Abstract
Large-scale functional genomics approaches are fundamental to the characterization of mammalian transcriptomes annotated by genome sequencing projects. Although current high-throughput strategies systematically survey either transcriptional or biochemical networks, analogous genome-scale investigations that analyze gene function in mammalian cells have yet to be fully realized. Through transient overexpression analysis, we describe the parallel interrogation of ≈20,000 sequence annotated genes in cancer-related signaling pathways. For experimental validation of these genome data, we apply an integrative strategy to characterize previously unreported effectors of activator protein-1 (AP-1) mediated growth and mitogenic response pathways. These studies identify the ADP-ribosylation factor GTPase-activating protein Centaurin α1 and a Tudor domain-containing hypothetical protein as putative AP-1 regulatory oncogenes. These results provide insight into the composition of the AP-1 signaling machinery and validate this approach as a tractable platform for genome-wide functional analysis.
The understanding of gene function on molecular, cellular, and physiological levels is a critical step in the study of organismal biology. Despite intensive efforts, functional annotation has been assigned only to approximately one-third of the estimated 35,000 genes in the human genome (1–3). More recently, a number of functional genomics technologies have emerged that are facilitating the comprehensive elucidation of gene function. These tools have been developed to gather experimental data in a highly parallel and efficient manner and have been applied to the study of RNA dynamics (4, 5), protein expression (6, 7), and protein–protein interactions (8–11). Although these approaches are critical for constructing systems- and network-level descriptions of cellular and physiological processes, they predict gene activities by inference, rather than through direct determination of gene function.
Several current strategies have elucidated novel gene activities through directing cellular expression of arrayed genes in discrete physical loci (12–16). Although these approaches may enable the examination of gene function at a genome scale, cDNAs used in these recent studies were derived from a restricted or undefined set of genes. Systematic study of mammalian gene function at a genome level, however, would require the incorporation of both comprehensive sequence-annotated nucleic acid arrays and highly parallel experimental strategies.
Here, we describe an approach that used an addressable library of >20,000 cDNAs (≈14,000 distinct genes) in the interrogation of the activator protein-1 (AP-1) signal transduction pathway. We characterized the proliferation-associated activities of identified AP-1 pathway modulators through a series of genetic, biochemical, and phenotypic validation methodologies. Together, these results demonstrate the feasibility of rapid and large-scale functional characterization of mammalian signal transduction pathways.
Materials and Methods
Library and Array Construction. cDNAs used in this study represented all genes in a collection of 350,000 5′ end-sequenced clones showing >95% identity over >200 base pairs to a known gene or predicted gene. Collectively, this set contained ≈14,200 putative full-length human genes in 20,704 clones, which comprised the Novartis Functional Genomics Area Collection (NFGA 14k set). Further characterization of this collection can be found in Iourgenko et al. (17). These cDNAs were arrayed into 384-well plate format and were subsequently used for screening. All genes in the NFGA 14k set were cloned into pCMVsport6.0 (Invitrogen). Plasmid DNA was prepared for screening by using plasmid miniprep consumables (Macherey–Nagel) run on a Roboprep 2500 prep robot (MWG Biotech, Ebersberg, Germany), normalized, and spotted into assay plates for screening. All screening hits were reannotated by sequence verification and subsequent blastn analysis.
High-Throughput (Retro) Transfection and Genome Functionalization Through Arrayed cDNA Transduction Analysis (GFAcT). Arrayed 384-well matrices containing 62.5 ng of cDNA per well were incubated with 20 μl of serum-free medium containing FuGENE 6 (Roche, Indianapolis) and indicated reporter plasmids, obtained from a commercial source (Clontech) unless otherwise noted, for ≈20 min. Approximately 37 ng per well of reporters containing AP-1 [phorbol 12-myristate 13-acetate (PMA)] (tandem 6× tgactaa), p53 (tandem 14× tgcctggacttgcctggcc), and 60 ng per well of erythropoietin (Epo) (18) responsive elements were used. Trypsinized cells [human embryonic kidney (HEK)293, HCT116, and HepG2, respectively] were delivered to wells in 20 μl of serum-containing medium, and 384-well plates were placed in a humidified chamber (5% CO2, 37°C). After ≈48 h, 40 μl of the luciferase assay reagent Bright-Glo (Promega) was added to each well, and luminescence was read within 10 min by using an Acquest Plate Reader (LJL Biosystems, Sunnyvale, CA). All fluid delivery was done by using Multidrop (Titertek, Huntsville, AL). Relative light activity was normalized on a per-plate basis, and genes were ranked according to median activation. Detailed specifications of this process can be found at http://function.gnf.org.
Data Analysis and Clustering of Activators. Readings for putative modulators across various AP-1-related reporters (Clontech) in HEK293 cells were determined as described above. Data were subsequently normalized to a negative control [pCMV-AsRed (Clontech)], baseline-subtracted for activation of a parental reporter vector pTAL (Clontech), and filtered for background activity. Resulting data were analyzed by linkage across arrays (reporters) and genes by using cluster software (Stanford University, Stanford, CA) and were visualized by using tree-view (Stanford University). All results shown depict the average of at least three experimental data points.
Small Inhibitor RNA (siRNA) Suppression Analysis. Approximately 8,000 HEK293 cells were plated into 96-well tissue culture plates and transfected with 25 ng of indicated siRNA per well (Fig. 3A; see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org) by using Lipofectamine2000 reagent (Invitrogen). After 24 h of incubation, medium was replaced, and cells were transfected by using FuGENE 6 (Roche) with indicated cDNA along with AP-1(PMA)-luciferase (148 ng per well) and constitutive β-galactosidase (5 ng per well) reporter vectors. After 48 h of incubation, single-well luciferase and β-galactosidase activities were determined by using Dual-Lite (Applied Biosystems) luminescence detection reagents. Data were normalized for transfection efficiencies, and activities for each series of AP-1 activators were compared with baseline values in wells with negative control siRNAs (pGL2, scrambled). Data were analyzed by using a hierarchical clustering algorithm (cluster, Stanford University) and visualized in treeview (Stanford University). Sequences of siRNAs are listed in Table 1.
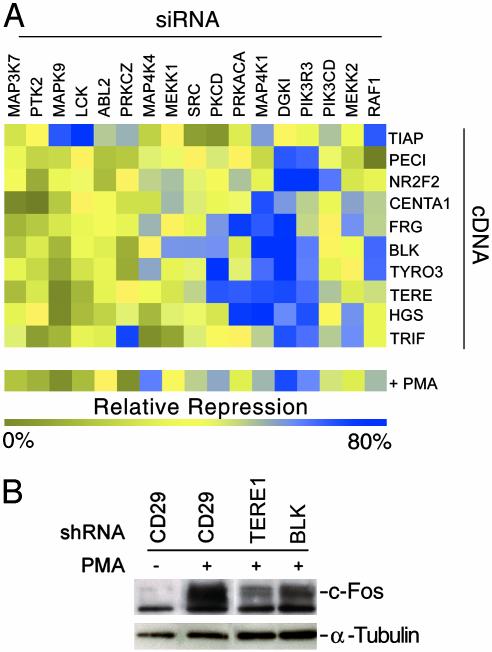
Fig. 3.
siRNA-mediated functional mapping of AP-1 growth effectors. (A) HEK293 cells were transfected with indicated siRNAs (columns) and subsequently transfected in a pairwise manner with indicated cDNAs (rows), along with an AP-1(PMA)-dependent luciferase reporter construct and a constitutive β-galactosidase reporter vector. After 48 h of incubation, fractional luciferase/β-galactosidase values were determined for each well and further normalized to the activities of three nonspecific siRNA control wells for each transfected cDNA. The resulting data were analyzed by using hierarchical clustering, visualized in treeview (Stanford University), and depict the average of at least three experiments. Effects of siRNA molecules on PMA-stimulated AP-1 activities are also shown. (B) shRNAs targeting predicted growth, and proliferation modulators transitional epithelia response 1 and BLK, and a nonspecific control (CD29), were transfected into HEK293 cells. Cultures were treated with PMA (100 ng/ml) for 24 h as indicated, and cell lysates were interrogated for c-Fos induction through Western blot analysis. Levels of α-tubulin protein were also detected to serve as loading controls.
Short Hairpin RNA (shRNA) and cDNA Subcloning. shRNA oligonucleotides were designed and cloned as described (19). After sequence verification, constructs were used in transient transfection analysis. Sequences of shRNAs are listed in Table 1. Down-regulation of endogenous transcript levels was verified by semiquantitative RT-PCR analysis. Transfer of cDNAs into replication-competent avian sarcoma leukosis virus LTR with splice acceptor (RCAS) retroviral vector (20) was mediated by Gateway LR Clonase (Invitrogen). Recombinant viral vectors were sequence verified and used to generate viral supernatants.
Western Blot and Antibody Reagents. Isolated cellular extracts were prepared, separated, and transferred onto nitrocellulose membranes as described (21). Antibodies used in these studies were purchased directly from suppliers and include c-Fos (Santa Cruz Biotechnology), c-Jun (Pharmingen), anti-rabbit IgG(H+L) horseradish peroxidase (HRP) (Promega), anti-mouse IgG HRP (Promega), Erk 1 and 2 (Santa Cruz Biotechnology), and γ tubulin (Santa Cruz Biotechnology).
Cell Culture and Growth Curves. HCT116 cells were cultured in McCoy 5A (GIBCO/BRL) media with 10% FBS, and 5 × 105/ml were plated in 384-well plates. HepG2 cells were cultured in DMEM (GIBCO/BRL) supplemented with 3% FBS at 37°C, 5% CO2, and plating densities of 1.8 × 106/ml were used for screening. HEK293 cells plated at a final density of 3.75 × 105 cells per ml were cultured in DMEM (GIBCO/BRL), supplemented with 10% FBS, at 37°C, 5% CO2. Primary chicken embryo fibroblast (CEF) cells were maintained and infected with AP-1 activator clones in subtype A RCAS virus and passaged three times, as described (22). Individual AP-1 activator-expressing CEF lines were then seeded in six-well plates at a density of 0.5 × 106 cells per well. The next day (day 0), cells were counted in triplicate for each respective clone, and counts were performed thereafter on days indicated.
Soft Agar and Focus Formation Assays. Soft agar and focus assays were performed as described (22, 23). CEF (5,000 cells per ml) were maintained in soft agar for 4 weeks by adding 1 ml of fresh nutrient agar every 4–6 days. For focus formation assays, 0.5 × 106 CEF cells were seeded and infected with viral supernatant. Cells were then overlaid with agar medium and maintained in culture for 10–14 days. Foci were visualized by crystal violet staining (GIBCO/BRL).
Results and Discussion
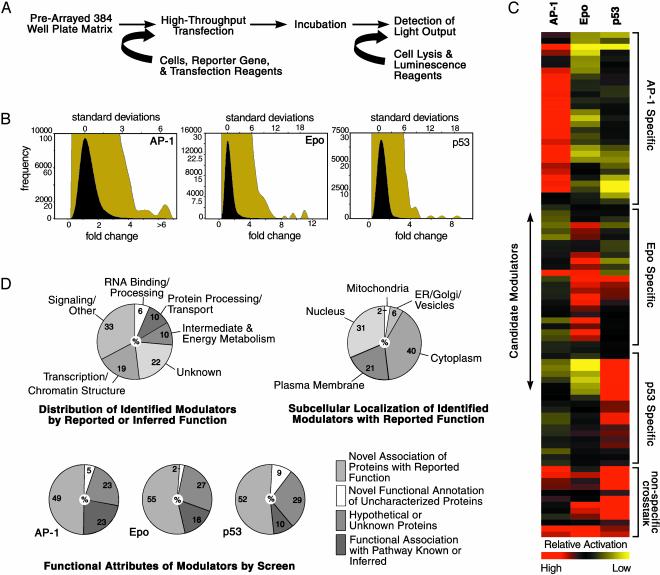
To identify modulators of AP-1 signaling, we profiled the activity of ≈20,000 arrayed full-length cDNAs on a PMA-responsive reporter construct. Sequence-annotated cDNAs were arrayed in 384-well plate format such that each well contained a single addressable gene. By means of a high-throughput transfection process (see Materials and Methods), individual genes, along with the AP-1(PMA) reporter construct, were introduced into mammalian cells. Single-well luminescence outputs were used to survey encoded-protein activities across the genome-scale matrix and identify pathway modulators (Fig. 1A). We refer to this process as GFAcT analysis. We detected ≈129 (0.64%) cDNAs that increased AP-1 reporter gene activities to a level >3 standard deviations from the experimental mean (Fig. 1B and Tables 2 and 3, which are published as supporting information on the PNAS web site; data not shown). To evaluate the specificity of gene activities elucidated through GFAcT analysis, we further used the cDNA matrix to ascertain inducers of p53 function and Epo gene transcription (Fig. 1B, Table 2, and data not shown). We subsequently crossprofiled 15–30 of the most potent modulators identified from each respective pathway screen (Fig. 1C and Table 4, which is published as supporting information on the PNAS web site, and data not shown). A small percentage of the encoded proteins demonstrated overlapping reporter activities, suggesting nonspecific activity or pathway cross-talk. However, the majority of the identified cDNAs showed preferential activation of a single reporter, indicating that this strategy uncovered distinct pathway modulators.
Fig. 1.
Genomewide functional annotation of cancer-related signalsomes. (A) Approximately 20,000 cDNAs were cotransfected in parallel into 384-well tissue culture plates with appropriate luciferase reporter plasmids. After a 48-h incubation period, cells underwent homogenous lysis, and relative luciferase activation levels were determined. (B) Frequency distributions of reported activities from assays by using AP1(PMA)-, p53-, and Epo-dependent reporters in HEK293, HCT116, and HepG2 cells, respectively, are shown on normal (black curve) and enlarged (olive curve) scales. Results depict the average of screens assayed in duplicate and are plotted against fold change and standard deviations from the respective experimental means. (C) Approximately 15–30 activators identified in each screen were assayed for specificity by evaluating activities across AP-1, p53, and Epo-responsive reporters. Results were subsequently normalized and assessed by a hierarchical clustering algorithm. Relative activation by modulators (rows) in each reporter assay (columns) is depicted by a red (higher than median activity) to yellow (lower than median activity) continuum color scheme. Clusters of pathway-specific and likely cross-modulatory activities are indicated. (D) Statistics of identified pathway effectors based on characteristics reported or inferred from the literature are shown. Numbers inside circles represent the percentage of total validated pathway activators.
In addition to elucidating on the order of 102 gene activities, our analysis also, as expected, identified modulators that have a known association with their respective pathways. For instance, c-FOS, c-JUN, GADD45 β and γ, TRAF6, ERB-B-2, FOSL1, ATF3, MAP3K11, RICK, and PKCζ were among the top 0.5% of activators in the AP-1 screen (Table 3 and data not shown). Furthermore, of the canonical FOS and JUN family members represented in the GFAcT cDNA matrix (FOS, FRA-1, FRA-2, JUNB, and JUND), only JUNB did not score in the top 3% of screened activators (Table 3 and data not shown). Surprisingly, this relatively unbiased survey also revealed a high proportion of functionally novel activities within the set of identified Epo, p53, and AP-1 effectors (Fig. 1D), suggesting that a significant portion of the cellular machinery that governs these biologically important pathways remains uncharacterized.
AP-1 activity is regulated by a variety of extracellular stimuli, including growth factors, cytokines, cell–matrix interactions, and genotoxic stress, among others (24). Although all of the activating genes were identified through their abilities to modulate AP-1(PMA)-dependent reporter gene transcription, we used a response-pathway mapping strategy to further classify novel AP-1 inducers on the basis of comparative activation of related reporter constructs. Specifically, we profiled relative activities of AP-1 modulators on reporter elements responsive to growth and proliferation signals {serum response element (SRE), NFATc response elements (NFAT), and PMA response elements [AP-1(PMA)]} (25, 26), and also those that preferentially monitor inflammation, IFN, or stress stimuli [cAMP response element-binding protein (CREB) response elements (CRE), IFN response elements (ISRE), and AP-1 response elements (AP-1)] (27, 28). Interestingly, hierarchical cluster analysis (29) of these data resulted in the co-segregation of reporter activities in relation to specific facets of AP-1 signaling (Fig. 2A, Table 5, which is published as supporting information on the PNAS web site). This allowed for distinctions between encoded proteins likely involved with genotoxic stress and (proinflammatory) cytokine signaling (ISRE, AP-1, CRE), and, conversely, those affiliated with growth factor and serum response [SRE, AP-1(PMA), NFATc response elements].
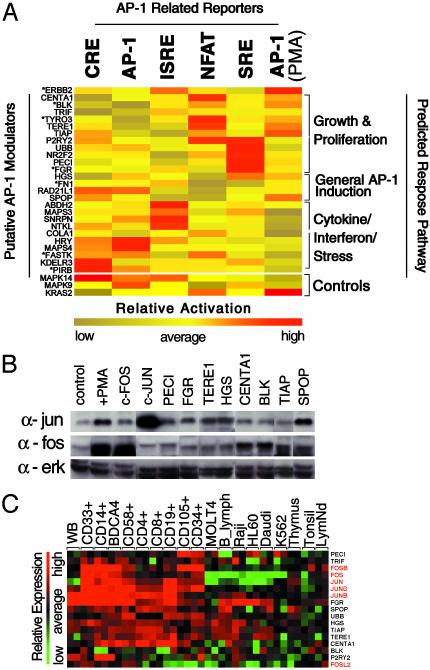
Fig. 2.
Functional validation of AP-1 modulators. (A) The activities of 25 activators of AP-1(PMA) signaling were profiled in triplicate against AP-1-related reporter constructs sensitive to growth and proliferation stimuli [AP-1(PMA), SRE, and NFATc response elements (plus 2 μm of ionomycin)] and stress/cytokine induction (cAMP response element-binding protein response elements, AP-1, IFN response elements), as described. To account for the differential dynamic range of activity inherent to each response element, reporter (columns) and gene (rows) activities were normalized and clustered by using a hierarchical algorithm and are depicted by a red (high relative activity) to yellow (low relative activity) continuum color scheme. Predicted response pathways are based on clustered gene activities on a subset of reporter constructs. Gene annotations reflect official gene symbols designated by locuslink (www.ncbi.nlm.nih.gov/LocusLink), with the exception of previously unannotated genes, which have been assigned interim gene symbols indicated in Table 5. Encoded proteins with known or inferred activities related to predicted pathways, respectively, are indicated by an asterisk (*). (B) Induction of endogenous c-Fos and c-Jun protein levels by transfection of “growth” and “general” responsive subsets of indicated AP-1 modulators, as determined in A, into HEK293 cells was assessed through Western blot analysis. Cell lysates from parental vector (pCMVSport6) transfected HEK293 cells were used as a negative control, and c-FOS- and c-JUN-transfected and PMA-treated cell extracts were used as positive controls. Levels of ERK1 and ERK2 were detected to account for loading. (C) Coexpression based on microarray analysis of canonical (red text) and identified (black text) AP-1 pathway members in primarily lymphoid cell types [WB (whole blood), CD33+ (myeloid, bone marrow), CD14+ (monocyte, peripheral blood), BDCA4+ (dendritic, peripheral blood), CD58+ (natural killer, peripheral blood), CD4+ (T cell, peripheral blood), CD8+ (T cell, peripheral blood), CD19+ (B cell, peripheral blood), CD105+ (endothelial, bone marrow), CD34+ (hematopoietic progenitor, bone marrow), MOLT4 (lymphoblastic leukemia), B_lymph (lymphoblastic leukemia), Raji (Burkitt's lymphoma), HL60 (promyelocytic leukemia), Daudi (Burkitt's lymphoma), K562 (chronic myelogenous leukemia), thymus, tonsil (adult, pooled), and LymNd (lymph node, adult)] are shown by a red (high relative expression) to green (low relative expression) continuum color scheme. Relative mRNA levels shown were derived from comparisons of >50 random primary tissue and cell types (http://expression.gnf.org).
We further investigated the mechanisms by which the latter subset of genes potentially regulates AP-1 growth and proliferation responsive pathways. Consistent with their postulated roles, transfection of most cDNAs from these predicted subsets resulted in the coordinate induction of c-Fos and/or c-Jun protein levels, as demonstrated by Western blot analysis (Fig. 2B). However, expression of a few encoded proteins in this grouping, such as TYRO3, P2RY2, and UBB, did not result in detectable elevated levels of the prototypic AP-1 transcription factors (data not shown), which may indicate that they modulate AP-1 activity through alternate family members or through as-yet-unknown mechanisms. Although we surveyed the regulation of only a limited number of AP-1 proteins by a subset of effectors, these results indicate that significant numbers of encoded proteins identified through GFAcT analysis are likely bona fide modulators of endogenous AP-1 activity. Intriguingly, analysis of human tissue mRNA levels revealed that many of these identified growth-associated AP-1 modulators and canonical AP-1 transcription factors were coincidentally expressed in cells of lymphoid origin (Fig. 2C and Fig. 5, which is published as supporting information on the PNAS web site), suggesting a functional alignment between these molecules in proliferating cell types.
To refine the mode of action by which putative growth regulatory proteins modulate the AP-1 pathway, we assessed their relative inductive activities in the presence of siRNA molecules directed against known components of AP-1 signaling. AP-1(PMA) reporter gene activities resulting from pairwise cDNA (novel activators) and siRNA (AP-1 pathway members) transfections were normalized to appropriate controls and analyzed through single-linkage hierarchical clustering (Fig. 3A and Table 1). For comparison, effects of the inhibitory RNAs on PMA-induced AP-1 reporter gene activity were also determined. siRNAs encoding PI-3 kinase (PIK3R3, PIK3CD), PKC (PKCD, PRKACA, DGKI, MEKK2), and RAS (SRC, RAF1) pathway members were most proficient in abrogating AP-1 induction by the bulk of the elucidated putative growth regulators. This demarcates a majority of identified AP-1 activators as functioning upstream of these proteins and suggests that they regulate AP-1 activity through modulation of PI-3 kinase, PKC, and/or RAS pathway members. For example, the activity of B lymphocyte kinase (BLK) on the AP-1 reporter gene was extinguished by siRNAs coding for ERK pathway effectors DGKI, MEKK1, SRC, RAF1, and PIK3R3 (blue), but not by siRNAs directed against JNK-related pathway molecules MAP3K7, MAPK9, and MAP4K4 (yellow). Although we cannot exclude the possibility of unintended (“off-target”) siRNA effects on this analysis (see Table 1), these activities are consistent with studies that have demonstrated the importance of PI-3 kinase, PKC, and RAS function for AP-1 response to proliferation stimuli (30, 31). In sum, this genetic suppression analysis approach led to the identification of AP-1 pathway components required for the inductive activities of novel modulators and further defined the junction at which these molecules intersect the AP-1 signaling cascade.
We additionally examined the roles of the predicted growth-associated AP-1 modulators in mitogenic response. By using DNA expression plasmids encoding shRNAs (19) directed against a subset of eight of the growth-related pathway activators, we screened for the inhibition of PMA-induced AP-1 activity through reporter gene analysis (data not shown). On the basis of their potent antagonism of PMA-induced reporter activity, we selected shRNAs encoding sequences homologous to the BLK and transitional epithelia response 1 (TERE1) genes for further characterization. Through Western blot analysis, we demonstrated that transfection of these shRNAs into PMA-stimulated cells resulted in a significant decrease in c-Fos induction in response to phorbol ester (Fig. 3B). These results reveal that these encoded proteins play critical roles in AP-1 regulation by mitogenic stimuli in a human embryonic kidney cell line. Examination of Blk and Tere1 protein complexes by MS analysis suggests that these proteins are possibly mediating AP-1 function through the indirect modulation of SRC and RAF-1 activities and of intracellular biosynthetic intermediate (i.e., ATP, serine derivatives) levels, respectively (Fig. 6, which is published as supporting information on the PNAS web site).
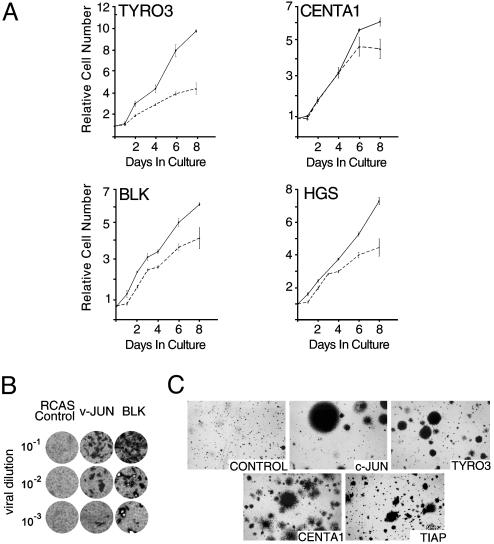
Given the role of AP-1-related proteins in proliferation and cancer-related phenotypes, we alternately assessed the ability of these molecules to impart a growth advantage to avian cells. Proliferative control in primary CEF cells is particularly susceptible to modulation by AP-1 effectors (31, 32). Growth analysis of CEF ectopically expressing a subset of putative AP-1 growth regulatory genes demonstrated that these molecules can increase rates of proliferation in this primary cell type (Fig. 4A). We further investigated the oncogenic potential of these genes in transformation assays. Previous reports have shown that expression of TYRO3 can mediate focus formation in an immortalized cell type (RAT1 fibroblasts), and an activated mutant of BLK has also previously been demonstrated to possess oncogenic activities (33, 34). Here, we have shown that expression of wild-type BLK induced focal microtumors on monolayers of CEF cells (Fig. 4B), and ectopic TYRO3 activity was sufficient to promote anchorage-independent growth of CEF (Fig. 4C). Infection of CEF with retroviruses encoding Centaurin-α 1 (CENTA1) and the hypothetical protein KIAA1847, which we have designated TIAP (Tudor-domain containing inducer of AP-1), also resulted in anchorage independence (Fig. 4C). Colonies derived from ectopic expression of these genes have more diffuse borders in contrast to the compact colonies induced by c-JUN or TYRO3. These results confirm the in vivo proliferative activities of several predicted growth-associated AP-1 activators identified through GFAcT analysis and suggest that the oncogenic activities of BLK, TYRO3, CENTA1, and TIAP are likely mediated, at least in part, through modulation of AP-1 activity.
Fig. 4.
Identification of candidate oncogenes associated with AP-1 proliferation. (A) Primary CEF expressing predicted modulators of AP-1-mediated proliferation were plated at a density of ≈5 × 105 in six-well plates, and cell counts were initially taken after 24 h (day 0) and at the indicated time points. Ectopically expressed genes (solid line) are indicated on each graph and compared with RCAS only control (dashed line). Results are shown as the function of initial seeding density (day 0) and depict the average of three experiments. (B) CEF cells were infected with BLK, c-JUN, or control RCAS retroviruses, and after 24 h, cells were overlaid with agar medium and maintained in culture for 10–14 days. (C) CEF cells were infected with retroviruses encoding c-JUN, TYRO3, Centaurin-α 1, TIAP, and RCAS control, as described above. Cells were maintained in soft agar for 4 weeks.
Pathway and cell-based functional annotation is an important methodology in deciphering the biological activities of genes identified by the genome sequencing projects. We describe here a method for genome-scale functional annotation of human genes with respect to AP-1, p53, and hypoxic (Epo) signaling. This approach is currently limited to the study of individual gene function through ectopic expression. Although overexpression of genes may activate legitimate signaling molecules in the absence of appropriate stimuli, in certain cases it can also result in the induction of physiologically irrelevant phenotypes. We have endeavored to exclude artifacts of overexpression by characterizing a subset of AP-1 activators in expanded and independent tests.
Twenty-five of the most potent AP-1 modulators identified by GFAcT analysis were further classified through reporter-based pathway mapping, identifying those genes that are likely to mediate AP-1-related proliferative activities. siRNA-based functional suppression analysis was used to demonstrate that this subset of modulators probably work through RAS/PKC/PI-3K pathways, and further characterization of these proliferation-associated genes revealed the requirement of BLK and transitional epithelia response 1 for unabridged AP-1 response to mitogenic stimulation. Finally, cellular analysis confirmed that many of the identified genes can function to augment proliferation in a primary cell type and led to the identification of Centaurin-α 1 and TIAP as mediators of cellular transformation.
We demonstrate that highly parallel genomics strategies, such as mRNA profiling (4), MS (9, 10), and siRNA-based functional annotation (35), can be used concomitantly to characterize genome-scale data. Application of such integrative genomics approaches will allow for increasingly sophisticated models that extend our knowledge of the complex relationships between gene products, signal transduction pathways, and their underlying biological functions.
Supplementary Material
Acknowledgments
We acknowledge Nikunj Somia, Pedro Aza-Blanc, and Michael Cooke for scientific input; and Christopher Cooper, Myleen Medina, Qing Li, Tracie Robinson, and Abel Gutierrez for technical contributions. We thank Drs. Josephine Harada and Trey Sato for helpful comments on the manuscript. This work is supported by funding from the Novartis Research Foundation and the National Cancer Institute (to P.K.V.). This is manuscript number 15966-MEM of The Scripps Research Institute.
Abbreviations: AP-1, activator protein 1; PMA, phorbol 12-myristate 13-acetate; GFAcT, genome functionalization through arrayed cDNA transduction; siRNA, small inhibitor RNA; shRNA, short hairpin RNA; CEF, chicken embryo fibroblast; Epo, erythropoietin; SRE, serum response element; BLK, B lymphocyte kinase; HEK, human embryonic kidney; RCAS, replication-competent avian sarcoma leukosis virus LTR with splice acceptor.
References
- 1.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 2.Pruitt, K. D., Tatusova, T. & Maglott, D. R. (2003) Nucleic Acids Res. 31, 34–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 4.Lockhart, D. J., Dong, H., Byrne, M. C., Follettie, M. T., Gallo, M. V., Chee, M. S., Mittmann, M., Wang, C., Kobayashi, M., Horton, H., et al. (1996) Nat. Biotechnol. 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 5.DeRisi, J. L., Iyer, V. R. & Brown, P. O. (1997) Science 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 6.Gao, C., Mao, S., Lo, C. H., Wirsching, P., Lerner, R. A. & Janda, K. D. (1999) Proc. Natl. Acad. Sci. USA 96, 6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haab, B. B., Dunham, M. J. & Brown, P. O. (2001) Genome Biol. 2, RESEARCH0004. Epub 2001 Jan 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu, H., Pan, S., Gu, S., Bradbury, E. M. & Chen, X. (2002) Rapid Commun. Mass Spectrom. 16, 2115–2123. [DOI] [PubMed] [Google Scholar]
- 9.Gavin, A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, C. M., et al. (2002) Nature 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 10.Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. (2002) Nature 415, 180–183. [DOI] [PubMed] [Google Scholar]
- 11.MacBeath, G. & Schreiber, S. L. (2000) Science 289, 1760–1763. [DOI] [PubMed] [Google Scholar]
- 12.Ziauddin, J. & Sabatini, D. M. (2001) Nature 411, 107–110. [DOI] [PubMed] [Google Scholar]
- 13.Fiscella, M., Perry, J. W., Teng, B., Bloom, M., Zhang, C., Leung, K., Pukac, L., Florence, K., Concepcion, A., Liu, B., et al. (2003) Nat. Biotechnol. 21, 302–307. [DOI] [PubMed] [Google Scholar]
- 14.Chen, C., Grzegorzewski, K. J., Barash, S., Zhao, Q., Schneider, H., Wang, Q., Singh, M., Pukac, L., Bell, A. C., Duan, R., et al. (2003) Nat. Biotechnol. 21, 294–301. [DOI] [PubMed] [Google Scholar]
- 15.Michiels, F., van Es, H., van Rompaey, L., Merchiers, P., Francken, B., Pittois, K., van der Schueren, J., Brys, R., Vandersmissen, J., Beirinckx, F., et al. (2002) Nat. Biotechnol. 20, 1154–1157. [DOI] [PubMed] [Google Scholar]
- 16.Matsuda, A., Suzuki, Y., Honda, G., Muramatsu, S., Matsuzaki, O., Nagano, Y., Doi, T., Shimotohno, K., Harada, T., Nishida, E., et al. (2003) Oncogene 22, 3307–3318. [DOI] [PubMed] [Google Scholar]
- 17.Iourgenko, V., Zhang, W., Mickanin, C., Daly, I., Jiang, C., Hexham, J. M., Orth, A. P., Miraglia, L., Meltzer, J., Garza, D., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 12147–12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanchard, K. L., Acquaviva, A. M., Galson, D. L. & Bunn, H. F. (1992) Mol. Cell. Biol. 12, 5373–5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Science 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 20.Loftus, S. K., Larson, D. M., Watkins-Chow, D., Church, D. M. & Pavan, W. J. (2001) DNA Res. 8, 221–226. [DOI] [PubMed] [Google Scholar]
- 21.Chao, S. H., Walker, J. R., Chanda, S. K., Gray, N. S. & Caldwell, J. S. (2003) Mol. Cell. Biol. 23, 831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sonderegger, C. K. & Vogt, P. K. (2003) Oncogene 22, 1749–1757. [DOI] [PubMed] [Google Scholar]
- 23.Blazek, E., Wasmer, S., Kruse, U., Aronheim, A., Aoki, M. & Vogt, P. K. (2003) Oncogene 22, 2151–2159. [DOI] [PubMed] [Google Scholar]
- 24.Shaulian, E. & Karin, M. (2002) Nat. Cell Biol. 4, E131–E136. [DOI] [PubMed] [Google Scholar]
- 25.Bohmann, D. & Tjian, R. (1989) Cell 59, 709–717. [DOI] [PubMed] [Google Scholar]
- 26.Ransone, L. J. & Verma, I. M. (1990) Annu. Rev. Cell Biol. 6, 539–557. [DOI] [PubMed] [Google Scholar]
- 27.Cahill, M. A., Janknecht, R. & Nordheim, A. (1996) Curr. Biol. 6, 16–19. [DOI] [PubMed] [Google Scholar]
- 28.Roesler, W. J., Vandenbark, G. R. & Hanson, R. W. (1988) J. Biol. Chem. 263, 9063–9066. [PubMed] [Google Scholar]
- 29.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitmarsh, A. J. & Davis, R. J. (1996) J. Mol. Med. 74, 589–607. [DOI] [PubMed] [Google Scholar]
- 31.Shaulian, E. & Karin, M. (2001) Oncogene 20, 2390–2400. [DOI] [PubMed] [Google Scholar]
- 32.Cohen, S. B., Waha, A., Gelman, I. H. & Vogt, P. K. (2001) Oncogene 20, 141–146. [DOI] [PubMed] [Google Scholar]
- 33.Malek, S. N., Dordai, D. I., Reim, J., Dintzis, H. & Desiderio, S. (1998) Proc. Natl. Acad. Sci. USA 95, 7351–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor, I. C., Roy, S., Yaswen, P., Stampfer, M. R. & Varmus, H. E. (1995) J. Biol. Chem. 270, 6872–6880. [DOI] [PubMed] [Google Scholar]
- 35.Pothof, J., Van Haaften, G., Thijssen, K., Kamath, R. S., Fraser, A. G., Ahringer, J., Plasterk, R. H. & Tijsterman, M. (2003) Genes Dev. 17, 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.