Abstract
We have identified a specific karyopherin docking complex within the yeast nuclear pore complex (NPC) that contains two novel, structurally related nucleoporins, Nup53p and Nup59p, and the NPC core protein Nup170p. This complex was affinity purified from cells expressing a functional Nup53p–protein A chimera. The localization of Nup53p, Nup59p, and Nup170p within the NPC by immunoelectron microscopy suggests that the Nup53p-containing complex is positioned on both the cytoplasmic and nucleoplasmic faces of the NPC core. In association with the isolated complex, we have also identified the nuclear transport factor Kap121p (Pse1p). Using in vitro binding assays, we showed that each of the nucleoporins interacts with one another. However, the association of Kap121p with the complex is mediated by its interaction with Nup53p. Moreover, Kap121p is the only β-type karyopherin that binds Nup53p suggesting that Nup53p acts as a specific Kap121p docking site. Kap121p can be released from Nup53p by the GTP bound form of the small GTPase Ran. The physiological relevance of the interaction between Nup53p and Kap121p was further underscored by the observation that NUP53 mutations alter the subcellular distribution of Kap121p and the Kap121p- mediated import of a ribosomal L25 reporter protein. Interestingly, Nup53p is specifically phosphorylated during mitosis. This phenomenon is correlated with a transient decrease in perinuclear-associated Kap121p.
Keywords: nucleoporins, nuclear transport, karyopherin, importin, cell cycle
The surface of the nuclear envelope (NE)1 is decorated by a non-random distribution of nuclear pore complexes (NPCs) (for review see Maul, 1977; Davis, 1995; Fabre and Hurt, 1997). The NPC consists of a symmetrical (822 symmetry) core structure composed of a ring–spoke assembly surrounding a structure termed the central transporter (Hinshaw et al., 1992; Akey and Radermacher, 1993; Yang et al., 1998). Attached to the core are a set of filaments extending into the cytoplasm from the corresponding face of the core and a morphologically distinct set extending from the core into the nucleoplasm and forming a structure referred to as the nuclear basket. It is likely that the NPC plays a role in the organization of the nucleoplasm and, potentially, the regulation of gene expression (Blobel, 1985). However, its most clearly defined and extensively studied function is the regulation of transport between the cytoplasm and the nucleoplasm.
Much of what we currently understand about nuclear transport has come from studies examining the import of proteins containing the “classical” nuclear localization signal (cNLS) composed of short peptides containing basic amino acid residues (for review see Dingwall and Laskey, 1991). cNLS-containing substrates are recognized by a receptor complex composed of two proteins, karyopherin α (Radu et al., 1995a ) (also referred to as importin α [Görlich et al., 1994] and the NLS-receptor [Adam and Gerace, 1991; Adam and Adam, 1994]) and karyopherin β (Radu et al., 1995a ) (also referred to as importin β [Görlich et al., 1995a ] and p97 [Chi et al., 1995]). Electron microscopy studies of cNLS-coupled gold particles suggest that the β/α/cNLS-cargo complex docks at multiple sites along the cytoplasmic filaments (Feldherr et al., 1984; Richardson et al., 1988; Akey and Goldfarb, 1989; Pante and Aebi, 1996). By what has been proposed to be a series of docking and release steps, the β/α/cNLS-cargo complex moves from the cytoplasmic filaments through the NPC core to the nucleoplasmic filaments before release of the cNLS-containing cargo and karyopherin α into the nucleus. Karyopherin β is not released into the nucleoplasm but appears to be directly recycled to the cytoplasm (Görlich et al., 1995b ; Moroianu et al., 1995).
The initial binding of the β/α/cNLS complex to the NPC is energy independent, however subsequent steps that accompany translocation do require energy. How energy is used to drive translocation is not clearly understood, but (at least in part) it is linked to the function of a small GTPase, Ran (for review see Cole and Hammell, 1998; Mattaj and Englmeier, 1998; Melchior and Gerace, 1998; Moore, 1998). It is believed that the predominant form of Ran in the cytosol is Ran-GDP and in the nucleus it is Ran-GTP. The gradient formed by these two pools of Ran has been proposed to play an important role in vectorial transport across the NPC (Cole and Hammell, 1998). This is exemplified by the ability of Ran-GTP, but not Ran-GDP, to stimulate the disassembly of the β/α/cNLS complex and to release the β/α complex from a bound NPC protein by binding to karyopherin β (Rexach and Blobel, 1995; Percipalle et al., 1997). Such observations and others have led to the proposal that Ran may act as a molecular switch in the binding and release steps that occur during transport through the NPC (Rexach and Blobel, 1995; Floer et al., 1997) and in the final release of cargo into the nucleoplasm (Görlich et al., 1996). Whether Ran-GTP alone provides a sufficient energy source for translocation through the NPC remains unknown.
The cNLS represents the first of a growing list of NLSs and NESs that have been identified (for review see Mattaj and Englmeier, 1998). In several of the cases examined so far, these signals are directly recognized and transported through the NPC by soluble transport factors structurally related to karyopherin β (for review see Wozniak et al., 1998). In yeast, as many as 14 potential β-type karyopherins have been identified. Individual members of this family have recently been implicated in the transport of specific classes of cargo. These cargo include the mRNP proteins (Aitchison et al., 1996; Pollard et al., 1996; Bonifaci et al., 1997; Fridell et al., 1997; Pemberton et al., 1997; Senger et al., 1998), a subset of ribosomal proteins (Rout et al., 1997; Yaseen and Blobel, 1997; Schlenstedt et al., 1997; Jäkel and Görlich, 1998) and tRNA processing proteins (Rosenblum et al., 1997). β-Type karyopherins have also been shown to facilitate the export of tRNA (Arts et al., 1998; Kutay et al., 1998), karyopherin α (Kutay et al., 1997), and proteins containing leucine-rich NES sequences (Fornerod et al., 1997a ; Kudo et al., 1997; Ossareh-Nazari et al., 1997; Stade et al., 1997; Kehlenbach et al., 1998). While these results suggest individual β-karyopherins are responsible for the transport of specific cargo, it is also apparent that in the absence of certain β-karyopherins others can compensate for their loss. For example, in yeast the β-karyopherin Kap123p imports the ribosomal protein L25 (Rout et al., 1997; Schlenstedt et al., 1997). In the absence of Kap123p, the mislocalization of an L25-NLS reporter protein can be corrected by a similar β-karyopherin, Kap121p. Similarly, the lethal phenotype of a KAP121 null mutation can be rescued by the overexpression of SXM1 (Seedorf and Silver, 1997).
The role that individual NPC proteins, termed nucleoporins, play in transport as they interface with karyopherins and their cargo remains largely undefined. On the basis of biochemical and genetic analyses in both yeast and vertebrates, a group of nucleoporins that contain multiple repeats of short peptides consisting of the amino acid residues GLFG, FXFG, or FG alone have been suggested to play a direct role in transport (for review see Fabre and Hurt, 1997; Doye and Hurt, 1997; Ohno et al., 1998). Several members of this group have been shown to physically interact with members of the β-karyopherin family and karyopherin α (Radu et al., 1995a ,b; Moroianu et al., 1995; Iovine et al., 1995; Rexach and Blobel, 1995; Aitchison et al., 1996; Hu et al., 1996; Nehrbass and Blobel 1996; Bonifaci et al., 1997; Fornerod et al., 1997b ; Iovine and Wente, 1997; Moroianu et al., 1997; Pemberton et al., 1997; Percipalle et al., 1997; Rosenblum et al., 1997; Rout et al., 1997; Shah et al., 1998). Moreover, mutations in a number of the repeat-containing nucleoporins alter the subcellular distribution of cNLS-containing proteins and poly A+ RNA (reviewed in Fabre and Hurt, 1997; Mattaj and Englmeier, 1998). Defective transport phenotypes, however, are not restricted to mutations in repeat-containing nucleoporins. A number of other nucleoporins that lack FG repeats are also required for nuclear transport. The majority of these have been identified in yeast. This includes several yeast nucleoporins which when mutated block the export of poly A+ RNA (reviewed in Schneiter et al., 1995; Fabre and Hurt, 1997). Mutations in other non repeat nucleoporins, including Nic96p (Grandi et al., 1995 a), exhibit cNLS-mediated import defects (also see Fabre and Hurt, 1997).
Included in the non-repeat nucleoporins are a set of proteins that share the common feature of being the most abundant of the yeast nucleoporins. Together these proteins, including Nic96p, the pore membrane protein Pom152p, Nup188p, Nup192p, and two structurally related proteins, Nup157p and Nup170p, account for ∼25% of the mass of an isolated yeast NPC (Wozniak et al., 1994; Aitchison et al., 1995; Nehrbass et al., 1996; Zabel et al., 1996; Wozniak, R., unpublished data; M. Rout, personal communication; also see Fabre and Hurt, 1997). The abundance of these proteins has led to the suggestion that they are components of the repetitive substructures that form the eightfold symmetrical core of the NPC. Here they may function as the framework of the NPC core and a foundation on which other nucleoporins, including repeat-containing nucleoporins, are organized. Consistent with this idea, many of these abundant nucleoporins are functionally linked to one another. For example, Pom152p physically and/or genetically interacts with Nup170p, Nup188p, and Nic96p (Aitchison et al., 1995; Nehrbass et al., 1996; Zabel et al., 1996). Nic96p and, potentially, Nup170p are in turn physically linked to repeat-containing nucleoporins (Grandi et al., 1993, 1995; Kenna et al., 1996).
In this study we have begun to further examine the physical and functional relationships between the abundant yeast nucleoporins, other constituents of the NPC and the transport machinery. Through the use of a genetic screen we have identified two previously uncharacterized yeast nucleoporins that we have termed Nup53p and Nup59p. Both of these proteins physically interact with Nup170p in a complex that is accessible on both faces of the NPC. Nup53p acts as a specific, high-affinity binding site for the β karyopherin Kap121p.
Materials and Methods
Yeast Strains and Media
All yeast strains used in this study are listed in Table I or referenced below. All strains were grown as described (Sherman et al., 1986) in YPD (1% yeast extract, 2% bactopeptone, and 2% glucose), synthetic minimal media (SM) supplemented with the necessary amino acids or nucleotides and 2% glucose. 5-Fluoroorotic acid (5-FOA) (Toronto Research Chemicals, Toronto, Ontario, Canada) plates were prepared in SM media as described (Ausubel et al., 1992). All strains were grown at 30°C unless otherwise specified. Procedures for yeast manipulations were conducted as described in Sherman et al. (1986) and transformation of plasmids into yeast was as described in Delorme (1989). Competitive growth assays were performed on the yeast strain NP53/NP59-2.1 and a wild-type sister haploid strain as previously described (Rout et al., 1997).
Table I.
Yeast Strains
| Strains | Genotype | Derivation | ||
|---|---|---|---|---|
| W303 | Mata/Matα ade2-1/ade2-1 ura3-1/ura3-1 his3-11,15/his3-11,15 | see Aitchison et al., 1995 | ||
| trp1-1/trp1-1 leu2-3,112/leu2-3,112 can1-100/can1-100 | ||||
| DF5 | Mata/Matα ura3-52/ura3-52 his3-Δ200/his3-Δ200 trp1-1/trp1-1 | |||
| leu2-3,112/leu2-3,112 lys2-801/lys2-801 | ||||
| NP53Δ | Mata/Matα ura3-52/ura3-52 his3-Δ200/his3-Δ200 trp1-1/trp1-1 | see text | ||
| leu2-3,112/leu2-3,112 lys2-801/lys2-801 nup53::HIS3/+ | ||||
| NP53-B1 | Mata ura3-52 his3-Δ200 trp1-1 leu2-3,112 lys2-801 nup53::HIS3 | Segregant of sporulated NP53Δ | ||
| NP53-A2 | Matα ura3-52 his3-Δ200 trp1-1 leu2-3,112 lys2-801 nup53::HIS3 | Segregant of sporulated NP53Δ | ||
| NP59Δ | Mata/Matα ade2-1/ade2-1 ura3-1/ura3-1 his3-11,15/his3-11,15 | see text | ||
| trp1-1/trp1-1 leu2-3,112/leu2-3,112 can1-100/can1-100 nup59::HIS3/+ | ||||
| NP59-21 | Matα ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 nup59::HIS3 | Segregant of sporulated NP59Δ | ||
| NP59-23 | Mata ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 nup59::HIS3 | Segregant of sporulated NP59Δ | ||
| NP53/NP170-11 | ade2 ura3 his3 trp1 leu2 nup53::HIS3 nup170::HIS3 | Segregant of sporulated NP53/NP170 | ||
| pRS316A-NUP170 (ADE3 URA3) | containing pRS316A-NUP170 | |||
| NP59/NP170-7c | ade2 ura3 his3 trp1 leu2 can1 nup59::HIS3 nup170::HIS3 | Segregant of sporulated NP59/NP170 | ||
| pRS316A-NUP170 (ADE3 URA3) | containing pRS316A-NUP170 | |||
| NP53/NP188-6a | ura3 his3 trp1 leu2 nup53::HIS3 nup188::HIS3 pRS316A-NUP188 | Segregant of sporulated NP53/NP188 | ||
| (ADE3 URA3) | containing pRS316A-NUP188 | |||
| NP59/NP188-7a | ade2 ade3 ura3 his3 trp1 leu2 can1 nup59::HIS3 nup188::HIS3 | Segregant of sporulated NP59/NP188 | ||
| pRS316A-NUP59 (ADE3 URA3) | containing pRS316A-NUP159 | |||
| NP53/NP157-77a | ura3 his3 trp1 leu2 nup53::HIS3 nup157::URA3 | Segregant of sporulated NP53/NP157 | ||
| NP59/NP157-π3 | ade3 ura3 his3 trp1 leu2 nup59::HIS3 nup157::URA3 | Segregant of sporulated NP59/NP157 | ||
| NP59/PM152-AC | ade2 ura3 his3 trp1 leu2 can1 nup59::HIS3 pom152::HIS3 | Segregant of sporulated NP59/PM152 | ||
| pRS316A-NUP59 (ADE3 URA3) | containing pRS316A-NUP59 | |||
| NP53/NP59-2.1 | ade2 ura3 his3 trp1 leu2 nup53:HIS3 nup59::HIS3 | Segregant of sporulated NP53/NP59 | ||
| NP53PA | Mata/Matα ade2-1/ade2-1 ura3-1/ura3-1 his3-11,15/his3-11,15 | Integrative transformation of W303 | ||
| trp1-1/trp1-1 leu2-3,112/leu2-3,112 can1-100/can1-100 | with Protein A-HIS3-URA3 at the | |||
| nup53-pA/+ (URA3 HIS3) | 3′ end of NUP53 | |||
| NP53pPA | Mata ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 | W303 haploid transformed with | ||
| pRS315-NUP53-pA (LEU2) | pRS315-NUP53-pA | |||
| NP59pPA | Matα ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 | NP59-21 transformed with pRS315-NUP59-pA | ||
| nup59::HIS3 pRS315-NUP59-pA (LEU2) | ||||
| cdc15-2 | Mata ade1 ade2 ura1 his7 lys2 tyr1 gal1 cdc15-2 | see Hartwell, 1971 | ||
| cdc15-2B | Matα cdc15-2 | Segregant derived from cross of cdc15-2 with PSE1-A | ||
| cdc15-2-53 | Matα cdc15-2 pRS315-NUP53-pA (LEU2) | cdc15-2B transformed with pRS315-NUP53-pA | ||
| cdc15-2-59 | Matα cdc15-2 pRS315-NUP59-pA (LEU2) | cdc15-2B transformed with pRS315-NUP59-pA | ||
| cdc15/cdc15-2B | Mata/Matα cdc15-2/cdc15-2 | Cross of cdc15-2 and cdc15-2B | ||
| cdc15/cdc15-2B-53 | Mata/Matα cdc15-2/cdc15-2 pRS315-NUP53-pA (LEU2) | cdc15-2B transformed with pRS315-NUP59-pA | ||
| cdc152/cdc15-2B-121 | Mata/Matα cdc15-2/cdc15-2 pPS1069 (TRP1) | cdc15-2B transformed with pPS1069 (KAP121-GFP) | ||
| NP53/KP123 | ura3-52 his3-Δ leu2-3,112 lys2-801 kap123::URA3 nup53::HIS3 | Segregant derived from cross of | ||
| NP53-A2 and 123Δ-14-1 (Rout et al., 1997) | ||||
| NP53/KP123 | ura3-52 his3-Δ leu2-3,112 lys2-801 kap123::URA3 nup53::HIS3 | Segregant derived from cross of NP59-21 and | ||
| 123Δ-14-1 (Rout et al., 1997) |
Plasmids
The plasmids used in this work are as follows: pRS315, CEN/LEU2 (Sikorski and Heiter, 1989); pRS316, CEN/URA3 (Sikorski and Heiter, 1989); pBluescript II SK (pBS) (Stratagene, La Jolla, CA); pSB32, CEN/ LEU2-based yeast genomic DNA library (provided by J. Rine, University of California, Berkeley, CA); pCH1122, CEN/URA3/ADE3 (Kranz and Holm, 1990); pCH1122-POM152 (Aitchison et al., 1995); p4047, an isolate of the pSB32 library containing a 3.8-kb fragment encoding the NUP59 gene; pScBH-NUP59, a 2.2-kb ScaI–BamHI fragment of p4047, containing the NUP59 open reading frame (ORF) and extending from nucleotide −164 to 2,066 (where +1 is the A of the initiation codon) inserted into EcoRV–BamHI cut pRS315; pBS-NUP59, a PCR amplification of the NUP59 ORF extending from nucleotide +1 to 2,930 inserted into a EcoRV site of pBS; pH6-NUP53, a 2.3-kb PCR product of NUP53 ORF extending from nucleotide −521 to 1,756 inserted into PstI–BamHI cut pRS315; pBS-NUP53, a 1.8-kb PCR product of NUP53 ORF extending from nucleotide +1 to 1,756 inserted into an EcoRV site of pBS; pGEX-NUP53, BamHI–BamHI fragment of pBS-NUP53 inserted into a BamHI site at the 3′ end of the glutathione-S-transferase (GST) ORF in pGEX-3X (Pharmacia Biotech Sverige, Uppsala, Sweden); pGEX-NUP59, BamHI–BamHI fragment of pBS-NUP59 inserted into a BamHI site in pGEX-4T1 (Pharmacia Biotech Sverige); pGEX-KAP121, a PCR product containing the KAP121 ORF, amplified from genomic DNA and containing an in frame XhoI site and a 3′ BamHI inserted into the same sites in pGEX-4T1 (Pharmacia Biotech Sverige); pRS316A, a 3.5-kb PCR product of the yeast ADE3 gene, using genomic DNA as a template, and inserted into the SacI site of pRS316; pRS316A-NUP170, (Aitchison et al., 1995); pRS316A-NUP188 (Nehrbass et al., 1996). pRS316A-NUP59, a 2.2-kb ScaI–BamHI fragment of pScBH-NUP59 inserted into the same sites of pRS316A. pNMD5-GFP; the NMD5 ORF was subcloned into the yeast plasmid pGFP-C-FUS (URA3) (Niedenthal et al., 1996) in frame with the GFP ORF to produce an NMD5-GFP fusion gene (Baker, R., and J. Aitchison, unpublished data). Plasmids used for gene disruptions and protein A fusion constructs are described below.
Complementation and Characterization of POM152 Synthetic Lethal Mutants Allelic to NUP59
Mutants dependent on a plasmid-born copy of POM152 were isolated using a colony sectoring assay as described in Aitchison et al. (1995). This screen identified four complementation groups, three of which have been previously characterized (Aitchison et al., 1995; Nehrbass et al., 1996). The remaining group contained four mutants including one designated psl40. These mutants were characterized as previously described. psl40 was transformed with a pSB32-based yeast genomic library and three independent sec+/5-FOA resistant colonies were isolated. Restriction analyses of the complementing plasmids showed that they contained overlapping inserts. The complementing regions within these plasmids were defined by deletion analysis and tested by reintroduction into psl40. The shortest complementing insert of 3.8 kb, plasmid p4047, was sequenced. This insert contained a single ORF (YDL088c) encoding NUP59. The plasmid p4047 complemented all of the mutants in this group. Each of these mutants was mated with NP59/PM152-AC (nup59Δ/pom152Δ, see below). The resulting diploid strains failed to grow on 5-FOA containing media, suggesting that the mutants are allelic to NUP59.
Construction of NUP59-pA and NUP53-pA Chimeric Genes
A NUP59-protein A chimeric gene was assembled by inserting an 800-bp DNA fragment encoding the IgG binding domains of Staphylococcus aureus protein A near the 3′ end of the NUP59 ORF. The DNA fragment encoding protein A was synthesized by PCR using the pRIT2T plasmid (Pharmacia Biotech Sverige) as a template and primers containing flanking SalI sites. The Expand Long Template PCR system (Boehringer Mannheim, Laval, Quebec, Canada) was used for this and all subsequent PCRs discussed in this work. This DNA fragment was inserted in frame into a SalI site of the NUP59 ORF 378 bp upstream of the stop codon in pBS-NUP59. The chimeric construct was then inserted into a pRS315 (LEU2). The resulting plasmid (pRS315-NUP59-pA), while lacking the last 124– amino acid residues of the NUP59 ORF, was functional in all mutant backgrounds tested.
Genomic and plasmid-born copies of the NUP53 gene were tagged with the same IgG binding domains of protein A. The genomic copy of NUP53 was tagged using an integrative transformation procedure previously described in Aitchison et al. (1995). The PCR product used for the transformation was synthesized using primers designed on the basis of the guidelines outlined in this reference. The PCR product was transformed into diploid W303 cells and His+ Ura+ colonies were selected. A strain synthesizing the Nup53-pA fusion was identified by Western blotting of whole cell lysates. In addition, a plasmid-born NUP53-pA chimera was constructed by ligating two PCR products encoding NUP53 (nucleotides −233 to 1,424 plus flanking BamHI sites) and protein A (an 800-bp fragment encoding the IgG binding domains plus an in frame 5′ BamHI site and a 3′ BamHI site) into the plasmid pRS315 (LEU2). The resulting plasmid (pRS315-NUP53-pA) functionally complements the synthetically lethal phenotypes of the nup170Δnup53Δ and nup188Δnup53Δ strains (data not shown).
Construction of nup59 and nup53 Null Mutations and Double Disruption Strains
Deletion of the NUP53 and NUP59 genes was performed by integrative transformation using the procedure of Rothstein (1991). The NUP53 deletion constructs were prepared in pBS-NUP53 by replacing nucleotides −87 to 1,649 with the HIS3 selectable marker gene. Similarly, HIS3 was used to replace nucleotides −95 to 1,684 of the NUP59 gene in the plasmid pBS-Nup59. The resulting disruption constructs were transformed into diploid yeast strains DF5 (for nup53::HIS3) or W303 (for nup59:: HIS3) and integrants were selected on SM-His plates. His+ transformants were analyzed by Southern blotting to identify heterozygous diploids carrying the nup53::HIS3 and nup59::HIS3 alleles. Cells containing a null mutation were sporulated and tetrads were dissected on YPD plates. In both cases, all spores were viable and the HIS3 marker segregated with the expected 2:2 ratio. Southern blotting of the His+ haploid segregants confirmed the absence of the wild-type genes, demonstrating that the nup53Δ and nup59Δ haploids were viable.
Double null mutants of nup53Δ and nup59Δ and each in combination with nup157Δ, nup170Δ, nup188Δ, and pom152Δ were constructed as follows. The nup53Δ (NP53-A2 or NP53-B1) and nup59Δ (NP59-23 or NP59-21) haploid strains were crossed with one another and the null haploids nup170Δ (NP170-11.1; Aitchison et al., 1995), nup188Δ (NP188-2.4; Nehrbass et al., 1996), pom152Δ (PM152-75; Wozniak et al., 1994), and nup157Δ (NP157-2.1; Aitchison et al., 1995) and diploids were selected by complementation of auxotrophic markers. The viability of double null haploid strains resulting from sporulation of the diploids was ensured by maintaining a complementing URA3-containing plasmid in all diploid strains as follows: the nup53Δnup170Δ and nup59Δnup170Δ strains contain pRS316A-NUP170, nup53Δnup188Δ contains pRS316A-NUP188, nup53Δpom152Δ contains pCH1122-POM152 and the nup59Δnup53Δ, nup59Δpom152Δ, and nup59Δnup188Δ strains contain pRS316A-NUP59. The nup59Δnup157Δ and nup53Δnup157Δ strains were dissected without a covering plasmid. The diploid strains were then sporulated and tetrads were dissected. The analysis used to identify the resulting double null haploids and the determination of their viability was performed as previously described in Aitchison et al. (1995). All lethal combinations were dependent on the covering URA3-containing plasmid and thus failed to grow on media containing 5-FOA at 30°C. Tetrads from the nup59Δnup157Δ and nup53Δnup157Δ diploid strains, lacking the covering plasmid, produced four viable segregates. The genotype of each of the double null haploids was confirmed by Southern blotting.
Isolation of the Nup53-pA–containing Complex
Strains expressing NUP53-pA (NP53PA) and UGB-pA (Wozniak, R.W., and J. Aitchison, unpublished data) were grown in 250 ml of YPD at 30°C to an OD600 of 1.0. All subsequent steps were conducted at 4°C unless otherwise specified. Cells were collected by centrifugation, washed with water, resuspended in 15 ml of lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1 mM MgCl2, 0.2 mM PMSF, 2 μg/ml leupeptin, 2 μg/ml aprotinin, 0.4 μg/ml pepstatin A) and lysed using a French press (1,000 ψ) in a prechilled chamber. The lysate was diluted with an equal volume of lysis buffer containing 40% dimethyl sulfoxide and 2% Triton X-100. The lysate was cleared by centrifugation at 11,300 g for 15 min followed by further clarification of the supernatant by centrifugation at 311,000 g for 45 min. The cleared supernatant was incubated in batch with 200 μl of IgG– Sepharose beads (Pharmacia Biotech Sverige) preequilibrated in wash buffer (150 mM NaCl, 0.1 mM MgCl2, 0.1% Tween-20, 50 mM Tris-HCl, pH 7.5, 0.2 mM PMSF, 2 μg/ml leupeptin, 2 μg/ml aprotinin, and 0.4 μg/ml pepstatin A) for 1 h. The bound complex was washed extensively in wash buffer and eluted with 0.5 M acetic acid, pH 3.4, at room temperature or, in the case of the eluate shown in Fig. 4 C (α-Nup59p), with 250 mM MgCl2. The eluates were concentrated by lyophilization and resuspended in SDS-PAGE sample buffer. To identify the 115-kD copurifying polypeptide, eluates from nine immunoisolation experiments were pooled and run on an SDS–polyacrylamide gel and visualized by Coomassie blue staining. The 115 kD species was then excised from the gel. The in-gel polypeptide was digested with endopeptidase Lys-C and peptides were purified and sequenced by the Protein/DNA Technology Center at The Rockefeller University (New York, NY). Sequence was obtained from two peptides that correspond to amino acid residues 84–89 and 90–96 of Kap121p/Pse1p (YMR308c; Chow et al., 1992).
Figure 4.
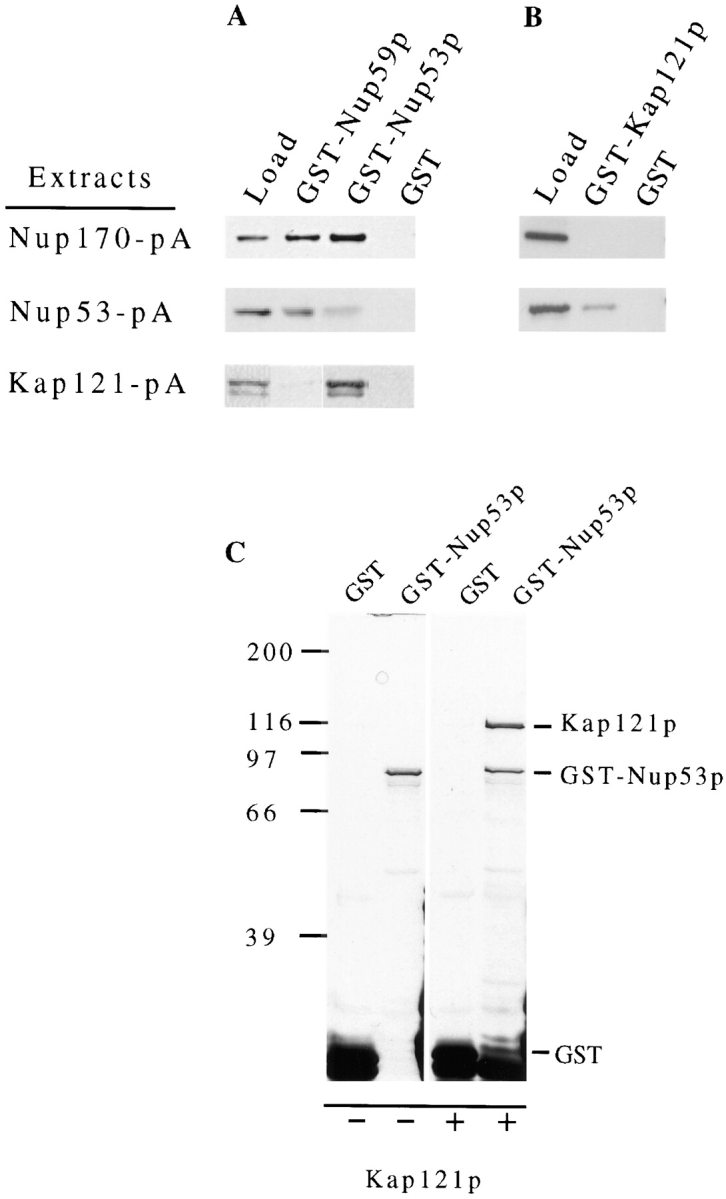
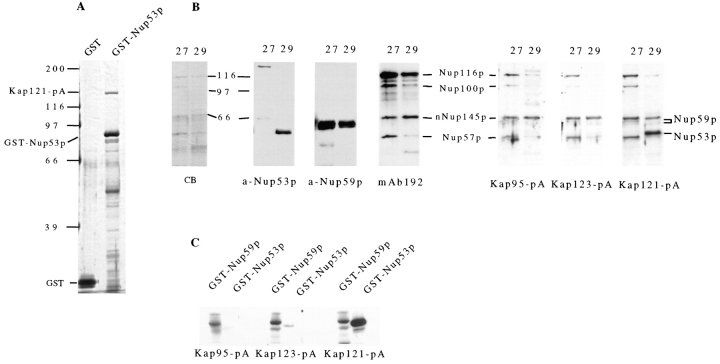
Analysis of the protein–protein interactions between members of the Nup53p-containing NPC subcomplex. GST either alone (GST) or fused to Nup53p (GST-Nup53p), Nup59p (GST-Nup59p), or Kap121p (GST-Kap121p) was synthesized in E. coli and purified on GT–Sepharose beads. These bead were then incubated with extracts derived from yeast NEs containing either Nup53-pA or Nup170-pA or with yeast cytosol containing Kap121-pA. After washing, polypeptides bound to the beads were eluted, separated by SDS-PAGE, and then analyzed by Western blotting. The protein A fusions were detected with HRP-conjugated donkey anti–rabbit IgG and ECL. The relevant regions of the autoradiograms are shown. The lane marked “Load” is derived from an aliquot of the protein A chimeric extract used. The results of two separate experiments are shown examining the binding of Nup53-pA, Nup170-pA, and Kap121-pA to GST-Nup53p, GST-Nup59p, and GST alone (A) or of Nup53-pA and Nup170-pA to GST-Kap121p and GST alone (B). The ability of Nup53p to directly bind Kap121p was further tested using recombinant proteins. In C, GST-Nup53p and GST alone were separately bound to GT–Sepharose beads and then incubated with buffer alone (−) or purified, recombinant Kap121p (+). After washing, bound polypeptides were eluted, resolved by SDS-PAGE and detected by silver staining. The positions of Kap121p, GST-Nup53p, and GST are indicated. Molecular mass markers in kD are indicated on the left.
Isolation of Recombinant Proteins, Production of Antibodies, and Immunoblotting Procedures
The plasmids pGEX-4T1, pGEX-NUP53, pGEX-NUP59, and pGEX-KAP121 were introduced into the Escherichia coli strain BLR (DE3) (Novagen, Inc., Madison, WI). Induction and purification of the fusion proteins were conducted according to the manufacturer's specifications (Pharmacia Biotech Sverige) with the following modifications. Expression of the fusion genes was induced at 37°C with 1 mM IPTG for 2 h. Cells were lysed by sonication in PBS (40 OD600) containing a protease inhibitor cocktail (0.1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 0.5 μg/ml pepstatin A). After sonication, Triton X-100 was added to a final concentration of 1% (vol/vol) and the samples were clarified by centrifugation at 13,000 g for 15 min at 4°C. The supernatants were then allowed to bind glutathione (GT)–Sepharose beads (Pharmacia Biotech Sverige) (10 μl of beads/ml of lysate). Fusion proteins were eluted with 50 mM Tris-HCl, pH 8.0, containing 10 mM GT and the protease inhibitor cocktail. GST– Nup53p was further purified by anion exchange chromatography using a Mono Q fast flow Sepharose column (Pharmacia Biotech Sverige) preequilibrated with 50 mM Tris-HCl, pH 8.0. The fusion protein was recovered in the flowthrough. Eluates of the GT–Sepharose and the flow through of the Mono Q fast-flow Sepharose were then dialyzed against 50 mM Tris-HCl, pH 8.0, and concentrated using an Ultrafree 0.5 ml centrifugal filter (Millipore, Bedford, MA). For binding studies using recombinant Kap121p, Kap121p was released from GST-Kap121p by cleavage with thrombin. GST–Kap121p (10 μg) bound to GT–Sepharose, prepared as described above, was treated with 1.5 NIH U of thrombin (Sigma, St. Louis, MO) in PBS for 3 h at room temperature. The released Kap121p was used directly for binding to GST-Nup53p (see below) which is insensitive to thrombin.
The purified GST-Nup59p, GST-Nup53p, and GST-Kap121p fusion proteins were used to elicit an immune response in rabbits. Polyclonal antibodies directed against Nup59p and Nup170p (Nehrbass et al., 1996) were affinity purified from sera using a previously described procedure (Harlow and Lane, 1988) modified as follows. Purified fractions of GST-Nup53p and GST-Nup170p (Marelli, M., and R.W. Wozniak, unpublished data) were separated by SDS-PAGE and transferred to nitrocellulose. The full-length fusions were detected by amido black staining (Sigma), excised and blocked in PBS containing 5% skim milk and 0.1% Tween-20 (Sigma). Strips were incubated with anti-sera for 1 h at room temperature and then washed extensively with PBS containing 0.1% Tween-20. Bound antibodies were eluted with 0.1 M glycine, pH 2.5, neutralized with 0.1 M Tris-HCl, pH 8.0, and used undiluted for Western blots.
The detection of various proteins by immunoblotting was conducted as follows. All blots were blocked with PBS containing 5% dried milk and 0.1% Tween-20. Protein A–tagged fusions were detected with rabbit IgG (ICN Pharmaceuticals, Aurora, OH), the repeat-containing nucleoporins Nup116p, Nup100p, Nup57p, and the NH2 terminus of Nup145p were detected with mAb192 (see Wente et al., 1992), Nup170p was detected with polyclonal anti-Nup170p antibodies (Nehrbass et al., 1996), and GST and GST fusions were detected with polyclonal anti-GST antibodies (Molecular Probes, Eugene, OR). Polyclonal antisera produced as described above were used to detect Nup53p, Nup59p, and Kap121p. Antibody binding was visualized with HRP-conjugated secondary antibodies and ECL using procedures outlined by the manufacturer (Amersham Corp., Oakville, Ontario, Canada).
In Vitro Binding Assays
In vitro binding assays were performed using the recombinant proteins GST-Nup53p, GST-Nup59p, GST-Kap121p, and GST purified as described above. NEs derived from 10 OD260 units of nuclei expressing protein A–tagged Nup53p and Nup170p were extracted with 150 μl of 20 mM Tris-HCl, pH 7.5, 1 M NaCl, 0.1 mM MgCl2, 1 mM DTT, 1% Triton X-100, 0.2 mM PMSF, and 0.4 μg/ml pepstatin A for 15 min on ice. The extracted NEs were then centrifuged at 100,000 g for 30 min at 4°C. 150 μl of the supernatant was diluted with 850 μl of extraction buffer lacking NaCl and Triton X-100. 200 μl of this diluted extract was incubated with 10 μl of GT–Sepharose beads (Pharmacia Biotech Sverige) previously loaded with 1–3 μg of the indicated recombinant GST fusion protein in siliconized, 0.5 ml tubes and incubated for 1 h at 4°C. Alternatively, the beads were incubated with 400 μl of cytosol, diluted twofold with PBS containing 0.1% Tween-20 and 0.1% casaminoacids (Difco Laboratories, Detroit, MI), prepared as previously described by Rout et al. (1997) from a yeast strain expressing Kap121-pA (PSE1-A). After binding, the beads were washed three times with buffer A (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1 mM MgCl2, 1 mM DTT, 0.1% Tween-20, 0.2 mM PMSF, 0.4 μg/ml pepstatin A) for Nup53-pA and Nup170-pA samples or buffer B (20 mM Hepes, pH 6.8, 150 mM KOAc, 2 mM Mg(OAc)2, 2 mM DTT, 0.1% Tween-20, 0.2 mM PMSF, and 0.4 μg/ml pepstatin A) for Kap121-pA samples. Bound proteins were eluted with SDS sample buffer. For these experiments the load fractions shown in Fig. 4 were derived from 10% of the total extract used and the eluates represent 30% (Fig. 4 A) and 10% (Fig. 4 B) of the total eluate analyzed. Similar approaches were also used in examining the binding of recombinant Kap121p (purified as described above) to GST-Nup53p and GST. In these experiments, the binding step was performed with 1 μg of Kap121p in 0.5 ml of buffer B.
Ran-induced release of Kap121p from Nup53p and the Nup53p-containing complex was examined as follows. Yeast Ran (Gsp1p; provided by M. Floer, The Rockefeller University, New York, NY) was loaded with GTP, GDP, and GTPγS as described (Rexach and Blobel, 1995). The Kap121-pA/GST-Nup53p complex was assembled on GT–Sepharose beads as described above. The Nup53p-containing complex, bound to IgG Sepharose, was isolated as described above and washed with 0.5 mM ATP in binding buffer for 30 min at room temperature before treatment with Ran. The beads (20 μl) were then incubated with 1 mM GTP alone or 0.8 μg of Ran-GTP, Ran-GDP, or Ran-GTPγS in buffer B for 30 min at 23°C. The meniscus, containing released proteins, was collected and prepared for SDS-PAGE by the addition of an equal volume of 2× sample buffer. Beads were then washed and resuspended in an equal volume of sample buffer. Polypeptides in bound and released fractions were resolved by SDS-PAGE and analyzed by silver staining or Western blotting.
Overlay Assays
Polypeptides in an enriched fraction of yeast NPCs were fractionated by SDS-hydroxylapatite chromatography as previously described (Wozniak et al., 1994). Polypeptides in fractions 27 and 29 were separated by SDS-PAGE and transferred to nitrocellulose (for a comparison of these fractions to that of the complete profile, see the corresponding fractions in Fig. 3 of Wozniak et al. [1994]). Alternatively, purified recombinant GST-Nup53p and GST–Nup59p were used. Nitrocellulose membranes were blocked and probed with a cytosolic fraction isolated from yeast cells synthesizing protein A–tagged Kap121p, Kap123p, and Kap95p as previously described by Aitchison et al. (1996). Kap95-pA–containing cytosol was provided by M. Rout (The Rockefeller University, New York, NY). Binding of the protein A fusions was detected with HRP-conjugated donkey anti–rabbit antibodies and ECL (Amersham Corp.).
Figure 3.
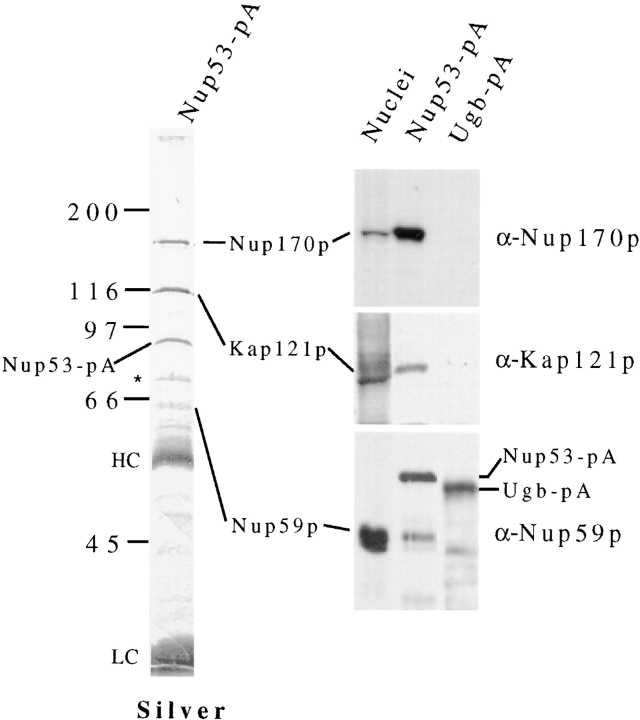
Affinity purification of a Nup53-pA containing NPC subcomplex. Lysates derived from cells synthesizing Nup53-pA (NP53pA) or as a control, a protein A–tagged nucleolar protein, Ugb-pA, were applied to an IgG–Sepharose column. Bound material was eluted and the released polypeptides were analyzed by SDS-PAGE. The polypeptide profile of the Nup53-pA eluate was visualized by silver staining. The positions of the Nup53-pA and Ugb-pA fusions are marked. Contaminating heavy (HC) and light chain (LC) IgG fragments are indicated. Four polypeptides were visible that copurified specifically with Nup53-pA. Western blots were performed on polypeptides present in a yeast nuclear fraction, the Nup53-pA eluate, and the Ugb-pA eluate. These fractions were probed with either anti-Nup170p (α-Nup170p), anti-Nup59p (α-Nup59p), or anti-Kap121p (α-Kap121p) antibodies. Relevant regions of the autoradiograms are shown on the right. Three of four species in the Nup53-pA eluate were identified as Nup170p, Kap121p, and Nup59p. Note, the species comigrating with Nup59p is specifically recognized by the anti-Nup59p antibodies. Additional species observed with the Nup59p antibodies in the Nup53-pA and Ugb-pA eluates are derived from the protein A fusions (data not shown). The fourth protein, marked by a single asterisk, was variably present in this fraction. The molecular mass markers expressed in kilodaltons are shown on the left.
Immunofluorescence Microscopy and Detection of the GFP Chimeras
Immunofluorescence microscopy of strains synthesizing protein A tagged Nup53p and Nup59p was performed as described by Kilmartin and Adams (1984) with modifications in (Wente et al., 1992; Aitchison et al., 1995). The protein A fusions were detected using rabbit IgG followed by Cy3-conjugated donkey anti–rabbit antibodies (Jackson ImmunoResearch, West Grove, PA). The L25 NLS–β-galactosidase reporter was detected in logarithmically growing cells using a mouse mAb directed against β-galactosidase (Boehringer Mannheim, Laval, Quebec, Canada) and rhodamine-conjugated goat anti–mouse antibodies (Jackson ImmunoResearch). mAb414 binding was detected with rhodamine-conjugated goat anti–mouse antibodies.
Yeast strains containing pPS1069 (KAP121-GFP [TRP]; Seedorf and Silver, 1997; provided by M. Seedorf and P. Silver, Dana Farber Cancer Institute, Boston, MA) or pUN100-GFP-NUP49 (GFP-NUP49 [URA3]; Belgareh and Doye, 1997; provided by V. Doye, Institut Curie, Paris, France) were grown in selection media lacking tryptophan to early log phase at the indicated temperatures. Strains containing pNMD5–GFP were grown overnight to mid log phase in selection media lacking uracil and methionine to induce the expression of the NMD5-GFP fusion gene. After washing the cells once with water, the cellular distribution of Kap121-GFP or Nmd5-GFP was directly visualized in the fluorescein channel.
To examine the distribution of Kap121-GFP and Nmd5-GFP at different stages of the cell cycle in wild-type DF5 cells the following procedure was used. A DF5 strain containing pPS1069 (KAP121-GFP TRP) or pNMD5-GFP was grown to mid log phase in synthetic media as described above. Cells were then collected by centrifugation, resuspended in water, and briefly sonicated. Nonbudded, small budded (<70% of the diameter of the mother cell), and large budded (>70% of the diameter of the mother cell) cells were randomly identified by phase microscopy and then visually scored for the presence or absence of a NE-associated Kap121-GFP or Nmd5-GFP signal in the fluorescein channel.
Immunoelectron Microscopy
NEs for immunoelectron microscopy studies were prepared as described by Strambio-de-Castillia (1995) from strains synthesizing Nup53-pA (NP53PA), Nup59-pA (NP59pPA), Nup157-pA (NP157pA; Aitchison et al., 1995), or Nup170-pA (NP170pA; Aitchison et al., 1995). Preembedding labeling of NEs was performed as described by Kraemer et al. (1995) with modifications (Nehrbass et al., 1996). 5 nm (Nup53-pA) or 10 nm (Nup59-pA, Nup157-pA, and Nup170-pA) gold particles conjugated to anti–rabbit IgG (Sigma) was used to detect binding.
Expression of NUP53-pA and NUP59-pA in cdc15-2 Strains
The S. cerevisiae cell cycle mutant cdc15-2 (Hartwell, 1971) was separately transformed with the plasmids pRS315-NUP53-pA and pRS315-NUP59-pA. The resulting strains, cdc15-2-53 and cdc15-2-59, were arrested in M phase as follows. Cells were grown in SM-Leu to ∼0.5 OD600 per ml at the permissive temperature of 23°C, transferred to YPD media for an additional 2–3 h, and then shifted to the non-permissive temperature of 37°C for 3.5 h. At this point, >95% of the cells were arrested in M phase as determined by bud size and the position of the nuclei. The cell cycle block was released by shifting the cultures back to 23°C. 1-ml aliquots were harvested from asynchronous cultures and arrested cultures at various time points after released. Cells were collected by centrifugation, resuspended in SDS-sample buffer and sonicated. For calf intestinal alkaline phosphatase (CIAP; Sigma) treatments, these samples were diluted with water to 0.5% SDS and digested with for 0.15 U/μl of enzyme at 37°C for 45 min. Polypeptides in the extracts then were separated by SDS-PAGE, transferred to nitrocellulose, and the protein A fusions visualized by Western blotting and ECL.
Sequences Alignments
Double alignments of Nup53p (YMR153w) and Nup59p (YDL008c) and multiple alignments of the indicated regions of Nup53p (amino acids residues 164–384), Schizosaccharomyces pombe (120–280), Chlamydomonas elegans (142–229), human (87–280), mouse (87–259), Xenopus laevis MP44 (80–267) (Stukenberg et al., 1997), Arabidopsis thaliana (106–287) were done using CLUSTALW. All the listed sequences are from the GenBank database. Sequences for S. pombe (these sequence data are available from GenBank/EMBL/DDBJ under accession No. Z98975), C. elegans (accession No. Q09601), A. thaliana (accession No. AC001645), X. laevis (MP44; accession No. U95098) were found as single sequences. Three EST sequences from human (accession Nos. H08487, AA160569, and W25130) and mouse (accession Nos. AA266826, AA413806, and w97460) were aligned and a composite sequence was produced using MegAlign software from DNAStar.
Results
The Identification of Nup53p and Nup59p
We have previously used a synthetic lethal screen to identify proteins that interact with the nuclear pore membrane protein Pom152p (Aitchison et al., 1995; Nehrbass et al., 1996). Three of the four complementation groups identified by this screen have been characterized and shown to contain mutants allelic to three nucleoporin genes, NUP170, NUP188, and NIC96. Like Pom152p, the products of each of these genes are abundant constituents of the NPC. A fourth complementation group has been identified that contains four mutants that are allelic to a gene encoding a 528–amino acid residue polypeptide (termed YDL088c). This gene has also been previously identified as a multicopy anti-suppressor of a temperature-sensitive mutant of DNA polymerase δ and was termed ASM4 (Giot et al., 1995). However, the function of ASM4 was undefined.
A comparison of the YDL088c ORF with the remainder of the yeast genome identified a second ORF, YMR153w, of 475 amino acid residues that exhibits a high degree of sequence similarity (30% identity, 48% similarity; Fig. 1 A). Further comparisons of these two sequences to available data bases identified structurally similar ORFs in S. pombe, human, mouse, X. laevis, C. elegans, and A. thaliana suggesting these sequences are highly conserved in all eukaryotes. In each case, the metazoan sequences are most similar to the YMR153w ORF. This is highlighted by a block of amino-acid residues within the central third of the polypeptides (Fig. 1 B).
Figure 1.
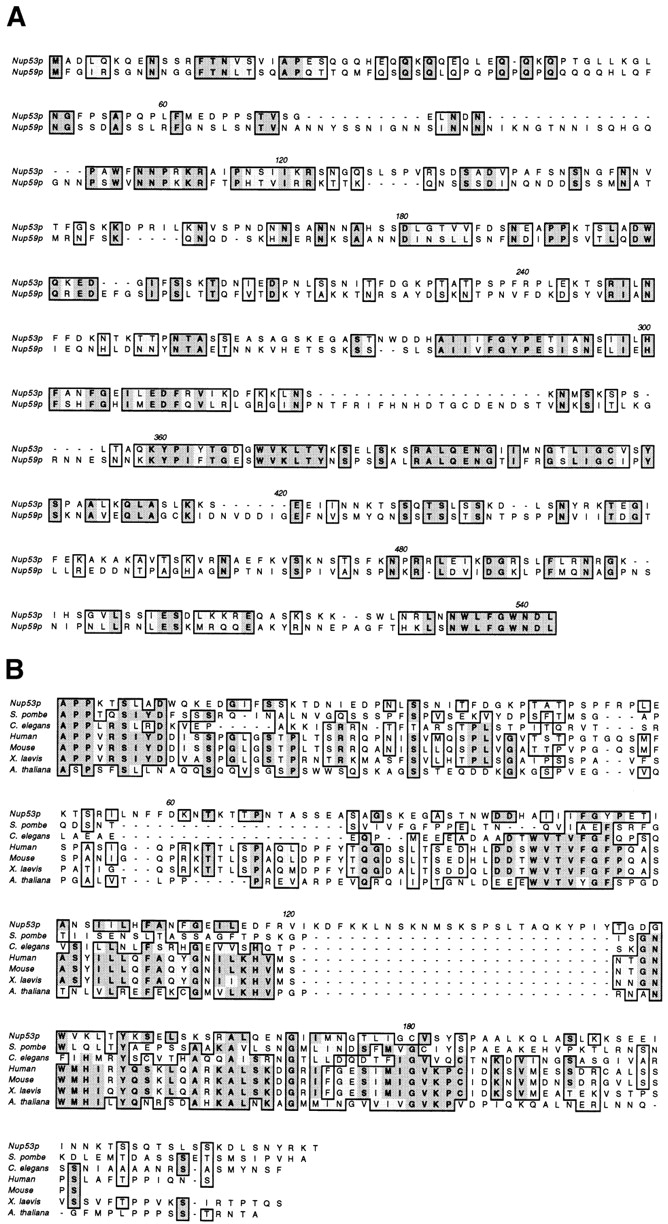
Sequence alignment of the yeast ORFs YMR153w, YDL088c, and multiple potential homologues. Shown in A is the sequence alignment of YMR153w (Nup53p) and YDL088c (Nup59p) produced using CLUSTALW. A database search with Nup53p identified potential homologues in yeast and metazoan species including S. pombe, human, mouse, X. laevis (MP44), C. elegans, and A. thaliana. The central third of these ORFs shows the highest degree of cross species similarity. An alignment of these regions was performed with CLUSTALW and is shown in B. Similar and identical residues are shaded and boxed. Residues that are identical in four or more sequences are shown in bold. Position numbers are indicated above the sequence alignment.
To determine the subcellular localization of the YDL088c and YMR153w gene products, both genes were tagged by the insertion of a DNA fragment encoding the IgG-binding domains of Staphylococcus aureus protein A at (YMR153w) or near (YDL088c) the 3′ end of their ORFs. These chimeric genes were each expressed in yeast and the resulting fusion proteins were localized by indirect immunofluorescence (Fig. 2 A). In both cases, the fluorescent signal was visible along the nuclear surface in a punctate pattern characteristic of that observed for nucleoporins. That these proteins are nucleoporins is further supported by their copurification with isolated NEs (data not shown), their presence in an enriched fraction of yeast NPCs (see Fig. 5), and by immunoelectron microscopy of isolated NEs (see Fig. 9). On the basis of these data, we have named the products of the YMR153w and YDL088c ORFs Nup53p and Nup59p, respectively, in accordance with established nucleoporin nomenclature and representing their deduced molecular masses.
Figure 2.
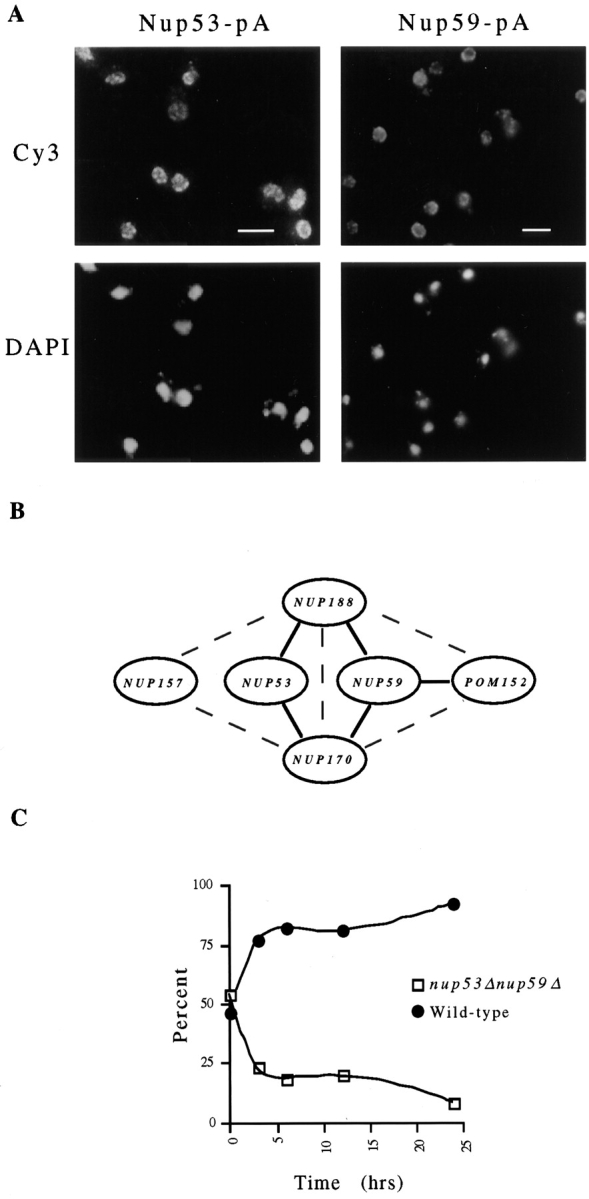
(A) Subcellular distribution of Nup53p and Nup59p. Yeast strains (NP53PA and NP59pPA) synthesizing protein A–tagged Nup53p (Nup53-pA) or Nup59p (Nup59-pA) were fixed, permeabilized and probed with rabbit anti–mouse IgG followed by donkey anti–rabbit IgG conjugated to the Cy3. The position of the nuclear DNA is visualized by DAPI staining. In each case a punctate perinuclear pattern typical of nuclear pore labeling is observed. (B) Analysis of the genetic interactions between NUP53, NUP59, and the genes encoding several abundant nucleoporins. Haploid strains containing double disruptions of nup53Δ and nup59Δ and paired combinations of each with nup170Δ, nup188Δ, nup157Δ, and pom152Δ were constructed and their viability assessed at 30°C. A diagram summarizing the results of these experiments is shown in (B). Solid lines connecting pairs of genes denotes a lethal combination of their null mutations. The gray dashed lines connect pairs of genes that have previously been shown to interact genetically (Aitchison et al., 1995; Nehrbass et al., 1996). The double null combination of nup53Δnup59Δ is viable. Shown in C are the results of a competitive growth assay examining the growth of the nup53Δnup59Δ strain (NP53/NP59-2.1) relative to a wild-type haploid strain. YPD cultures were seeded at 0.1 OD600 per ml with ∼60% NP53/NP59-2.1 and ∼40% wild-type cells (t = 0). At various times thereafter, the percentage of each strain in the culture was determined and is displayed graphically. Bar, 5 μm.
Figure 5.
Nup53p specifically binds Kap121p. (A) Recombinant GST-Nup53p and GST alone were bound to GT–Sepharose beads. Each was then incubated with total cytosol isolated from a strain synthesizing Kap121-pA (PSE1-A). Bound polypeptides were resolved by SDS-PAGE and silver stained. The positions of Kap121-pA, GST-Nup53p, and GST are indicated. Kap121-pA binds specifically to GST-Nup53p. Its identity was confirmed by Western blotting (data not shown). No other protein species with an apparent molecular mass greater than GST-Nup53p were detectable. (B) Polypeptides in an enriched fraction of yeast NPCs were separated by SDS-hydroxylapatite chromatography. Proteins in two elution fractions, 27 and 29, were separated by SDS-PAGE and either stained with Coomassie blue (CB) or transferred to nitrocellulose. Nup53p (a-Nup53p), Nup59p (a-Nup59p), and several repeat-containing nucleoporins including Nup116p, Nup100p, Nup57p, and the NH2 terminus of Nup145p (nNup145p) (mAb192) were detected in these fractions by Western blotting with the indicated antibody. The positions of these nucleoporins are indicated. Overlay assays were performed on the same fractions (B) and recombinant GST-Nup53p and GST-Nup59p (C) with total cytosol derived from yeast strains synthesizing Kap95-pA, Kap123-pA, and Kap121-pA. Binding of these fusions to the nucleoporins was detected with HRP-conjugated donkey anti– rabbit IgG and ECL. Molecular mass markers in kD are indicated.
Figure 9.
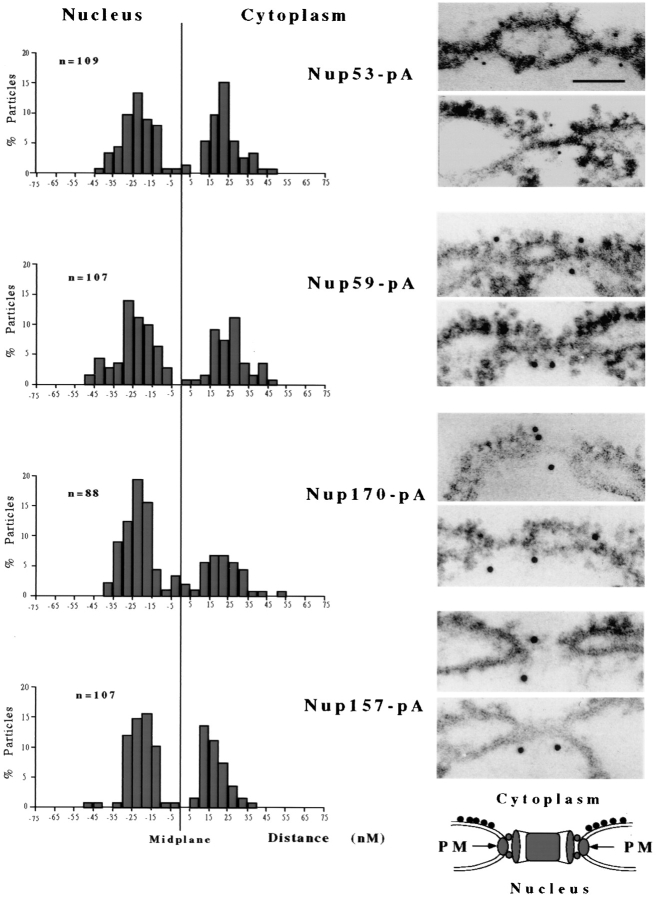
Nup53p, Nup59p, Nup170p, and Nup157p are symmetrically distributed on both faces of the NPC. The localization of protein A–tagged Nup53p, Nup59p, Nup170p, and Nup157p was examined in NEs isolated from four separate yeast strains. The chimeras were visualized by the binding of rabbit IgG and either 5 nm (Nup53-pA) or 10 nm (Nup59-pA, Nup157-pA, and Nup170-pA) gold-labeled secondary antibodies. On the right are shown two representative micrographs of each labeled nucleoporin. In each case, the NEs are orientated as shown in the model below the micrographs (PM, pore membrane). All four of these nucleoporins are accessible on both faces of the NPC in multiple copies per pore. On the left are shown the histogram quantification of the distribution of gold particles (n = total number counted) as measured from the midplane of the associated NPC to the center of the gold particle. Positive distances are assigned to particles on the cytoplasmic side of the NPC and negative distances to those on the nucleoplasmic side. Bar, 0.1 μm.
To examine the phenotypes of nup53Δ and nup59Δ null mutations we replaced the NUP53 and NUP59 ORFs with the HIS3 gene by integrative transformation of the diploid strains W303 and DF5. Sporulation and tetrad analysis of the resulting heterozygous diploid strains (NP53Δ and NP59Δ) yielded four viable haploids exhibiting a 2:2 segregation of the HIS3 marker (data not shown). These results indicate that the null mutations nup53Δ and nup59Δ are viable. Both the nup53Δ (NP53-B1) and the nup59Δ (NP59-21) haploid strains grew at or near wild-type rates at temperatures ranging from 16° to 37°C. This phenotype is similar to that observed in strains lacking NUP157, POM152, and the POM152-interacting genes NUP170 and NUP188 (Wozniak et al., 1994; Aitchison et al., 1995; Kenna et al., 1996; Nehrbass et al., 1996; Zabel et al., 1996). However, double null combinations of nup59Δ with either nup170Δ, nup188Δ, or pom152Δ were synthetically lethal. Similarly, nup53Δ in combination with nup170Δ and nup188Δ also exhibited a lethal phenotype. However, nup53Δnup157Δ, nup59Δnup157Δ, and nup53Δnup59Δ haploids were viable although the latter two exhibited growth defects at temperatures ranging from 23° to 37°C. These results are summarized in Fig. 2 B. The growth defect exhibited by the nup53Δnup59Δ strain (NP53/NP59-2.1) was clearly demonstrated when these cells were grown competitively with a wild-type strain (Fig. 2 C). While both strains were seeded in approximately equal amounts, the percentage of NP53/NP59-2.1 cells rapidly decreased with time (Fig. 2 C).
Isolation of an NPC Subcomplex Containing Nup53p, Nup59p, Nup170p, and the Karyopherin Kap121p
The physical and functional basis for the multiple genetic interactions outlined in Fig. 2 B remains largely undefined. To address this question we used COOH-terminal, protein A–tagged (pA) chimeras of several of these nucleoporins to examine their interactions with one another and other nucleoporins. Here we will focus primarily on experiments conducted with Nup53-pA. On the basis of its localization to the NPC and its ability to complement nup53Δnup170Δ and nup53Δnup188Δ mutant strains (data not shown), we concluded that the Nup53-pA chimera functionally replaced wild-type Nup53p. Cells expressing Nup53-pA (NP53PA) were lysed under non-denaturing conditions. Released Nup53-pA (>95% of the cellular pool) was affinity purified on IgG–Sepharose beads and the bound material analyzed by SDS-PAGE. Five polypeptides with apparent molecular masses of ∼170, 115, 70, and a closely spaced doublet at 60 kD co-eluted with Nup53-pA (Fig. 3). These species were not present in eluates derived from experiments conducted with a strain lacking the Nup53-pA fusion (data not shown) or a strain expressing a protein A–tagged nucleolar protein, Ugb-pA (Fig. 3; Aitchison, J., and R. Wozniak, unpublished data). Both the 170- and 115-kD species were consistently found in a near 1:1 stoichiometry with Nup53-pA both when detected by silver staining (Fig. 3) or Coomassie blue staining (data not shown). The presence of the 70- and 60-kD species, however, was more variable suggesting that they are less tightly associated with the Nup53-pA complex.
The molecular masses of two polypeptides in the Nup53p- containing complex suggested that they may be Nup170p and Nup59p. This was in fact established by Western blot analysis performed with anti-Nup170p and anti-Nup59p antibodies (Fig. 3). The 115-kD species was identified by direct microsequencing of proteolytic fragments as the β-karyopherin transport factor Kap121p/Pse1p (YMR308c; Chow et al., 1992; Rout et al., 1997). This was further confirmed by the binding of anti-Kap121p antibodies to the 115-kD protein (Fig. 3). Kap121p is one of two highly similar yeast proteins, the other being Kap123p, that are structurally related to Kap95p and as many as 11 other β-type karyopherin proteins found in yeast. Because of the structural similarity between Kap121p and Kap123p, we tested for the presence of Kap123p in the Nup53p-containing complex by Western blot analysis. No Kap123p was detected in this fraction (data not shown). In addition, we probed the Nup53p-containing complex with a battery of antibodies that bind to multiple GLFG and FXFG repeat-containing nucleoporins including Nup1p, which has previously been reported to bind Nup170p in vitro (Kenna et al., 1996), Nup116p, Nup100p, Nup57p, Nup49p, and the NH2 terminus of Nup145p. None of these repeat-containing nucleoporins were detected (data not shown). Similar analysis also failed to detect Nup188p, Nic96p, and Nup157p (data not shown). These data further support the conclusion that Nup53p specifically interacts with Nup170p, Nup59p, and Kap121p.
In Vitro Analysis of the Interactions between Components of the Nup53p-containing Complex
We further investigated the interactions between the identified members of the Nup53p-containing complex using in vitro binding analysis. The ORFs encoding three members of this complex, Nup53p, Nup59p, and Kap121p were fused to the 3′ end of the glutathione-S-transferase (GST) ORF and expressed in E. coli. Each of the resulting GST chimeras were purified on glutathione (GT)–Sepharose beads. The immobilized fusions were tested for their ability to bind protein A chimeras of Nup170p and Nup53p extracted from yeast nuclear envelopes (NEs) and cytosolic Kap121p. For these experiments, NEs were isolated from strains synthesizing either Nup53-pA or Nup170-pA. These NEs were extracted with high salt (1 M NaCl) and nonionic detergent to disrupt the NPCs and release the protein A fusions. The salt concentration of the extracts was then adjusted to 150 mM NaCl and the extracts allowed to bind to the immobilized GST fusions. For Kap121-pA, cytosolic fractions derived from cells expressing this fusion were diluted twofold and used directly for binding. As shown in Fig. 4, the Nup170-pA fusion specifically bound to GST-Nup53p and GST-Nup59p (A) but it failed to bind GST-Kap121p (B) or the GST alone control. Nup53-pA, however, did bind to GST-Kap121p, GST-Nup59p, and weakly to the GST form of itself but not to GST alone. The binding of Kap121p to Nup53p was also evident in experiments where Kap121-pA was incubated with GST-Nup53p (Fig. 4 A). However, Kap121-pA did not bind to GST alone and only weakly to GST-Nup59p. Our interpretation of these results is that both Nup53p and Nup59p interact with Nup170p and with each other. We also conclude that Nup53p acts as the major docking site for Kap121p in the Nup53p-containing complex. The direct interaction of Nup53p and Kap121p was further evaluated using recombinant proteins synthesized in E. coli. In these experiments GST-Nup53p and GST alone were immobilized on GT–Sepharose beads and then allowed to bind purified, recombinant Kap121p. Inspection of the bound fraction reveals that Kap121p specifically bound to GST-Nup53p (Fig. 4 C), demonstrating that Kap121p binds directly to Nup53p.
The Interaction of Nup53p with Kap121p Is Specific for This β-Karyopherin
We next examined whether Nup53p's ability to bind Kap121p was specific for this β-karyopherin. To address this question we used two experimental approaches. First, GST-Nup53p was immobilized on beads and incubated with total yeast cytosol isolated from a Kap121-pA expressing strain (PSE1-A) (Fig. 5 A). In addition to Kap121-pA, this cytosolic fraction is likely to contain various known and potential β-karyopherins (Aitchison et al., 1996; Pemberton et al., 1997; Rosenblum et al., 1997; Rout et al., 1997). All of the potential yeast β-karyopherins have a predicted molecular mass of between 95 and 142 kD, similar to wild-type Kap121p (Wozniak et al., 1998). By using cytosol containing the Kap121-pA chimera (∼146 kD), the binding of other β-karyopherins to GST-Nup53p could be visualized by SDS-PAGE as additional bands and distinguished from that of the more slowly migrating Kap121-pA. An analysis of the eluates of the GST-Nup53p and GST alone beads are shown in Fig. 5 A. A protein of the predicted size of Kap121-pA is clearly visible in the GST-Nup53p eluate. Its identity was confirmed by Western blotting (data not shown). No other polypeptides in the molecular mass range of ∼95–142 kD were detected in the GST-Nup53p eluate suggesting that, under these conditions, Kap121p is the only β-karyopherin that binds to Nup53p. Consistent with these observations, Western blots performed with antibodies directed against Kap123p and Kap104p failed to detect these proteins in this fraction (data not shown).
The specificity of the interactions between Nup53p and Kap121p was further evaluated by blot overlay assays similar to those previously used to identify interactions between several other β-type karyopherins and the GLFG and FXFG repeat–containing nucleoporins (Radu et al., 1995a ; Aitchison et al., 1996; Rout et al., 1997; also see Introduction). For these experiments, cytosol from yeast strains synthesizing Kap121-pA, Kap123-pA, and Kap95-pA (Rout et al., 1997) were used to probe yeast nucleoporins present within selected fractions derived from the SDS–hydroxylapatite chromatography of an enriched preparation of yeast NPCs (Wozniak et al., 1994). The fractions probed contain Nup53p, Nup59p, and several of the repeat-containing nucleoporins including Nup116p, Nup100p, Nup57p, and an NH2-terminal 65-kD fragment of Nup145p (Fig. 5 B). Each of these proteins was identified by Western blotting (Fig. 5 B) and, for Nup116p and Nup100p, by direct protein sequencing (Wozniak, R., unpublished data). While each of the karyopherins bound to the repeat-containing nucleoporins Nup116p, Nup100p, Nup57p, and the NH2 terminus of Nup145p, only Kap121-pA bound to Nup53p (Fig. 5 B; Kap121-pA). Strikingly, Kap95-pA and Kap123-pA showed only negligible binding to Nup53p. These results suggest that Kap121p binds specifically to Nup53p as well as to a subset of repeat-containing nucleoporins. The binding Kap95-pA and Kap123-pA to the repeat-containing nucleoporins is consistent with previous observations (Iovine et al., 1995; Aitchison et al., 1996; Iovine and Wente, 1997; Rout et al., 1997). It is interesting to note that Nup59p (visible as a doublet migrating at ∼64 kD; Fig. 5 B) also bound to each of the karyopherins but lacked the specificity for Kap121p exhibited by Nup53p. These results were further confirmed by performing similar overlay experiments on recombinant GST- Nup53p and GST-Nup59p. As shown in Fig. 5 C, Kap121-pA bound strongly to GST-Nup53p while all of the karyopherins tested bound with relatively equal strength to GST- Nup59p.
Ran-GTP Releases Kap121p from Nup53p
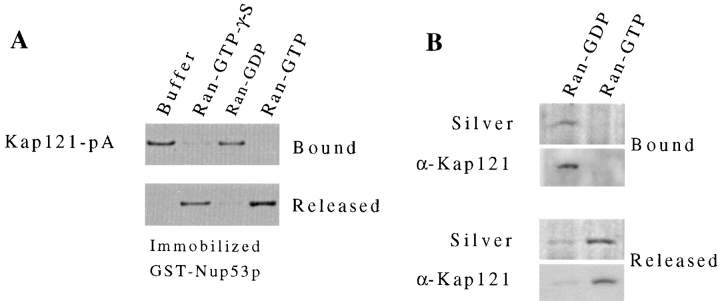
Rexach and Blobel (1995) have previously shown that an in vitro assembled complex of Kap95p/Kap60p (yeast karyopherins β/α) and Nup1p can be disassembled by the GTP-bound form of Ran. We have tested the ability of yeast Ran (Gsp1p) in both the GTP- and GDP-bound forms to release Kap121p from Nup53p. For these experiments, the GST-Nup53p/Kap121-pA complex was assembled on GT–Sepharose beads as described in the previous section. The complex was then treated with Ran preloaded with GDP, GTP, or the nonhydrolyzable GTP analogue GTP-γ-S (Fig. 6 A). Kap121-pA was specifically released from GST-Nup53p by Ran-GTP. GTP hydrolysis did not appear to be required as release was also observed with Ran-GTP-γ-S. No release was observed with GTP alone and only trace amounts were released with Ran-GDP (Fig. 6 A). These same nucleotide-specific Ran effects were observed on a recombinant Nup53p/Kap121p complex (data not shown). In addition, we have conducted similar experiments on the isolated Nup53p-containing complex (Fig. 6 B). When the complex, bound to IgG–Sepharose, was incubated with Ran-GTP, Kap121p was also released. After treatment of the complex with Ran-GDP, however, the majority of Kap121p remained bound. Thus, the mechanism of Kap121p release from Nup53p appears to be similar to that previously observed for the Kap95p/Nup1p complex.
Figure 6.
Ran-GTP induces the release of Kap121p from Nup53p. (A) A Kap121-pA/ GST-Nup53p complex was formed in vitro by incubating yeast cytosol containing Kap121-pA with GST-Nup53p immobilized on GT– Sepharose beads (see Fig. 5 A). This complex was then incubated for 30 min at room temperature with buffer alone or Ran preloaded with GTP, GDP, or GTP-γ-S. Kap121-pA that was released from the column and that which remained bound were separated by SDS-PAGE and detected by Western blotting using donkey anti–rabbit conjugated to HRP and ECL. Shown are the relevant regions of the resulting autoradiogram. (B) The ability of Ran-GTP to release Kap121p from the isolated Nup53-pA complex (see Fig. 3) was examined. Equal amounts of the complex were incubated with Ran-GTP or Ran-GDP. Kap121p remaining bound to the complex and in the released fractions was detected by silver staining (Silver) and Western blot analysis using anti-Kap121p antibodies (α-Kap121). Relevant regions of the gel and autoradiogram are shown.
Steady-State Levels of Kap121p at the NE Are Decreased in nup53Δnup59Δ Strains
As indicated above, nup53Δ and nup59Δ haploid strains are viable, as is the slower growing nup53Δnup59Δ haploid strain. We have introduced a plasmid-born copy of KAP121 tagged at its 3′ end with the coding region of the green fluorescent protein (KAP121-GFP; Seedorf and Silver, 1997) into each of these strains to investigate the effects of removing these nucleoporins on the subcellular distribution of Kap121p. It has been previously shown, by fluorescence microscopy, that in wild-type cells Kap121-GFP is concentrated at the NE and diffusely distributed throughout the cytoplasm and the nucleoplasm (Seedorf and Silver, 1997). We observed a similar pattern in DF5 strains (Fig. 7, Wild-type) and in the nup59Δ strain (Fig. 7). However, inspection of the nup53Δ strain revealed that the concentration of Kap121-GFP at the NE was visibly decreased relative to the wild-type strain. This reduction in signal was further exaggerated in the nup53Δnup59Δ strain where the perinuclear Kap121-GFP signal was greatly reduced or not visible (Fig. 7). In this strain, the Kap121-GFP signal was diffusely distributed throughout the cell. These changes do not appear to be due to massive alterations in the structure of the NPC or NE. No changes were observed in the distribution of GFP-Nup49p or the mAb414-reactive FXFG nucleoporins (Fig. 7) in the nup53Δnup59Δ strain. Introduction of a plasmid-born copy of NUP53 into the nup53Δnup59Δ strain restored the perinuclear Kap121-GFP signal (Fig. 7). Thus we conclude that Nup53p, as well as Nup59p, can function as binding sites for Kap121p in vivo.
Figure 7.
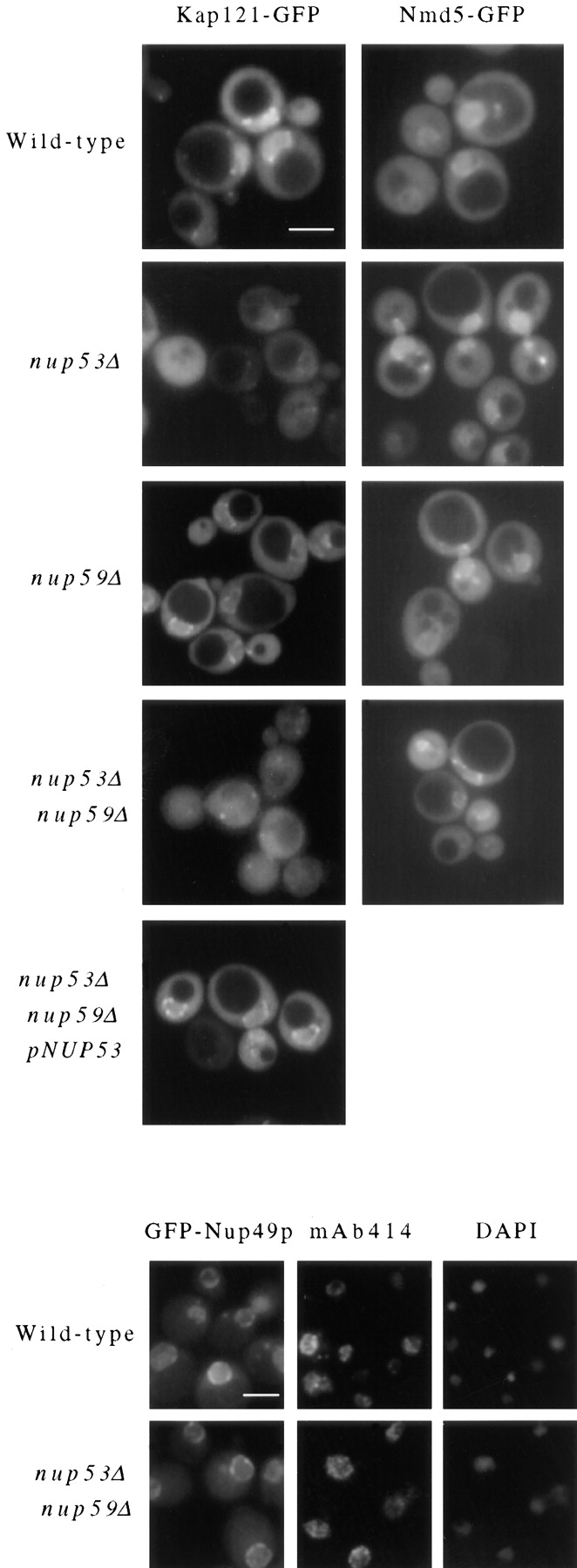
Cellular distribution of Kap121-GFP and Nmd5-GFP in nup53Δ and nup59Δ null mutants. A plasmid-born copy of the Kap121-GFP or Nmd5-GFP chimeric gene was introduced into four haploid yeast strains: wild-type DF5, nup53Δ (NP53-B1), nup59Δ (NP59-23), nup53Δnup59Δ (NP53/NP59-2.1), and, for Kap121-GFP, the nup53Δnup59Δ strain containing a plasmid-born copy of NUP53 (pRS315-NUP53). In each case, cells were grown to mid-log phase and examined directly by fluorescent microscopy. Similarly, the distribution of GFP-Nup49p was examined in wild-type DF5 and nup53Δnup59Δ strains. Immunofluorescence microscopy was also performed on these strains using mAb414 to detect the localization of FXFG-containing nucleoporins. The position of the nuclear DNA is visualized by DAPI staining. Bar, 5 μm.
We have performed similar experiments to examine the effects of these various mutants on the cellular distribution of another member of the β-karyopherin family, Nmd5p (Görlich et al., 1997; Baker, R., and J. Aitchison, unpublished data). As for KAP121, NMD5 was tagged by the addition of GFP to the COOH-terminal end of the its ORF and the chimera was expressed in the indicated strains. The subcellular distribution of Nmd5-GFP in wild-type cells was similar to that observed with Kap121-GFP (Baker, R., and J. Aitchison, unpublished data; Fig. 7). However, unlike Kap121-GFP, we observed no changes in the distribution of Nmd5-GFP in the any of the nup53Δ or nup59Δ strains.
Mutations in NUP53 Inhibit Kap121p-mediated Import
Substrates that are imported into the nucleus by Kap121p in wild-type cells remain to be identified. However, in strains lacking Kap123p, the import of a Kap123p substrate, the ribosomal protein L25, can be rescued by Kap121p (Rout et al., 1997). We have used this as a model system to examine the effects of nup53Δ and nup59Δ null mutations on Kap121p-mediated import. For these experiments, nup53Δkap123Δ (NP53/KP123) and nup59Δkap123Δ (NP59/KP123) haploid strains expressing a reporter gene encoding the L25 NLS linked to β-galactosidase (L25 NLS-β-gal) were isolated. The nuclear import of the L25 NLS-β-gal reporter protein was examined in these strains, as well as in wild-type, nup53Δ, and kap123Δ haploid strains, by indirect immunofluorescence microscopy. In wild-type (Fig. 8) and nup53Δ (data not shown) strains, the L25 NLS-β-gal reporter was localized exclusively to the nucleus. However, consistent with previous observations (Rout et al., 1997; Schlenstedt et al., 1997), in the kap123Δ strain we observed, in addition to nuclear staining, a distinct increase in the amount of the L25 NLS-β-gal reporter protein present in the cytosol. Strikingly, in the nup53Δkap123Δ strain the L25 NLS-β-gal reporter protein was dramatically mislocalized to the cytosol (Fig. 8). This effect was not observed in nup59Δkap123Δ strains that exhibits a staining pattern similar to that seen in the kap123Δ strain. These results further suggest that Nup53p plays a direct role in Kap121p-mediated import.
Figure 8.
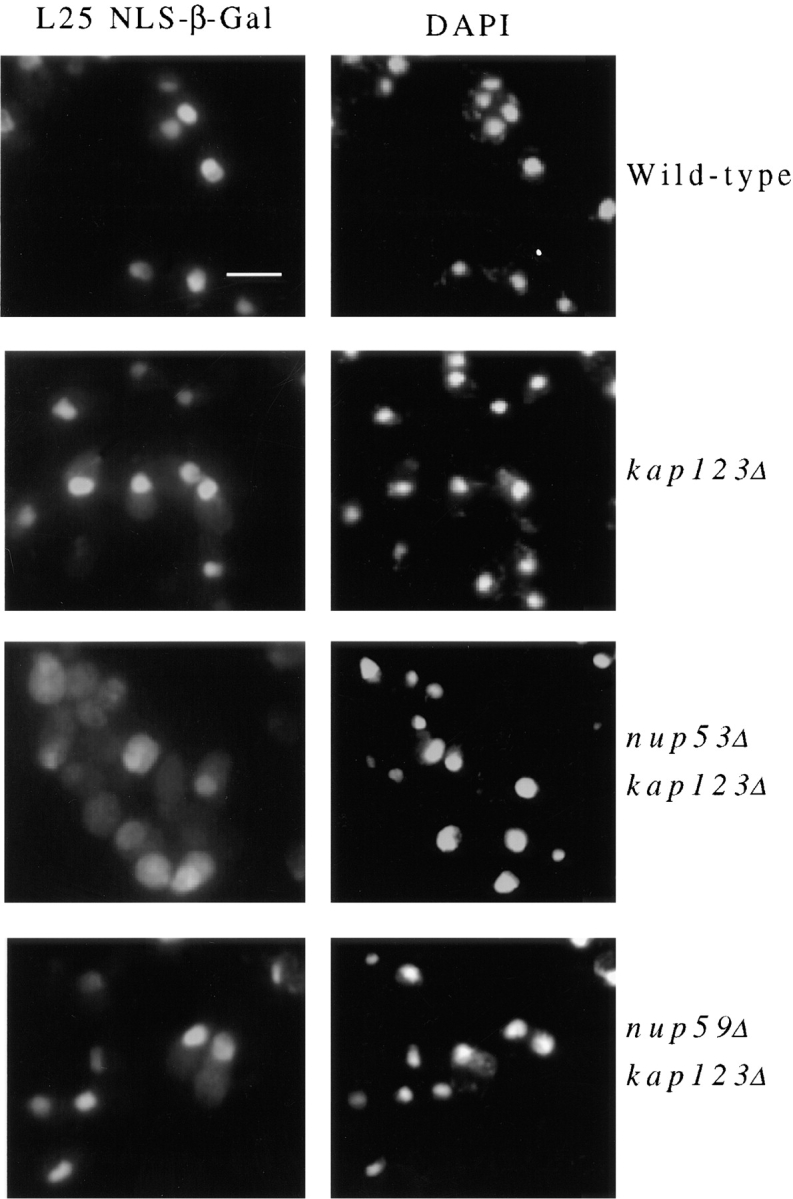
Deletion of the NUP53 gene inhibits the Kap121p-mediated import. A plasmid-born copy of an L25 NLS–β-galactosidase chimeric gene was introduced into four haploid yeast strains: wild-type DF5, kap123Δ (123Δ-14-1), nup53Δkap123Δ (NP53/KP123), and nup59Δkap123Δ (NP59/KP123). Cells were grown to mid-log phase, fixed, permeabilized, and probed with mouse anti–β-galactosidase mAb. Binding was detected with goat anti–mouse IgG conjugated to rhodamine. The position of the nuclear DNA is visualized by DAPI staining. Bar, 5 μm.
Components of the Nup53p-containing Complex Are Present on Both the Cytoplasmic and Nucleoplasmic Faces of the NPC
We have attempted to localize the Nup53p-containing complex by examining the distribution of Nup53p, Nup59p, and Nup170p within the NPC using a preembedding, immunolabeling technique on isolated NEs. This technique allows us to approximate the position of nucleoporins relative to the central plane of the NPC. It has previously been used to establish the symmetrical distribution of Nup188p on both faces of the NPC core (Nehrbass et al., 1996) and to localize Nup159p to the cytoplasmic face of the NPC (Kraemer et al., 1995). NEs isolated from strains synthesizing protein A–tagged chimeras of either Nup53p, Nup59p, Nup170p, or Nup157p were probed with purified rabbit antibodies and secondary antibodies coupled to gold particles. Samples were then embedded, sectioned, and viewed by electron microscopy. In all cases, gold particles were visible on both the cytoplasmic and nucleoplasmic faces of the NPC (Fig. 9). In sections that were cut perpendicular to the plane of the NE and through the NPC, we counted the number of gold particles and measured their distance from the midplane of the NPC. As shown in Fig. 9, for each of these nucleoporins, gold particles were distributed in nearly equal amounts on both the nucleoplasmic and cytoplasmic faces of the NPC. In all cases, the majority of the gold particles lie within 30 nm of the midplane of the NPC, with a peak distribution occurring between 15 and 25 nm. By comparison, in similar experiments performed with Nup188p (Nehrbass et al., 1996) and Nup159p (Kraemer et al., 1995), the greatest concentration of particles was observed at distinctly different distances of between 5–15 nm and 30–50 nm, respectively, from the midplane of the NPC. The similar labeling patterns observed with Nup53p, Nup59p, and Nup170p are consistent with their physical association within an NPC substructure.
Nup53p Is Phosphorylated during Mitosis
As indicated above (Fig. 1), a comparison of yeast Nup53p with protein sequence data bases identified a potential X. laevis homologue termed MP44. MP44 was identified by Stukenberg et al. (1997) on the basis of its ability to act as a substrate for mitosis-specific kinases present in Xenopus egg extracts. However, the location and function of MP44 was not addressed. To determine whether Nup53p and Nup59p are also mitotic phosphoproteins, we have introduced plasmid-born copies of the NUP53-pA and NUP59-pA genes into the yeast cell cycle mutant cdc15-2 (Hartwell et al., 1971; Pringle and Hartwell, 1981). In this temperature-sensitive strain, a shift to the nonpermissive temperature (37°C) arrests cells in late mitosis and maintains the M phase–specific kinase, p34cdc2, in an active state (Surana et al., 1993). Haploid cdc15-2 cells synthesizing either Nup53-pA (cdc15-2-53) or Nup59-pA (cdc15-2-59) were grown at the permissive temperature (23°C), shifted to the nonpermissive temperature (37°C) for 3.5 h, and then allowed to recover at 23°C. The states of Nup53-pA and Nup59-pA at various time points were analyzed by Western blotting (Fig. 10 A). At 23°C Nup53-pA was visible largely as a single band with an apparent molecular mass identical to that observed in asynchronous wild-type cells. However in M phase–arrested cultures, we observed a distinct reduction in the mobility of Nup53-pA (Fig. 10; t = 0). This mobility shift can be largely eliminated by treatment of the arrested extracts with calf intestinal alkaline phosphatase (Fig. 10 B), implying that the mass shift is due to the addition of phosphate. Moreover, the mass shift in Nup53-pA was reversed upon release of the mitotic block induced by shifting the cells to 23°C for 30 min or more (Fig. 10 A). In parallel experiments, however, we did not observe alterations in the mobility of the Nup59-pA doublet (Fig. 10 A).
Figure 10.
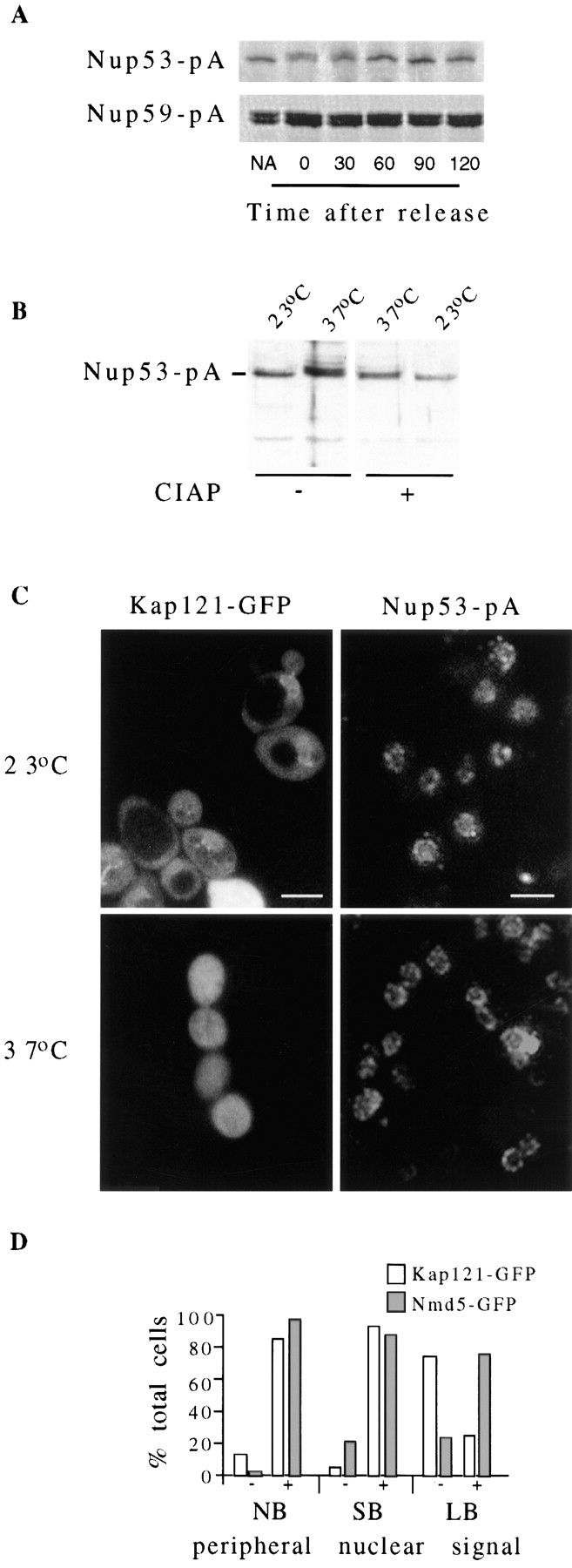
(A and B) Mitosis-specific phosphorylation of Nup53-pA. Cdc15-2 cells expressing NUP53-pA (cdc15-2-53) and NUP59-pA (cdc15-2-59) were grown at the permissive temperature of 23°C to early log phase (non-arrested) before being shifted to 37°C (non-permissive temperature) for 3.5 h. The arrested cultures were then released from M phase arrest by shifting the culture back to 23°C. Whole cell lysates from non-arrested (NA), cells arrested for 3.5 h (t = 0), and cells released from arrest for various times (t = 30–120 min) were isolated. Polypeptides in these samples were separated by SDS-PAGE and then analyzed by Western blotting to detect the Nup53-pA or Nup59-pA fusion proteins. As shown in A, an M phase–specific decrease in the electrophoretic mobility of the Nup53-pA, but not Nup59-pA, was observed in arrested cultures (t = 0). This change in mobility was reversed after the cultures were returned to the permissive temperature. Extracts from cdc15-2-53 cells grown at 23°C or arrested for 3.5 h at 37°C were treated with (+) or without (−) calf intestinal alkaline phosphatase (B, CIAP). The molecular mass of Nup53-pA was then evaluated as in A. In A and B, relevant regions of the gel and autoradiogram are shown. (C and D) Alterations in the localization of Kap121-GFP in arrested cdc15/ cdc15 and wild-type strains. The cellular distribution of Kap121-GFP in the diploid strain cdc15/cdc15-2B was examined by fluorescence microscopy at the permissive temperature of 23°C and 3.5 h after a shift to 37°C to arrest cells in M phase (C). The distribution of Nup53-pA in the cdc15/cdc15-2B-53 strain at the permissive temperature (23°C) and in the arrested cultures (37°C) was examined by immunofluorescence microscopy (right column). In D a histogram displays the results of experiments examining the subcellular distribution of Kap121-GFP in an asynchronous culture of wild-type DF5 cells. Approximately 300 nonbudded (NB), small-budded (<70% of the diameter of the mother cell; SB), and large-budded (>70% of the diameter of the mother cell; LB) cells were identified by phase microscopy and then scored for the presence of a NE-associated Kap121-GFP signal in the fluorescein channel. The relative percentage of cells showing (+) or not showing (−) a distinct perinuclear signal in each group of cells is shown. Bar, 5 μm.
The mitosis-specific phosphorylation of Nup53p raises the intriguing possibility that this modification alters its interactions with other nucleoporins and Kap121p. To begin to address this question, we have examined the subcellular distribution of Nup53-pA and Kap121-GFP in cdc15-2/ cdc15-2 homozygous diploid strains (cdc15/cdc15-2B-53 and cdc15/cdc15-2B-121) at permissive and non permissive temperatures. In both asynchronous and M phase–arrested cultures, Nup53-pA was located at the nuclear periphery in a distinct punctate pattern (Fig. 10 C), suggesting that phosphorylation does not affect its localization to the NPC. The distribution of Kap121-GFP in cdc15/cdc15-2B-121 cells at the permissive temperature also appeared as in wild-type cells, exhibiting both diffuse cytoplasmic and nucleoplasmic staining as well as a concentration along the nuclear periphery (Fig. 10 C, 23°C). However, in cells arrested in M phase after shifting to 37°C for 3.5 h (Fig. 10 C, 37°C), Kap121-GFP was visible throughout the cytoplasm and the nucleoplasm, but was no longer concentrated at the NE. Similar temperature shifts performed on wild-type DF5 cells synthesizing Kap121-GFP had no effect on its subcellular distribution (data not shown). The mitotic-specific redistribution of Kap121-GPF was also examined in an asynchronous culture of DF5 cells. Nonbudded, small-budded, and large-budded cells were randomly identified and then examined by fluorescent microscopy to evaluate the cellular distribution of Kap121-GFP. We observed that in most of the nonbudded (86%) and small-budded (82%) cells Kap121-GFP was visibly concentrated at the NE (Fig. 10 D). In contrast, only 12% of large-budded cells, the majority of which are in the late M phase (Byers, 1981), exhibited a NE signal. In most of these cells (88%), Kap121-GFP was not concentrated at the NE and is visible as a diffuse signal throughout the cytoplasm and nucleus, similar to the pattern observed in arrested cdc15-2 strains. By comparison, Nmd5-GFP was concentrated at the NE in the majority of cells regardless of their stage in the cell cycle (Fig. 10 D). These observations suggest that the localization of Kap121-GFP to the NE, but not Nup53-pA or Nmd5-GFP, is modulated during mitosis.
Discussion
Recent biochemical and structural analysis suggest that the yeast NPC is likely composed of ∼40 proteins (Rout and Blobel, 1993). Of these a subset of the major constituents has been proposed to form the repetitive structures of the NPC core (Aitchison et al., 1996). These proteins likely provide binding sites for the attachment of the cytoplasmic and nucleoplasmic filaments and a framework on which other nucleoporins, including the GLFG and FXFG repeat–containing nucleoporins, are spatially organized and presented to the soluble transport machinery. Such an ordered array of nucleoporins likely plays a fundamental role in vectorial transport through the NPC.
We have identified a subcomplex of the NPC that establishes a link between a putative core component of the NPC and the soluble transport machinery. This subcomplex contains the abundant nucleoporin, Nup170p, two structurally related nucleoporins, Nup53p and Nup59p, and the β-karyopherin Kap121p. On the basis of the near quantitative recovery of the Nup53-pA chimera used to isolate this complex, we conclude that it is largely derived from disassembled NPCs. In vitro binding studies suggest that Nup170p and Kap121p do not directly interact but instead are linked through Nup53p. Nup59p is also present in this complex, however in lesser and variable amounts suggesting it is more loosely bound. The conclusion is further supported by in vitro binding studies (Fig. 4) and two-hybrid analysis (Tcheperegine, S., and R. Wozniak, unpublished data) that show that Nup59p interacts with Nup53p and Nup170p. We have also investigated whether Nup1p is associated with the Nup53p-containing complex. In previous work conducted by Kenna et al. (1996) it was suggested that Nup1p may also interact with Nup170p. However, we failed to detect Nup1p in this isolated complex (data not shown).
The association of Kap121p with this complex of nucleoporins is most likely mediated by its binding to Nup53p. We have drawn this conclusion on the basis of their ability to specifically interact with one another under various experimental conditions. Strikingly, we observe that the interaction between Nup53p and Kap121p is specific for this β-karyopherin. This is underscored by experiments demonstrating that Nup53p only binds Kap121p when presented with a pool of karyopherins present in total yeast cytosol, including a highly similar and 10-fold more abundant karyopherin, Kap123p (Rout et al., 1997; Schlenstedt et al., 1997). Moreover, overlay assays used to examine the ability of different β-karyopherins to bind Nup53p also reflects this specificity (Fig. 5). In addition to these interactions, two other observations support the idea that Nup53p plays an important physiological role in the Kap121p-mediated transport pathway. First nup53Δ mutations, either alone or, more strikingly, in combination with nup59Δ mutations, alter the nuclear envelope association of Kap121p. Second, consistent with this phenomenon, the Kap121p-mediated import of an L25 NLS-β-galactosidase reporter is inhibited in the absence of Nup53p (Fig. 8).
It is likely that Nup59p also serves as a binding site for Kap121p as well as other β-karyopherins (Fig. 5). We detect weaker but specific interactions between Nup59p and Kap121p, Kap123p, and Kap95p using overlay assays. These results suggest that both Nup53p and Nup59p function in binding Kap121p at the NPC, although their apparent differences in affinity and specificity for Kap121p suggest they perform separate functions. The elimination of these sites may significantly alter Kap121p-mediated transport, a process that is presumed to be vital as deletion mutants of KAP121 are lethal (Rout et al., 1997; Seedorf and Silver, 1997). This would explain the growth defects and the competitive disadvantage experienced by the nup53Δ-nup59Δ double null strain relative to wild-type strains (Fig. 2 C).
The nup53Δnup59Δ mutant, however, is viable suggesting that Kap121p can either bypass these binding sites or use alternative pathways through the NPC. Either scenario would suggest that Kap121p can interact with other nucleoporins. Candidate proteins include the GLFG and FXFG repeat–containing nucleoporins. As shown in Fig. 5, Kap121p can bind to at least a subset of these proteins including Nup116p, Nup100p, Nup57p, and the NH2-terminal region of Nup145p. These results are not surprising. Various approaches, in both yeast and vertebrates, have established that a number of β-karyopherins, as well as other potential transport factors, bind to this group of nucleoporins (see Introduction). In yeast, this has been shown using a number of experimental approaches including blot overlay, in vitro binding, and two-hybrid assays (Rexach and Blobel, 1995; Iovine et al., 1995; Aitchison et al., 1996; Rout et al., 1997; Pemberton et al., 1997; Rosenblum et al., 1997; Iovine and Wente, 1997). It is intriguing to note, however, that individual karyopherins can bind separate but overlapping subsets of the repeat-containing nucleoporins. For example, using overlay assays, both Kap104p and Kap95p bind to Nup116p, but of the two only Kap95p appears to bind Nup1p (Aitchison et al., 1996). Moreover, the relative strength of the interactions between individual karyopherins and nucleoporins also varies (Aitchison et al., 1996; Rout et al., 1997; Pemberton et al., 1997; Rosenblum et al., 1997). This selectivity has also been observed in vertebrates using overlay assays (Bonifaci et al., 1997; Yaseen and Blobel, 1997) and by coimmunoprecipitation (Fornerod et al., 1997a ; Shah et al., 1998). Using the latter approach, stable associations have been identified between human Nup214p and Crm1p (Fornerod et al., 1997a ) and Xenopus Nup153p and karyopherin β implicating these as high affinity binding sites (Shah et al., 1998). These results, like those we have obtained with Nup53p and Kap121p, support the hypothesis that individual or groups of karyopherins may follow separate but overlapping pathways through the NPC. These pathways are likely to contain binding sites that are preferred by individual karyopherins and thus they could create distinct intermediate transport steps. Functionally this could provide a mechanism for improving the efficiency of transport and a means of regulating specific transport pathways at the level of the NPC (see below).
Upon close inspection of the amino acid sequence of Nup53p and Nup59p we identified sequence elements that suggest that these proteins may be distantly related members of the GLFG and FXFG repeat–containing family of nucleoporins. Both Nup53p and Nup59p contain several dispersed FG repeat sequences (four in Nup53p and six in Nup59p). Moreover, one of these lies within a stretch of amino acid residues that is highly conserved in all the potential Nup53p homologues (Fig. 1 B) suggesting these regions play an important functional role. However, the similarity to the repeat-containing family is largely restricted to these FG residues. Unlike all the other members of the repeat family, neither Nup53p nor Nup59p contains any repetitive GLFG or FXFG consensus sequences. Moreover, they are not recognized by monoclonal (mAb414, mAb192) or polyclonal (anti-GLFG) antibodies that recognize multiple members of this family (data not shown). Searches of sequence databases also failed to detect any similarities between Nup53p or Nup59p and the GLFG and FXFG repeat–containing proteins. However, the apparent functional similarity between these proteins, i.e., their ability to bind karyopherins, does suggest that they contain similar structural elements. The divergence of the Nup53p sequence from this consensus may in fact be responsible for its specific interaction with Kap121p.
Like several other members of the β-karyopherin family, Kap121p has previously been shown to bind Ran-GTP (Görlich et al., 1997). We have shown that Kap121p bound to Nup53p in various contexts, both within the isolated NPC subcomplex and using recombinant proteins, can be released by Ran-GTP, but not by Ran-GDP. These results are similar to those previously reported for the release of Kap95p from Nup1p (Rexach and Blobel, 1995). On the basis of these data, Rexach and Blobel proposed that Ran-GTP plays a direct role in the release of the Kap95p/ Kap60p/cNLS cargo from multiple nucleoporin binding sites as it moves through the NPC. Since Ran-GTP also dissociates the Kap95p/Kap60p/cNLS cargo complex (Rexach and Blobel, 1995), it has been suggested that this reaction is restricted to the terminal stages of transport on the nucleoplasmic face of the NPC where it would presumably be exposed to nuclear Ran-GTP (Görlich et al., 1996). Our immunolocalization studies suggest that the Nup53p-containing complex is accessible on both the cytoplasmic and nucleoplasmic faces of the NPC. The Ran-GTP–dependent release of Kap121p from these sites would thus require local concentrations of Ran-GTP on both faces of the NPC. Moreover, the symmetrical localization of Nup170p that has also been implicated as a binding site for Nup1p (Kenna et al. 1996), suggests that Nup1p may also be located on both faces of the NPC. These data are consistent with a model in which Ran-GTP dissociates karyopherins from nucleoporins at multiple locations within the NPC.
Somewhat surprisingly we observed that Nup53p is phosphorylated during mitosis. Moreover, yeast Nup53p is specifically phosphorylated during M phase in cycling Xenopus egg extracts (Shibuya, E., A. Chau, M. Marelli, and R. Wozniak, unpublished data). Several potential sites for the mitotic kinase p34cdc2 exist in Nup53p. Which of these sites are phosphorylated in Nup53p remains to be determined. Nup59p, whose mobility is not altered during mitosis in yeast, also contains several potential phosphorylation sites. Whether these sites are phosphorylated is currently under investigation.
Nup53p is the first yeast nucleoporin shown to be phosphorylated during mitosis. In metazoan cells several NPC proteins, including the pore membrane protein Gp210 and the repeat-containing nucleoporins Nup153p, Nup214p, Nup358p (Favreau et al., 1996), and a 97-kD protein (possibly Nup98; Macaulay et al., 1995) are phosphorylated (or hyperphosphorylated) during mitosis. In these cells it has been largely assumed that the mitotic-specific phosphorylation of NPC proteins plays a role in disassembling this structure during nuclear envelope breakdown. However, yeast do not disassemble their nuclear envelopes or NPCs during mitosis (Byers, 1981; Copeland and Snyder, 1993). Consistent with this observation, the subcellular distribution of Nup53p is not changed in M phase–arrested cdc15-2 strains (Fig. 10). An alternative function for this modification may be to alter Nup53p's association with specific nucleoporins or Kap121p. Intriguingly, the nuclear envelope concentration of Kap121-GFP is reduced in late M phase in both arrested cdc15/cdc15-2B-121 and wild-type cells. One plausible explanation for these data is that steady-state levels of NPC-bound Kap121p are altered either by the inhibition of binding to, or by a more rapid release from, Nup53p. A consequence of this could be the modulation of import (or export) of Kap121p cargo. The functional significance of this will await the identification of Kap121p's repertoire of substrates. One can envision that such an M phase–specific change in nuclear transport could also occur in higher eukaryotes as well, perhaps as a prelude to nuclear envelope breakdown or after its reassembly.
Acknowledgments
We thank the Protein/DNA Technology Center at the Rockefeller University (New York) for peptide sequencing, especially Joseph Fernandez; Honey Chan for assistance in performing the electron microscopy studies; Mike Rout for helpful discussions; Jennifer Smith for critical reading of the manuscript; Serguei Tcheperegine, Rosanna Baker, and Ramsey Saleem for technical assistance. We also thank Mike Rout, Monique Floer, Günter Blobel, Matthias Seedorf, Pamela Silver, and Valérie Doye for providing reagents listed in the text.
Abbreviations used in this paper
- GFP
green fluorescent protein
- GST
glutathione-S-transferase
- GT
glutathione
- NE
nuclear envelope
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- ORF
open reading frame
Footnotes
This work is dedicated to the memory of Natalie Ann.
R.W. Wozniak and J.D. Aitchison are Alberta Heritage Foundation for Medical Research Scholars. Support for this work is provided by an operating grant from the Medical Research Council of Canada.
References
- Adam SA, Gerace L. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- Adam EJ, Adam SA. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW. Two novel related yeast nucleoporins Nup170p and Nup157p: Complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Akey CW, Goldfarb DS. Protein import through the nuclear pore complex is a multistep process. J Cell Biol. 1989;109:971–982. doi: 10.1083/jcb.109.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey CW, Radermacher M. Architecture of the Xenopusnuclear pore complex revealed by three dimensional cryo-electron microscopy. J Cell Biol. 1993;122:1–19. doi: 10.1083/jcb.122.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts GJ, Fornerod M, Mattaj IW. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A., Smith, and K. Struhl. 1992. Short Protocols in Molecular Biology. Greene Publishing Associates, New York.
- Belgareh N, Doye V. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J Cell Biol. 1997;136:747–759. doi: 10.1083/jcb.136.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G. Gene gating: a hypothesis. Proc Natl Acad Sci USA. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin β2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, B. 1981. Cytology of the yeast cell cycle. In The Molecular Biology of the Yeast Saccharomyces cerevisiae: Life Cycle and Inheritance. Cold Spring Harbor Laboratories, Cold Spring Harbor, New York. 59–96.
- Chi NC, Adam E, Adam SA. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow TY-K, Ash JJ, Dignard D, Thomas DY. Screening and identification of a gene, PSE-1, that affects protein secretion in Saccharomyces cerevisiae. . J Cell Sci. 1992;101:709–719. doi: 10.1242/jcs.101.3.709. [DOI] [PubMed] [Google Scholar]
- Cole CN, Hammell CM. Nucleocytoplasmic transport: driving and directing transport. Curr Biol. 1998;8:368–372. doi: 10.1016/s0960-9822(98)70239-8. [DOI] [PubMed] [Google Scholar]
- Copeland CS, Snyder M. Nuclear pore complex antigens delineate nuclear envelope dynamics in vegetative and conjugating Saccharomyces cerevisiae. . Yeast. 1993;9:235–249. doi: 10.1002/yea.320090304. [DOI] [PubMed] [Google Scholar]
- Davis LI. The nuclear pore complex. Annu Rev Biochem. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- Delorme E. Transformation of Saccharomyces cerevisiaeby electroporation. Appl Environ Microbiol. 1989;55:2242–2246. doi: 10.1128/aem.55.9.2242-2246.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences—a consensus? . Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Doye V, Hurt E. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- Fabre E, Hurt EC. Yeast genetics to dissect the nuclear pore complex and nucleocytoplasmic trafficking. Annu Rev Genet. 1997;31:277–313. doi: 10.1146/annurev.genet.31.1.277. [DOI] [PubMed] [Google Scholar]
- Favreau C, Worman HJ, Wozniak RW, Frappier T, Courvalin J-C. Cell cycle-dependent phosphorylation of nucleoporins and nuclear pore membrane protein Gp210. Biochemistry. 1996;35:8035–8044. doi: 10.1021/bi9600660. [DOI] [PubMed] [Google Scholar]
- Feldherr CM, Kallenbach E, Schultz N. Movement of a karyophilic protein through the nuclear pores of oocytes. J Cell Biol. 1984;99:2216–2222. doi: 10.1083/jcb.99.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M, Blobel G, Rexach M. Disassembly of RanGTP-karyopherin β complex, an intermediate in nuclear protein import. J Biol Chem. 1997;272:19538–19546. doi: 10.1074/jbc.272.31.19538. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997a;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO (Eur Mol Biol Organ) J. 1997b;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridell RA, Truant R, Thorne L, Benson RE, Cullen BR. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-beta. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- Giot L, Simon M, Dubois C, Faye G. Suppressors of thermosensitive mutations in the DNA polymerase delta gene of Saccharomyces cerevisiae. . Mol Gen Genet. 1995;246:212–222. doi: 10.1007/BF00294684. [DOI] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Laskey RA, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Görlich D, Kostka S, Kraft R, Dingwall C, Laskey RA, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995a;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- Görlich D, Vogel F, Mills AD, Hartmann E, Laskey RA. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995b;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- Görlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO (Eur Mol Biol Organ) J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Doye V, Hurt EC. Purification of NSP1 reveals complex formation with “GLFG” nucleoporins and a novel nuclear pore protein NIC96. EMBO (Eur Mol Biol Organ) J. 1993;12:3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Schlaich N, Tekotte H, Hurt EC. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. EMBO (Eur Mol Biol Organ) J. 1995;14:76–87. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, F., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Hartwell LH. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- Hinshaw JE, Carragher BO, Milligan RA. Architecture and design of the nuclear pore complex. Cell. 1992;69:1133–1141. doi: 10.1016/0092-8674(92)90635-p. [DOI] [PubMed] [Google Scholar]
- Hu T, Guan T, Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine MK, Watkins JL, Wente SR. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J Cell Biol. 1995;131:1699–1713. doi: 10.1083/jcb.131.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine MK, Wente SR. A nuclear export signal in Kap95p is required for both recycling the import factor and interaction with the nucleoporin GLFG repeat regions of Nup116p and Nup100p. J Cell Biol. 1997;137:797–811. doi: 10.1083/jcb.137.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel S, Gorlich D. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO (Eur Mol Biol Organ) J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehlenbach RH, Dickmanns A, Gerace L. Nucleocytoplasmic shuttling factors including Ran and CRM1 mediate nuclear export of NFAT in vitro. J Cell Biol. 1998;141:863–874. doi: 10.1083/jcb.141.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna MA, Petranka JG, Reilly JL, Davis LI. Yeast N1e3p/ Nup170p is required for normal stoichiometry of FG nucleoporins within the nuclear pore complex. Mol Cell Biol. 1996;16:2025–2036. doi: 10.1128/mcb.16.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Adams AE. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. . J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer DM, Strambio-de-Castillia C, Blobel G, Rout MP. The essential yeast nucleoporin Nup159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J Biol Chem. 1995;270:19017–19021. doi: 10.1074/jbc.270.32.19017. [DOI] [PubMed] [Google Scholar]
- Kranz JE, Holm C. Cloning by function: an alternative approach for identifying yeast homologues of genes from other organisms. Proc Natl Acad Sci USA. 1990;87:6629–6633. doi: 10.1073/pnas.87.17.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Khochbin S, Nishi K, Kitano K, Yanagida M, Yoshida M, Horinouchi S. Molecular cloning and cell cycle-dependent expression of mammalian CRM1, a protein involved in nuclear export of proteins. J Biol Chem. 1997;272:29742–29751. doi: 10.1074/jbc.272.47.29742. [DOI] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bschoff RF, Scharzmaier P, Hartmann E, Görlich D. Identification of a tRNA-specific nuclear export factor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- Macaulay C, Meier E, Forbes DJ. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J Biol Chem. 1995;270:254–262. doi: 10.1074/jbc.270.1.254. [DOI] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Maul, G.G. 1977. The nuclear and the cytoplasmic pore complex: structure, dynamics, distribution, and evolution. Int. Rev. Cytol. 5(Suppl.):75–186. [PubMed]
- Melchior F, Gerace L. Two-way trafficking with Ran. Trends Cell Biol. 1998;8:175–179. doi: 10.1016/s0962-8924(98)01252-5. [DOI] [PubMed] [Google Scholar]
- Moore MS. Ran and nuclear transport. J Biol Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin α1β and α2β heterodimers: α1 or α2subunit binds nuclear localization signal and β subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J, Blobel G, Radu A. RanGTP-mediated nuclear export of karyopherin α involves its interaction with the nucleoporin Nup153. Proc Natl Acad Sci USA. 1997;94:9699–9704. doi: 10.1073/pnas.94.18.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U, Rout MP, Maguire S, Blobel G, Wozniak RW. The yeast nucleoporin Nup188p interacts genetically and physically with the core structures of the nuclear pore complex. J Cell Biol. 1996;133:1153–1162. doi: 10.1083/jcb.133.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U, Blobel G. Role of the nuclear transport factor p10 in nuclear import. Science. 1996;272:120–122. doi: 10.1126/science.272.5258.120. [DOI] [PubMed] [Google Scholar]
- Niedenthal RK, Riles L, Johnston M, Hiegemann JH. Green fluorescent protein as a marker for the gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Ohno M, Fornerod M, Mattaj IW. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Pante N, Aebi U. Sequential binding of import ligands to distinct nucleopore regions during their nuclear import. Science. 1996;273:1729–1732. doi: 10.1126/science.273.5282.1729. [DOI] [PubMed] [Google Scholar]
- Pemberton LF, Rosenblum JS, Blobel G. Messenger RNA-binding proteins are imported into the nucleus by distinct and parallel pathways. J Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P, Clarkson WD, Kent HM, Rhodes D, Stewart M. Molecular interactions between the importin α/β heterodimer and proteins involved in vertebrate nuclear protein import. J Mol Biol. 1997;266:722–732. doi: 10.1006/jmbi.1996.0801. [DOI] [PubMed] [Google Scholar]
- Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Pringle, J.R., and L.H. Hartwell. 1981. The Saccharomyces cerevisiae cell cycle. In The Molecular Biology of the Yeast Saccharomyces cerevisiae: Life Cycle and Inheritance. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York. 97–142.
- Radu A, Blobel G, Moore MS. Identification of a protein complex that is required for nuclear import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995a;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995b;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Mills AD, Dilworth SM, Laskey RA, Dingwall C. Nuclear protein migration involves two steps: rapid binding at the nuclear envelope followed by slower translocation through nuclear pores. Cell. 1988;52:655–664. doi: 10.1016/0092-8674(88)90403-5. [DOI] [PubMed] [Google Scholar]
- Rosenblum JS, Pemberton LF, Blobel G. A nuclear import pathway for a protein involved in tRNA maturation. J Cell Biol. 1997;139:1655–1661. doi: 10.1083/jcb.139.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Rout MP, Blobel G. Isolation of the yeast nuclear pore complex. J Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Blobel G, Aitchison JD. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- Schlenstedt G, Smirnova E, Deane R, Solsbacher J, Kutay U, Görlich D, Ponstingl H, Bishoff FR. Yrb4p, a yeast Ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO (Eur Mol Biol Organ) J. 1997;16:6237–6249. doi: 10.1093/emboj/16.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R, Kadowaki T, Tartakoff AM. mRNA transport in yeast: time to reinvestigate the functions of the nucleolus. Mol Biol Cell. 1995;6:357–370. doi: 10.1091/mbc.6.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf M, Silver PA. Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc Natl Acad Sci USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B, Simos G, Bischoff FR, Podtelejnikov A, Mann M, Hurt E. Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO (Eur Mol Biol Organ) J. 1998;17:2196–2207. doi: 10.1093/emboj/17.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Tugendreich S, Forbes D. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J Cell Biol. 1998;141:31–49. doi: 10.1083/jcb.141.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., G.R. Fink, and J.B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor. 186 pp.
- Sikorski RS, Heiter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Strambio-de-Castillia C, Blobel G, Rout MP. Isolation and characterization of nuclear envelopes from the yeast Saccharomyces. . J Cell Biol. 1995;131:19–31. doi: 10.1083/jcb.131.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenberg PT, Lustig KD, McGarry TJ, King RW, Kuang J, Kirschner MW. Systematic identification of mitotic phosphoproteins. Curr Biol. 1997;7:338–348. doi: 10.1016/s0960-9822(06)00157-6. [DOI] [PubMed] [Google Scholar]
- Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO (Eur Mol Biol Organ) J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Rout MP, Blobel G. A new family of yeast nuclear pore complex proteins. J Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RW, Blobel G, Rout MP. POM152 is an integral protein of the pore membrane domain of the yeast nuclear envelope. J Cell Biol. 1994;125:31–42. doi: 10.1083/jcb.125.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RW, Rout MP, Aitchison JD. Karyopherins and kissing cousins. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- Yaseen NR, Blobel G. Cloning and characterization of human karyopherin β3. Proc Natl Acad Sci USA. 1997;94:4451–4456. doi: 10.1073/pnas.94.9.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel U, Doye V, Tekotte H, Wepf R, Grandi P, Hurt EC. Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p. J Cell Biol. 1996;133:1141–1152. doi: 10.1083/jcb.133.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]