Abstract
We used DNA microarrays representing >12,000 human genes to characterize gene expression patterns in skin biopsies from individuals with a diagnosis of systemic sclerosis with diffuse scleroderma. We found consistent differences in the patterns of gene expression between skin biopsies from individuals with scleroderma and those from normal, unaffected individuals. The biopsies from affected individuals showed nearly indistinguishable patterns of gene expression in clinically affected and clinically unaffected tissue, even though these were clearly distinguishable from the patterns found in similar tissue from unaffected individuals. Genes characteristically expressed in endothelial cells, B lymphocytes, and fibroblasts showed differential expression between scleroderma and normal biopsies. Analysis of lymphocyte populations in scleroderma skin biopsies by immunohistochemistry suggest the B lymphocyte signature observed on our arrays is from CD20+ B cells. These results provide evidence that scleroderma has systemic manifestations that affect multiple cell types and suggests genes that could be used as potential markers for the disease.
Scleroderma is a complex, heterogeneous, and sometimes fatal disease that affects ≈150,000 people in the United States (1). Although scleroderma pathogenesis is poorly understood, disease progression is known to involve the immune system, the vasculature, and extracellular matrix deposition; there are, however, no definitive markers or curative treatments (2). Scleroderma can occur in a localized form confined to the skin or a systemic form referred to as systemic sclerosis (SSc), which involves internal organs and the skin. Involvement of critical internal organs leads to death in some patients (3). The skin is affected in >90% of patients, and cutaneous involvement closely correlates with internal organ pathology and can be used as a surrogate index for prognosis (4, 5).
The disease is most prevalent in women with a median age of onset of 45 years; symptoms develop symmetrically and include swelling in the hands and Raynaud's phenomenon. The skin becomes tense, shiny, and painful. Skin changes progress and may involve the face, trunk, and lower extremities. Ultimately, the edematous skin becomes fibrotic and hardens (hence the term scleroderma, which means hard skin). These dramatic and distressing skin changes are generally accompanied by internal organ involvement. Gastroesophageal reflux disease, weight loss, interstitial lung disease, pulmonary arterial hypertension, and renal and cardiac manifestations may occur and require close monitoring and treatment.
In an attempt to better understand the disease, and in the hope of finding molecular markers for the disease, we have undertaken studies of gene expression in the skin of individuals affected with scleroderma. We used DNA microarrays to characterize gene expression patterns in skin biopsies from individuals with a diagnosis of SSc with diffuse scleroderma and compared those to the patterns of gene expression seen in biopsies from normal, unaffected individuals.
Materials and Methods
All subjects signed consent forms approved by the Committee on Human Research at the University of California, San Francisco. Patients met American College of Rheumatology classification criteria for SSc (6), further characterized as the diffuse subset (7). Four patients (two men and two women) underwent two sets of biopsies (Table 1). Three 5-mm punch biopsies from the lateral forearm, 8 cm proximal to the ulnar styoid, were taken for clinically involved skin, and three biopsies were taken from the buttock or back for clinically uninvolved skin. Four normal controls (one man and three women) underwent the same number of biopsies in the identical locations (forearms and backs); the exception was normal control 4 who only underwent a forearm biopsy. Two biopsies were immediately frozen, the third biopsy was bisected; half went into 10% formalin for routine histology processing and the other half was for fibroblast cell culture (8).
Table 1. Subjects clinical characteristics.
| Subject | Age/sex | Duration, yrs | Skin score | ANA | Organs | Medications |
|---|---|---|---|---|---|---|
| SSc 1 | 41/M | 1 | 43/66 | + | Lung | MTX, omeprazole |
| Speckled | Muscles | Rofecoxib | ||||
| SSc 2 | 54/M | 4 | 30/66 | — | GI | Omeprazole |
| RP | ||||||
| SSc 3 | 62/F | 1 | 33/66 | +1:640 | GI | Nifedipine |
| nucleolar | ||||||
| SSc 4 | 58/F | 2 | 27/66 | +1:640 | Lung | Nifedipine, lansoprazole |
| Speckled | RP, GI |
The clinical data for the four subjects from which skin biopsies were taken are shown. In each case the age, sex, and duration of disease as defined by onset of first non-Raynaud's phenomenon symptoms, modified Rodnam skin scores on a 66-point scale, anti-nuclear antibody (ANA) titers, internal organ involvement, and medication taken by each patient are indicated. The age and sex of normal controls are as follows: Nor1, 39, male; Nor2, 58, female; Nor3, 58, female; and Nor4, 41, female. GI, gastrointestinal tract; RP, Raynaud's phenomenon; MTX, methotrexate; M, male; F, female.
Preparation of total RNA (10–24 μg of total RNA from two 5-mm punch biopsies), cRNA synthesis, and hybridization to 12,000-element Affymetrix Hu95A microarrays are described in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org, as are cell growth in culture, RNA extraction, and analysis (9) on 42,512-element cDNA microarrays (representing 28,384 Unigene clusters) manufactured in the Stanford Microarray Facility (www.microarray.org). The full data set is available at http://genome-www.stanford.edu/scleroderma. Figures in both red/green and blue/yellow color scheme and more detailed materials and methods are available in Supporting Text and Figs. 5–7, which are published as supporting information on the PNAS web site.
Results
We studied gene expression in skin biopsies from four patients with a diagnosis of SSc with diffuse scleroderma and from four demographically matched normal volunteers (Table 1). Eight subjects were biopsied at identical locations; biopsies were taken from the forearm (clinically affected) in all eight subjects and from the back (clinically unaffected) of seven subjects (all four scleroderma subjects and three normal controls). Total RNA prepared from each biopsy was analyzed on Affymetrix oligonucleotide arrays representing 12,000 human genes. In total, we analyzed 15 oligonucleotide microarrays; 12 signal amplification replicates (see Supporting Text) were added, generating a total of 27 data columns. Each of these columns was treated independently rather than averaging the data columns.
The data were processed to display the changes in gene expression as the log2 of the ratio of the intensity relative to its average intensity. A threshold was set such that intensities of 20 or below were set to an arbitrary value of 20. We rescaled the data obtained from the Affymetrix oligonucleotide microarrays such that each array had the same overall intensity. The intensity value for each gene was converted to a ratio by dividing the intensity value for each gene by its average expression profile across all 27 data columns to obtain a ratio value (10), and the data were analyzed by hierarchical clustering (11).
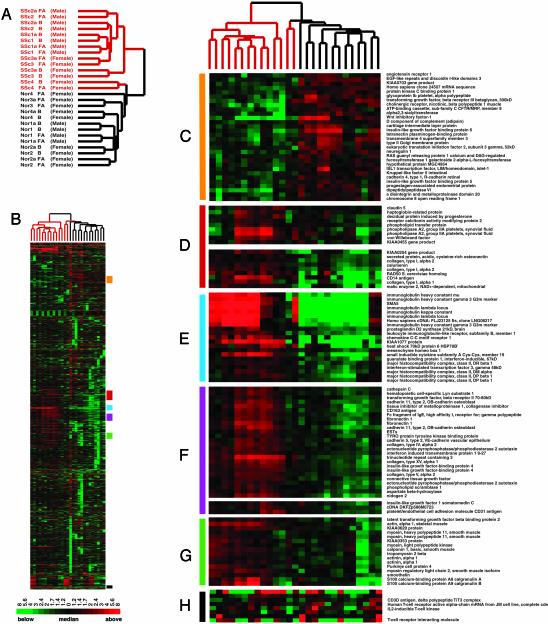
Overview of Gene Expression Profiles. We identified 2,776 genes whose expression varied from its mean value for this set of samples by >2-fold in at least three of the 27 experiments and analyzed them by hierarchical clustering in both the gene and experiment dimension (Fig. 1). The dendrogram representing the results from clustering the tissues samples according to similarity in their patterns of expression of these 2,776 genes clearly shows that tissues from the affected individuals are easily distinguished by their patterns of gene expression from the cognate normal tissues. With the exception of the two samples from patient SSc4, all of the scleroderma samples cluster onto one branch, whereas the normal samples cluster onto a different branch. Surprisingly, the clinically unaffected back samples cluster side-by-side with the clinically affected forearm tissue as a result of having very similar gene expression patterns (Fig. 1A). These results indicate that the region of the skin considered uninvolved actually is affected and that the disease in at least three of these four patients was systemic at the time the biopsies were made. Although the back tissues were identified as clinically uninvolved, many “unaffected” skin biopsies show sclerosis by histological analysis. The agreement between our microarray results and histological analysis suggests that these objective techniques will more accurately define involved and uninvolved tissue and provides optimism that our microarrays can detect changes well before clinical signs appear.
Fig. 1.
Gene expression in scleroderma skin biopsies. (A) The dendrogram representing four scleroderma (SSc) subjects is colored red, and the dendrogram for the four normal (nor) control subjects has been colored black. Samples taken from affected forearm (FA) or unaffected back (B) of SSc subjects and normal control subjects are indicated. (B) Overview of gene expression patterns indicating the select clusters of genes shown in more detail. (C) Normal cluster. (D) Collagen I cluster. (E) B lymphocyte cluster. (F) Cell adhesion and extracellular matrix. (G) Smooth muscle cluster. (H) T cell cluster.
Also shown in Fig. 1 are selected portions of the clustered data that differ between samples from affected and unaffected individuals. Prominent among these are clusters of genes highly expressed in tissue from affected individuals but not normal individuals, including groups of genes characteristically expressed in endothelial cells (Fig. 1 D and F) and B lymphocytes (Fig. 1E), a set of genes associated with synthesis of the extracellular matrix (Fig. 1F), a set of genes suggestive of smooth muscle (Fig. 1G), and genes characteristically expressed in T cells (Fig. 1H).
Genes that are expressed in normal tissue but only weakly in scleroderma have no readily identifiable biological theme (Fig. 1C). Expression of these genes could not be assigned to any specific cell type (see below) and examination of the gene ontology biological process and cellular component annotations (12) from LocusLink (13) did not indicate a unifying biological theme as is often evident in groups of coexpressed genes. The only plausible theme we could discern, based on the cellular component annotations, is enrichment for genes that are secreted and plasma membrane-associated (Fig. 5). The reason for or the significance of this apparent enrichment is unclear.
Cell Lineage Contributions to the Gene Expression Patterns in Scleroderma. Skin is composed of diverse cell types that perform different functions (fibroblasts, epithelial cells, endothelial cells, smooth muscle cells, adipocytes, and leukocytes, etc.) and, as expected, we observe gene expression patterns characteristic of a variety of different cell types. To help provide an interpretive framework for the gene expression patterns observed in scleroderma skin biopsies, we measured the gene expression in 11 different cell lines grown in culture that represent cell types likely to be present in skin: dermal microvascular endothelial cells, aortic smooth muscle cells, Hs578T myofibroblast-like cells, B lymphocytes from a follicular lymphoma (HF-1), HeLa epithelial cells, and dermal microvascular endothelial cells. In addition, we studied dermal fibroblasts grown to confluence in 10% serum from forearm and back skin biopsies (normal and affected individuals) and normal fibroblasts under two alternate conditions: resting cells (0.1% serum) on tissue culture plates and resting cells on collagen I-coated plates. Additionally, the patterns of gene expression in a normal lymph node and follicular lymphoma lymph node biopsies were included in the analysis. These measurements were made with cDNA microarrays by using a common reference (Stratagene) as described (9).
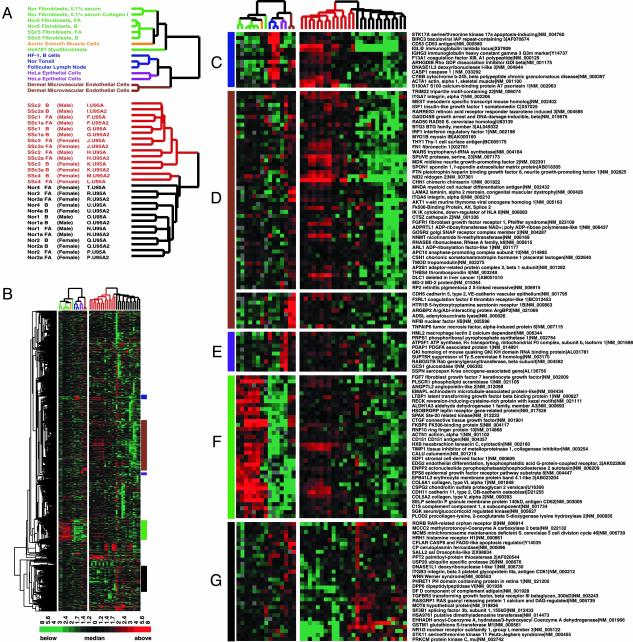
The data from these cell line experiments were analyzed together with the data from the scleroderma and normal biopsies as follows. The genes jointly represented in the two different kinds of microarray were associated by UniGene cluster ID. The expression levels measured for each gene were centered on their median value independently in the skin biopsy data and in the cell line data (Fig. 2). The biopsy samples and cell culture samples were clustered separately, based on their patterns of gene expression with the same set of 1,123 genes (Fig. 2 A). The clustering of the biopsies recapitulates the major themes of Fig. 1 A, with minor differences. To simplify the visual interpretation of the results, the branches of the dendrogram are color-coded by cell type: normal and scleroderma fibroblasts (green), muscle cells (orange), lymphoid cells and tissue (blue), epithelial cells (purple), and endothelial cells (brown). The cell culture samples cluster by cell type as has been observed in previous experiments examining the gene expression patterns of cells in culture (14).
Fig. 2.
Cell type-specific gene expression in scleroderma skin biopsies. Cell lines were used to define the cell lineage-specific gene expression observed in the skin biopsies. Gene expression data for SSc and normal (Nor) skin biopsies were collapsed by UniGene cluster ID. (A) The dendrogram for fibroblast cell lines is labeled green, muscle cell line is orange, lymphoid tissue and cell lines are blue, epithelial cells are purple, and endothelial cells are brown. (B) Overview of the 1,123 genes that change >2-fold on at least four arrays in the skin biopsy data; the colored bars to the right identify the location of the insets displayed in C–G. (C) B lymphocyte gene expression cluster. (D) Endothelial gene expression cluster. (E) Epithelial gene expression cluster. (F) Fibroblasts gene expression cluster. (G) Normal gene expression cluster.
This analysis allows us to associate the expression of some prominent clusters of genes differentially expressed in the biopsies with particular kinds of cells. For example, many of the genes that are differentially expressed between scleroderma tissue and normal tissue appear to be characteristic of endothelial cells (Fig. 2D) or fibroblasts (Fig. 2F). VE-cadherin, Thy1, von Willebrand factor, and CD31 expressed in endothelial cells (Figs. 2D and 1 C–E); platelet-derived growth factor α-associated protein 1 and macrophage lectin 2 was associated with epithelial cells (Fig. 2E); and the collagens and extracellular matrix components expressed in fibroblast and smooth muscle cells (Fig. 2F). Importantly, genes that are differentially expressed in the skin biopsies and also expressed in fibroblasts (Fig. 2F) were not differentially expressed in scleroderma-derived fibroblasts and normal-derived fibroblasts grown in culture. Most of the genes that are expressed at a higher level in normal skin relative to scleroderma skin are not easily associated with any single cell type represented in this data set (Fig. 2G).
Immunoglobulins and CD53 genes associated with B lymphocytes (including plasma cells; Fig. 2C) show prominent expression among the scleroderma samples. The strong association of a B lymphocyte signature with the scleroderma samples was unexpected; whereas it was observed before that lymphocytes are often present in skin affected by scleroderma, these were previously believed to be primarily T cells (15) although earlier reports suggested the mononuclear cell infiltrates included not only T lymphocytes, but also plasma cells and macrophages (16). In contrast to these expectations, neither expression of the T cell receptor nor of CD3 were correlated with disease state in this analysis (see Fig. 1H).
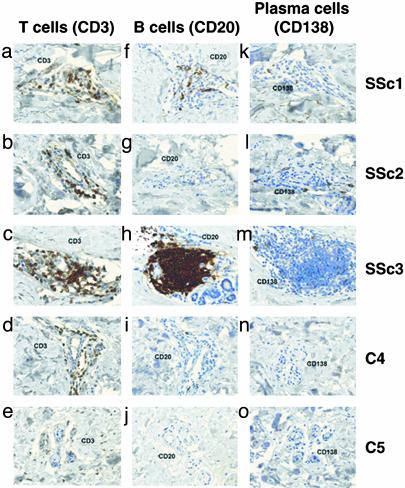
To further investigate the identity and localization of lymphocytes in these samples, immunohistochemistry was performed for T cells (CD3), B cells (CD20), and plasma cells (CD138) (Fig. 3). Shown are sections of skin biopsies from a subset of our patient cohort. Although, CD3+ T cells are present in the all three sections from the SSc patients, there is only a slight increase in cell number relative to the controls (consistent with our gene expression results). In contrast, a very notable finding was a large cluster of CD20+ B cells in a peri-eccrine distribution in patient SSc3. This population was not enriched for plasma cells (as indicated by the lack of CD138 staining). This finding could explain the preferential B cell and Ig gene expression profile over other immune cells in the SSc biopsies.
Fig. 3.
Immunohistochemistry for lymphocyte subsets in scleroderma skin. Lymphocyte subsets in forearm skin biopsies of three SSc patients (SSc1, SSc2, and SSc3) and two normal controls (C4 and C5) were investigated by immunohistochemistry. Paraffin sections were stained for T cells (CD3, a–e), B cells (CD20, f and g), and plasma cells (CD138, k–o). (Magnification: ×200.)
Although an increased number of CD20+ B cells was observed only in SSc3, it should be noted that the number of lymphocytes between two sections of the same biopsy and between two biopsy specimens on the same forearm could differ. A previous study of scleroderma skin biopsies suggested that there was some variability among the histologic findings between different biopsy sites (15). Adjacent biopsies, or different sections from the same biopsy, may contain different numbers of CD20+ B cells that are difficult to detect in serial sections.
Characterization of Differential Gene Expression in Skin Biopsies by Supervised Analysis. One benefit of a large-scale gene expression study is the ability to identify markers for the disease under study (17, 18). We used a nonparametric (model-free) Wilcoxon rank sum test to select genes that show significant differential expression between the scleroderma samples and the normal samples (19). P values are corrected for multiple testing by using Bonferroni correction.
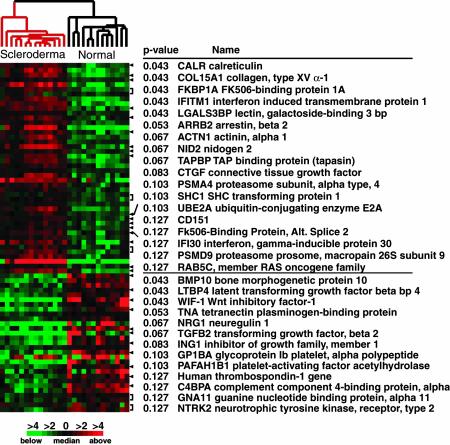
Genes whose Bonferroni-corrected P values were <0.1 are shown in Fig. 4 (the detailed position and P values for all genes <0.1 can be found in Fig. 6). Genes that were more highly expressed in scleroderma tissue include calreticulin, a chaperone protein that is involved in folding of glycosylated proteins in the endoplasmic reticulum (20, 21); collagen IV and XV; FK506-binding protein 1A (FKBP1A), a cis-trans prolyl isomerase involved in protein folding that binds the immunosuppressive drugs FK506 and rapamycin (22, 23); and arrestin-beta 2, involved in agonist-mediated desensitization of G protein-coupled receptors (24). This set of genes, which could be markers for the disease, may be able to detect changes well before clinical symptoms appear.
Fig. 4.
Genes differentially expressed between scleroderma and normal skin biopsies. A Wilcoxon rank sum test (19) was used to select genes that showed significant differences in expression between scleroderma skin biopsies and normal skin biopsies. Shown are the P values for each gene that have been corrected for multiple testing (P < 0.13). The full list of genes differentially expressed between scleroderma and normal biopsies is available in Fig. 6.
Gene Expression in Fibroblasts from Scleroderma Skin Biopsies. Because fibroblasts have commonly been used as a model system for scleroderma, we attempted to ascertain whether we could detect differences in the gene expression of fibroblasts from patients with scleroderma, morphea and normal controls (Table 2, which is published as supporting information on the PNAS web site). Fibroblasts were grown from either back or forearm biopsies from six patients with a diagnosis of SSc with diffuse scleroderma (10 samples; Table 2) and from back or forearm biopsies of three normal controls (four samples). Four additional sets of fibroblasts were derived from affected and unaffected skin biopsies of patients with a diagnosis of morphea (four samples); 18 fibroblast cell cultures were analyzed in total.
In contrast to our results with the whole skin biopsies, we could not discern obvious differences in the patterns of gene expression between fibroblasts derived from different disease tissues. We used a heuristic selection for genes that change 2-fold from their median value in at least three of the microarrays analyzed; 818 genes were selected, and the samples were analyzed by hierarchical clustering (data not shown). Unlike the results with skin biopsies, when the cell lines were ordered by their patterns of gene expression with hierarchical clustering, no clear pattern of clustering according to disease status could be discerned. To determine whether we could detect differences in the fibroblasts by using supervised analysis methods the fibroblasts were analyzed with the Wilcoxon rank sum test (RST) (19). Using the RST (Bonferroni-corrected), we were unable to detect differentially expressed genes with significant P values. Similar results were obtained with Statistical Analysis of Microarrays (SAM) (25).
It is important to note that we have extracted only a fraction of the information inherent to this data set. The entire data set is now available for any purpose from http://genome-www.stanford.edu/scleroderma. We anticipate that comparison of these data with future studies will be illuminating, as has been the experience with studies of gene expression in cancer.
Discussion
Systematic analysis of the patterns of gene expression in biopsies from four scleroderma patients and four normal individuals reveals what appears to be consistent differences in gene expression. Surprisingly, these differences were apparent irrespective of the clinical appearance of the skin at the biopsy site. We conclude that scleroderma has systemic manifestations relatively early in the course of the disease that can be measured in what is considered, healthy clinically unaffected skin. It should be noted that each of the four patients had some internal organ involvement (Table 1), a clinical indication that their disease was systemic at the time the biopsies were taken.
At the level of cell-type composition in the skin biopsies, we found clear differences. It is clear that some of these differences involve infiltration of cell types absent or less prevalent in unaffected skin: namely, B lymphocytes. Our data indicate that these may be primarily CD20+ B cells. An early study of infiltrates in scleroderma skin by Fleischmajer and coworkers (16) noted perivascular, perieccrine, or diffuse infiltrates in 50% of systemic scleroderma patients studied. Although the types of inflammatory cells present are reported to be lymphocytes (T and B cell), plasma cells and macrophages, the cell population was not determined by immunohistochemistry (16). A later study also found infiltrates in 50% of scleroderma forearm, skin biopsies (15). Investigation of the lymphocyte subsets by immunhistochemistry with mAbs indicated the infiltrates were primarily T cells (15). The reason for our difference in results is not clear. Interestingly, a recent study of B lymphocyte signaling in the tight-skin mouse, an animal model of fibrosis, suggests a pathological role for B cells in the skin fibrosis observed there (26). A study of gene expression in renal allograft rejection, which is thought to be T cell mediated, also produced the unexpected finding of dense clusters of CD20+ B cells; the presence of B cells was strongly correlated with severe graft rejection (27). Our sample size is not large enough to draw definitive conclusions, but rather suggest this issue should be revisited with a much larger study on the role of B cells in scleroderma.
In the case of other cell types, such as fibroblasts and endothelial cells, we believe it is unlikely that the increase in cell type-specific gene expression results from an increase in the number of cells. Increases in the number of fibroblasts can be determined by light microscopy as illustrated by scleromyxedema (28) and the more recently defined nephrogenic fibrosing dermopathy (29), which are characterized by dermal fibroblast proliferation. No such change is observed in our scleroderma biopsies (Fig. 7).
We found, as expected, significant increases in expression of genes associated with extracellular matrix. The genes involved are clearly expressed in fibroblasts, but many may be expressed strongly in other cell types as well. We did not find a disease-related difference in gene expression in fibroblasts that grow out of the skin biopsies. Previous studies of gene expression in scleroderma have focused on the differences among fibroblasts grown from biopsies in culture (8, 30). Despite the evidence for diversity in gene expression patterns among fibroblasts in the body that remains evident in culture (31), we found no such difference in scleroderma. This result is similar to our experience with studies of cancer; tissue samples consistently show larger differences in gene expression then those observed among cell lines from similar tumors (e.g., breast tumors) (32). It is possible that variation in culture conditions (e.g., the serum response) obscures any differences that may be present between the scleroderma and normal fibroblasts, although we think this is unlikely because we cannot detect differences between the two populations when the cells are grown in low serum conditions.
Finally, we believe that a major value of this study, one that will require extension and replication, is the ability to identify patterns of gene expression that readily distinguish normal from scleroderma skin, and possibly specific overexpression or underexpression of specific molecular markers that correlate with disease progression and prognosis. This approach has the advantage of identifying multiple genes representing multiple different cell types in a complex disease whose underlying pathogenesis is still unknown. From a research perspective, it may give rise to new hypotheses about pathogenesis that can be tested. From a clinical point of view, it is possible that the changes in the skin are detectable before the other systemic manifestations. If so, then early diagnosis and intervention may improve the lives of patients affected with the disease.
Supplementary Material
Acknowledgments
This paper is dedicated to the late Sharon Monsky, without whose passionate advocacy this study would never have been undertaken. We thank Xiaoquin Dou and Chandi Griffin for technical assistance, Dr. Sean Bohen for the gift of normal tonsil and follicular lymph node RNA, Dr. Wen-Kai Weng and Dr. Ronald Levy for the HF-1 B cell line, John C. Matese for assistance with the web supplement, and Dr. Howard Y. Chang for critical reading of the manuscript. The Affymetrix arrays were processed in the Core Genomics Laboratory of the General Clinical Research Center at San Francisco General Hospital (RR00083-41). M.L.W., D.R.F., J.-T.C., and M.K.C. were supported by funds from the Scleroderma Research Foundation. M.L.W. is supported by National Research Service Award Postdoctoral Fellowship HG00220 from the National Human Genome Research Institute. J.I.M. is a Howard Hughes Medical Institute Predoctoral Fellow. P.O.B. is an Investigator of the Howard Hughes Medical Institute.
Abbreviation: SSc, systemic sclerosis.
References
- 1.Mayes, M. D. (1998) Semin. Cutan. Med. Surg. 17, 22–26. [DOI] [PubMed] [Google Scholar]
- 2.Seibold, J. R. (2001) in Kelley's Textbook of Rheumatology, eds. Ruddy, S., Harris, E. D. & Sledge, C. B. (Saunders, Philadelphia), pp. 1211–1239.
- 3.Steen, V. D. & Medsger, T. A., Jr. (2000) Arthritis Rheum. 43, 2437–2444. [DOI] [PubMed] [Google Scholar]
- 4.Ferri, C., Valentini, G., Cozzi, F., Sebastiani, M., Michelassi, C., La Montagna, G., Bullo, A., Cazzato, M., Tirri, E., Storino, F., et al. (2002) Medicine (Baltimore) 81, 139–153. [DOI] [PubMed] [Google Scholar]
- 5.Scussel-Lonzetti, L., Joyal, F., Raynauld, J. P., Roussin, A., Rich, E., Goulet, J. R., Raymond, Y. & Senecal, J. L. (2002) Medicine (Baltimore) 81, 154–167. [DOI] [PubMed] [Google Scholar]
- 6.Masi, A. T. (1980) Arthritis Rheum. 23, 581–590. [DOI] [PubMed] [Google Scholar]
- 7.Leroy, E. C., Black, C., Fleischmajer, R., Jablonska, S., Krieg, T., Medsger, T. A., Jr., Rowell, N. & Wollheim, F. (1988) J. Rheumatol. 15, 202–205. [PubMed] [Google Scholar]
- 8.Leask, A., Abraham, D. J., Finlay, D. R., Holmes, A., Pennington, D., Shi-Wen, X., Chen, Y., Venstrom, K., Dou, X., Ponticos, M., et al. (2002) Arthritis Rheum. 46, 1857–1865. [DOI] [PubMed] [Google Scholar]
- 9.Whitfield, M. L., Sherlock, G., Saldanha, A. J., Murray, J. I., Ball, C. A., Alexander, K. E., Matese, J. C., Perou, C. M., Hurt, M. M., Brown, P. O., et al. (2002) Mol. Biol. Cell 13, 1977–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spellman, P. T., Sherlock, G., Zhang, M. Q., Iyer, V. R., Anders, K., Eisen, M. B., Brown, P. O., Botstein, D. & Futcher, B. (1998) Mol. Biol. Cell 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., et al. (2000) Nat. Genet. 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pruitt, K. D. & Maglott, D. R. (2001) Nucleic Acids Res. 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross, D. T., Scherf, U., Eisen, M. B., Perou, C. M., Rees, C., Spellman, P., Iyer, V., Jeffrey, S. S., van de Rijn, M., Waltham, M., et al. (2000) Nat. Genet. 24, 227–235. [DOI] [PubMed] [Google Scholar]
- 15.Roumm, A. D., Whiteside, T. L., Medsger, T. A., Jr., & Rodnan, G. P. (1984) Arthritis Rheum. 27, 645–653. [DOI] [PubMed] [Google Scholar]
- 16.Fleischmajer, R., Perlish, J. S. & Reeves, J. R. (1977) Arthritis Rheum. 20, 975–984. [DOI] [PubMed] [Google Scholar]
- 17.Sorlie, T., Perou, C. M., Tibshirani, R., Aas, T., Geisler, S., Johnsen, H., Hastie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alizadeh, A. A., Eisen, M. B., Davis, R. E., Ma, C., Lossos, I. S., Rosenwald, A., Boldrick, J. C., Sabet, H., Tran, T., Yu, X., et al. (2000) Nature 403, 503–511. [DOI] [PubMed] [Google Scholar]
- 19.Troyanskaya, O. G., Garber, M. E., Brown, P. O., Botstein, D. & Altman, R. B. (2002) Bioinformatics 18, 1454–1461. [DOI] [PubMed] [Google Scholar]
- 20.Coppolino, M. G. & Dedhar, S. (1998) Int. J. Biochem. Cell Biol. 30, 553–558. [DOI] [PubMed] [Google Scholar]
- 21.Cabral, C. M., Liu, Y. & Sifers, R. N. (2001) Trends Biochem. Sci. 26, 619–624. [DOI] [PubMed] [Google Scholar]
- 22.Standaert, R. F., Galat, A., Verdine, G. L. & Schreiber, S. L. (1990) Nature 346, 671–674. [DOI] [PubMed] [Google Scholar]
- 23.Maki, N., Sekiguchi, F., Nishimaki, J., Miwa, K., Hayano, T., Takahashi, N. & Suzuki, M. (1990) Proc. Natl. Acad. Sci. USA 87, 5440–5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefkowitz, R. J. (1998) J. Biol. Chem. 273, 18677–18680. [DOI] [PubMed] [Google Scholar]
- 25.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito, E., Fujimoto, M., Hasegawa, M., Komura, K., Hamaguchi, Y., Kaburagi, Y., Nagaoka, T., Takehara, K., Tedder, T. F. & Sato, S. (2002) J. Clin. Invest. 109, 1453–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarwal, M., Chua, M. S., Kambham, N., Hsieh, S. C., Satterwhite, T., Masek, M. & Salvatierra, O. (2003) N. Engl. J. Med. 349, 125–138. [DOI] [PubMed] [Google Scholar]
- 28.Pomann, J. & Rudner, E. (2003) Int. J. Dermatol. 42, 31–35. [DOI] [PubMed] [Google Scholar]
- 29.Cowper, S., Su, L., Bhawan, J., Robin, H. & LeBoit, P. (2001) Am. J. Dermatopathol. 23, 383–393. [DOI] [PubMed] [Google Scholar]
- 30.Zhou, X., Tan, F. K., Xiong, M., Milewicz, D. M., Feghali, C. A., Fritzler, M. J., Reveille, J. D. & Arnett, F. C. (2001) J. Immunol. 167, 7126–7133. [DOI] [PubMed] [Google Scholar]
- 31.Chang, H. Y., Chi, J. T., Dudoit, S., Bondre, C., van de Rijn, M., Botstein, D. & Brown, P. O. (2002) Proc. Natl. Acad. Sci. USA 99, 12877–12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perou, C. M., Jeffrey, S. S., van de Rijn, M., Rees, C. A., Eisen, M. B., Ross, D. T., Pergamenschikov, A., Williams, C. F., Zhu, S. X., Lee, J. C., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 9212–9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eisen, M. B. & Brown, P. O. (1999) Methods Enzymol. 303, 179–205. [DOI] [PubMed] [Google Scholar]
- 34.Fisk, H. A. & Winey, M. (2001) Cell 106, 95–104. [DOI] [PubMed] [Google Scholar]
- 35.Sherlock, G., Hernandez-Boussard, T., Kasarskis, A., Binkley, G., Matese, J. C., Dwight, S. S., Kaloper, M., Weng, S., Jin, H., Ball, C. A., et al. (2001) Nucleic Acids Res. 29, 152–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.