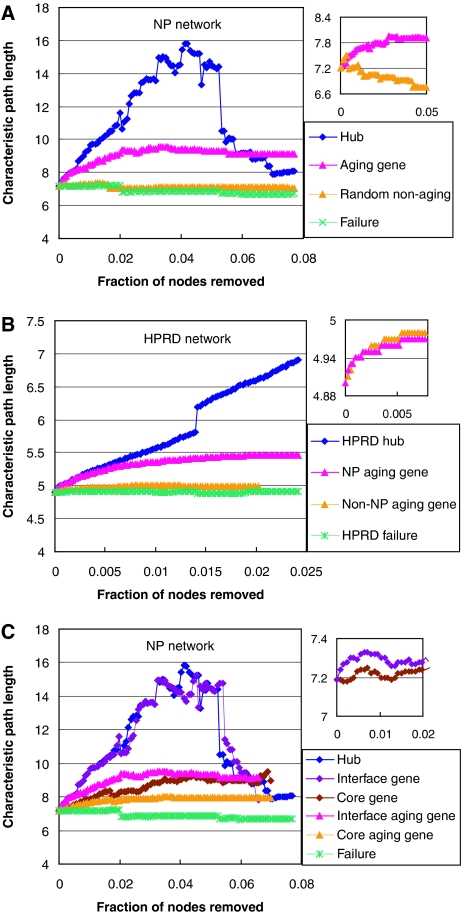
To test whether the aging genes are important to the network stability, we used an established test for network structure stability—the analysis of the changes in characteristic path length (CPL) in the largest connected component of the network after sequential node removal. If the nodes removed are important mediators for network communication, connections from one node to another within the network will take longer paths, hence an increase of CPL of the resulting network (Albert et al, 2000). Sequential removal of nodes according to decreasing interaction degrees is also called an ‘attack' (Albert et al, 2000). Attacking hubs in the network, as expected, increases the CPL very rapidly, whereas random removal of nodes from the network hardly changes the CPL (Figure 4A). Attacking the aging genes obviously has a much stronger effect on the network stability than removal of random proteins without sorting for interaction degrees (also called ‘failure' (Albert et al, 2000)) or attacking randomly selected non-aging genes, but is not as severe as specific attacks on the hubs (Figure 4A). Removal of aging genes belonging to the NP network has stronger effects on CPL than those that are not in the NP network, consistent with the NP network being an aging-specific subnetwork with genes important to the network or system stability overrepresented (Figure 4B). In agreement with the enrichment of aging genes and regulatory genes at the module interfaces, attacking the interface genes dramatically destabilizes the NP network when compared to those in the cores of the modules or to the removal of random genes (Figure 4C). A similarly drastic difference also exists between the aging genes on the interfaces and those in the cores (Figure 4C).
Figure 4.
Aging genes and interface genes are important to network stability. (A) Attacking the aging genes in the NP network increases the CPL of the network more rapidly than attacking randomly selected non-aging genes or removal of random nodes (‘failure'), but slower than specific attacks on hubs. Degree-matched attacks on aging and non-aging genes in the NP network are shown in the inset. (B) Attacking aging genes belonging to the NP network increases CPL of the HPRD network more rapidly than attacking aging genes not in the NP network or random removal of genes in the HPRD network. Degree-matched attacks on NP and non-NP aging genes in the HPRD network are shown in the inset. (C) Attacking the aging (or all) genes on the module interfaces increases CPL of the NP network more rapidly than attacking their counterparts in the cores. Degree-matched attacks on the core and interface genes are shown in the inset. Only the first 2% of the attacks are shown in the inset for the interface and core genes, the trend continues for the rest.
Other than interaction degrees, positions in the network can also affect whether a node is dispensable or indispensable, for instance, a centrally located node may be indispensable, whereas a peripherally located one is not. These properties can be inferred by CPL changes upon degree-matched attacks. Upon degree-matched attacks (Supplementary data), the differences of aging versus non-aging and interface versus core still exist albeit to a less extent (Figure 4A, C, insets), suggesting that other than higher interaction degrees, the aging genes and the interface genes also have more important network positions in the network compared to the non-aging genes and core genes. In contrast, the difference between NP aging genes and non-NP aging genes mostly disappear upon degree-matched attacks (Figure 4B, inset), suggesting that their differences are largely due to the difference in interaction degrees. ‘Betweeness'-matched attacks from highest to lowest betweeness nodes also left some differential impact on CPL unexplained (Supplementary Figure 7). The residual differences remained after controlling for degree or betweeness is likely a feature of the date hubs, which in addition to having relatively higher PPI degrees and betweeness values as compared with party hubs, also tends to connect different dynamic modules (Han et al, 2004; Bertin et al, 2007; Yu et al, 2007). The fly aging network displays similar properties (Supplementary Figure 8).
Discussion
Genetic screens and natural mutations have identified more than 200 genes that affect aging and/or longevity. These mutant alleles come from diverse functional categories. However, none of these biological functions or processes are isolated, but instead are embedded in a large web of physical and genetic interactions to achieve a balanced and coordinated state at the cellular and organism level. Here, different from many other meta-analyses, by combining gene expression data with the knowledge of PPI interaction networks, our approach allowed us to identify ‘active' protein interaction modules—subnetworks that are coherently expressed during aging. This approach enables us to analyze the temporal profiles and the correlations between these modules, as well as the connections between the topological properties of the genes in the modules (such as interaction degrees or betweeness) and their physiological roles (such as the effect on lifespan).
Our analysis revealed a modularized network view of the aging process, where aging is linked to the dynamic network stability. Such a view provides a potential molecular explanation for the stochastic effects of aging, that is, isogenic populations age at vastly different paces (Herndon et al, 2002; Rea et al, 2005; Somel et al, 2006), where the states of the network can be differentially affected by developmental and environmental factors.
We have also analyzed the C. elegans NP network derived based on Y2H and/or computationally predicted PPIs and aging transcriptome profiles, but did not find any obvious age-related modules. Possible explanations are as follows. (1) McCarroll's data set (McCarroll et al, 2004) contains only seven time points, too few for robust calculations of PCC. (2) Lund's data set contains enough data points, but the sample repeatability is low. In the original study, the authors had to pool multiple sample points together to get statistically reliable results, and found only a very small set of 164 genes change with age (Lund et al, 2002). In agreement with their results, we could only find very small modules in the NP network, only one of them having more than 100 genes, with no significantly enriched GO annotations in any of the modules. Furthermore, their overlaps to all the human and fly modules are too small to determine statistical significance. Most importantly, the modules' expression levels do not change significantly with age. (3) The PPI data set is relatively small for C. elegans (Li et al, 2004).
Although knocking down of a gene leading to shortened lifespan cannot fully establish that the gene is the causal to aging, it is encouraging that overexpressing one such gene, SP3, extends worm lifespan, as an expected opposite phenotype to RNAi knockdown of the gene (B Xian et al, unpublished results). It should also be noted that only knockdown of a gene leading to shortened lifespan is a more direct evidence for the network destabilization model for aging, although extending lifespan might be due to an indirect effect, such as antagonizing or balancing effect to the other group of genes, as the relationship of P and D modules.
The modular aging networks uncovered by our analysis provided an entry point to address many fundamental questions on aging at the systems level. The answers to these questions will provide guidance for finding preventive and interference strategies for the aging process and its associated diseases. For example, as the coordination and regulation of the modules have strong impact on aging, we may want to design drugs to target the regulatory circuitry. Also, as some changes are more reversible than others, we should make them high-priority drug targets.
Materials and methods
Data sets
The human brain NP analysis were based on the PPIs in the HPRD database (Peri et al, 2003) downloaded from www.hprd.org on November 22, 2004 and September 13, 2005 and two human Y2H data sets were included in an extended PPI network (Rual et al, 2005; Stelzl et al, 2005). Details are described by Xia et al (2006). Two Y2H screens were combined as the fly protein interactions data set (Giot et al, 2003; Formstecher et al, 2005). Microarray expression profiles were obtained from previously published studies on post-mortem human brains of subjects between ages 26 and 106 (Lu et al, 2004), Drosophila melanogaster aging and diet restriction (Pletcher et al, 2002), yeast metabolic cycle (Tu et al, 2005), gene expression in the heads of young and old flies (McCarroll et al, 2004), in young and old human muscles (GEO accession number GSE674 for female muscle) (Welle et al, 2003) and in young and old human primary skin cells (Kyng et al, 2003).
GO annotations was downloaded from ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/ on March 10, 2005.
Human G1/S and G2/M cell-cycle genes are derived from a microarray analysis of the HeLa cell-cycle gene expression (Whitfield et al, 2002) and downloaded from http://genome-www.stanford.edu/Human-CellCycle/Hela/. The fly orthologs of these genes were used to examine the percentage of the fly cell-cycle genes in each module.
Genes downloaded from http://genomics.senescence.info/genes/ (de Magalhaes et al, 2005) on Dec 12, 2005 were combined with those manually curated from several reviews to form the ‘aging gene' list. The list contains both genes derived from human progeria diseases and genes when mutated or up- or downregulated affect lifespan in model organisms, but not necessarily tested directly in human aging.
Orthologs
Fly orthologs in human, worm and yeast were identified as the best reciprocal BlastP hits with e-value cutoff of 10−6 based on RefSeq protein sequences downloaded on December 9, 2004.
Filtering GO terms, GO term enrichment calculation, expression clustering and modular NP network layout were performed as described previously (Xia et al, 2006).
RNAi experiment
RNAi of N2 worms were carried out as described by Kamath et al (2001) with minor modifications. We selected the bacteria from Ahringer's RNAi feeding bacteria library (Kamath et al, 2003). The same HT115 bacteria carrying empty L4440 construct were used as controls in all experiments.
Lifespan assay
10–20 gravid N2 worms were plated on RNAi plates to lay eggs for 2–4 h at 20°C and then removed. After the synchronized eggs grew to young adult stage, we distributed them (≈20 worms per plate) to RNAi plates containing 20 μg/ml FUDR to prevent the growth of the progeny (day=0). Three plates were scored for each gene in each experiment. Worms were scored every 2 days from the young adult stage until they showed no response to a gentle prodding with a platinum wire. The worms were transferred to fresh RNAi plates every 4 days to ensure the continued efficacy of RNAi knockdown. Worms that crawled off were excluded from the experiments. All experiments were independently performed at least twice. The P-value is calculated by a log-rank test on the Kaplan–Meier curves.
Degree-matched or betweeness-matched attacks
To examine whether factors other than the interactions degrees (or ‘betweeness') also contribute to the differential impacts on network topology between two different groups of genes, we ordered all the genes in the two groups in one list according to decreasing interaction degrees (or betweeness). Then starting from the first gene in the low average degree (or betweeness) group, we alternatively select genes of the low and high average degree (or betweeness) group next on the list, until the last gene on the list of the high average degree (or betweeness) group. The resulting two subgroups of genes to be compared have one-to-one matched degree (or betweeness) distributions. Then the nodes in each ordered list are sequentially attacked according to decreasing the PPI degrees (or betweeness values).
Supplementary Material
Supplementary Information
Supplementary Figures
Supplementary Table I
Supplementary Table II
Acknowledgments
We thank Dr Thomas E. Johnson and Dr James R. Cypser at the University of Colorado at Boulder for numerous helpful discussions of the data, Dr Ralph Greenspan (The Neuroscience Institute, San Diego), Dr Nicholas Baker (Albert Einstein College of Medicine), Dr Li Cai (Rutgers University) and Dr Yong Liu (CAS Institute of Nutrition) for helpful discussions and three anonymous reviewers for their valuable comments. We are also grateful to Dr Chonglin Yang for sharing C. elegans resources. This work was supported by NSFC Grant 30588001, National Basic Research Program of China (2006CB910700) and CAS funds to J-DJH.
References
- Albert R, Jeong H, Barabasi AL (2000) Error and attack tolerance of complex networks. Nature 406: 378–382 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin N, Simonis N, Dupuy D, Cusick ME, Han JD, Fraser HB, Roth FP, Vidal M (2007) Confirmation of organized modularity in the yeast interactome. PLoS Biol 5: e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes JP, Costa J, Toussaint O (2005) HAGR: the Human Ageing Genomic Resources. Nucleic Acids Res 33: D537–D543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch CE (1990) Longevity, Senescence, and the Genome. Chicago: University of Chicago Press [Google Scholar]
- Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, Jacq B, Arpin M, Bellaiche Y, Bellusci S, Benaroch P, Bornens M, Chanet R, Chavrier P, Delattre O, Doye V et al. (2005) Protein interaction mapping: a Drosophila case study. Genome Res 15: 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M et al. (2003) A protein interaction map of Drosophila melanogaster. Science 302: 1727–1736 [DOI] [PubMed] [Google Scholar]
- Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP, Vidal M (2004) Evidence for dynamically organized modularity in the yeast protein–protein interaction network. Nature 430: 88–93 [DOI] [PubMed] [Google Scholar]
- Hasty J, McMillen D, Collins JJ (2002) Engineered gene circuits. Nature 420: 224–230 [DOI] [PubMed] [Google Scholar]
- Hekimi S (2006) How genetic analysis tests theories of animal aging. Nat Genet 38: 985–991 [DOI] [PubMed] [Google Scholar]
- Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, Sakano Y, Paupard MC, Hall DH, Driscoll M (2002) Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature 419: 808–814 [DOI] [PubMed] [Google Scholar]
- Hood L (2003) Systems biology: integrating technology, biology, and computation. Mech Ageing Dev 124: 9–16 [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J (2001) Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2: RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood TB (2005) Understanding the odd science of aging. Cell 120: 437–447 [DOI] [PubMed] [Google Scholar]
- Kyng KJ, May A, Kolvraa S, Bohr VA (2003) Gene expression profiling in Werner syndrome closely resembles that of normal aging. Proc Natl Acad Sci USA 100: 12259–12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF et al. (2004) A map of the interactome network of the metazoan C. elegans. Science 303: 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA (2004) Gene regulation and DNA damage in the ageing human brain. Nature 429: 883–891 [DOI] [PubMed] [Google Scholar]
- Lund J, Tedesco P, Duke K, Wang J, Kim SK, Johnson TE (2002) Transcriptional profile of aging in C. elegans. Curr Biol 12: 1566–1573 [DOI] [PubMed] [Google Scholar]
- Lynch MA (2004) Long-term potentiation and memory. Physiol Rev 84: 87–136 [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin CS, Jan YN, Kenyon C, Bargmann CI, Li H (2004) Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat Genet 36: 197–204 [DOI] [PubMed] [Google Scholar]
- Ota H, Tokunaga E, Chang K, Hikasa M, Iijima K, Eto M, Kozaki K, Akishita M, Ouchi Y, Kaneki M (2006) Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras–MAPK signaling in human cancer cells. Oncogene 25: 176–185 [DOI] [PubMed] [Google Scholar]
- Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, Ibarrola N, Deshpande N, Shanker K, Shivashankar HN, Rashmi BP, Ramya MA, Zhao Z, Chandrika KN, Padma N, Harsha HC et al. (2003) Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res 13: 2363–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher SD, Macdonald SJ, Marguerie R, Certa U, Stearns SC, Goldstein DB, Partridge L (2002) Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr Biol 12: 712–723 [DOI] [PubMed] [Google Scholar]
- Rea SL, Wu D, Cypser JR, Vaupel JW, Johnson TE (2005) A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat Genet 37: 894–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S et al. (2005) Towards a proteome-scale map of the human protein–protein interaction network. Nature 437: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Sinclair DA (2005) Toward a unified theory of caloric restriction and longevity regulation. Mech Ageing Dev 126: 987–1002 [DOI] [PubMed] [Google Scholar]
- Somel M, Khaitovich P, Bahn S, Paabo S, Lachmann M (2006) Gene expression becomes heterogeneous with age. Curr Biol 16: R359–R360 [DOI] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B et al. (2005) A human protein–protein interaction network: a resource for annotating the proteome. Cell 122: 957–968 [DOI] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL (2005) Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science 310: 1152–1158 [DOI] [PubMed] [Google Scholar]
- Welle S, Brooks AI, Delehanty JM, Needler N, Thornton CA (2003) Gene expression profile of aging in human muscle. Physiol Genomics 14: 149–159 [DOI] [PubMed] [Google Scholar]
- Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, Botstein D (2002) Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell 13: 1977–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K, Dong D, Xue H, Zhu S, Wang J, Zhang Q, Hou L, Chen H, Tao R, Huang Z, Fu Z, Chen YG, Han JD (2006) Identification of the proliferation/differentiation switch in the cellular network of multicellular organisms. PLoS Comput Biol 2: e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M (2007) The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol 3: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Figures
Supplementary Table I
Supplementary Table II