Abstract
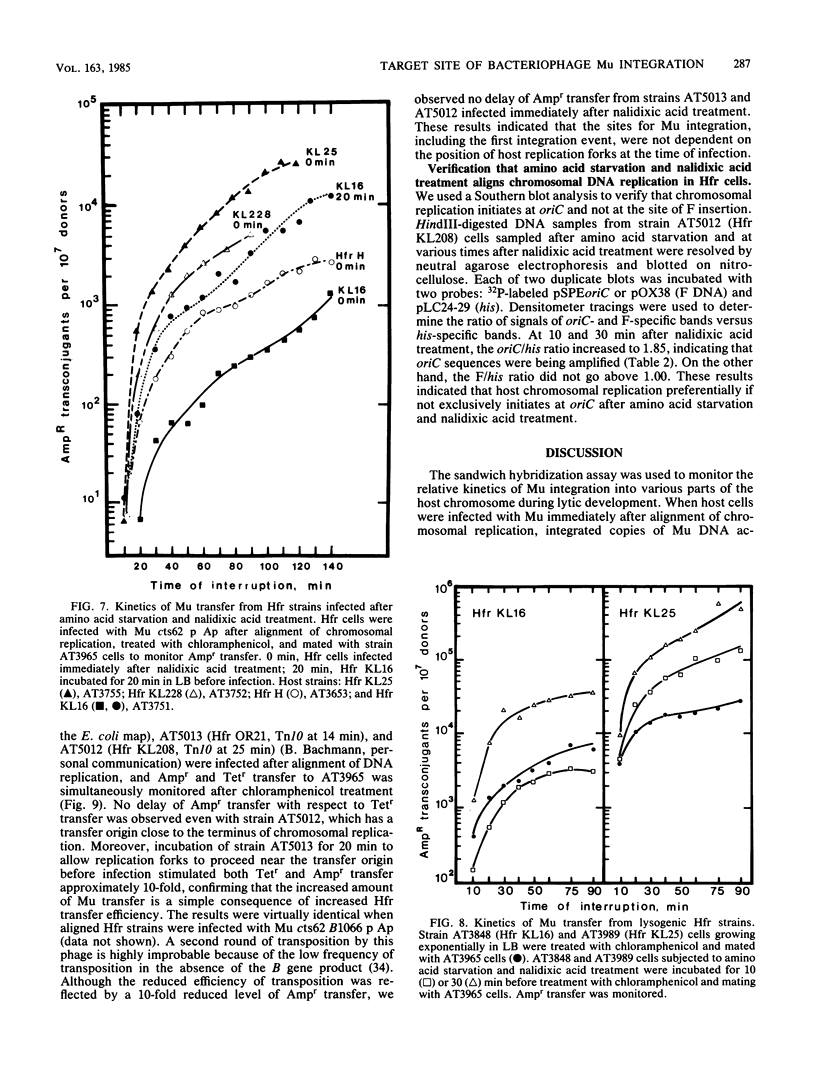
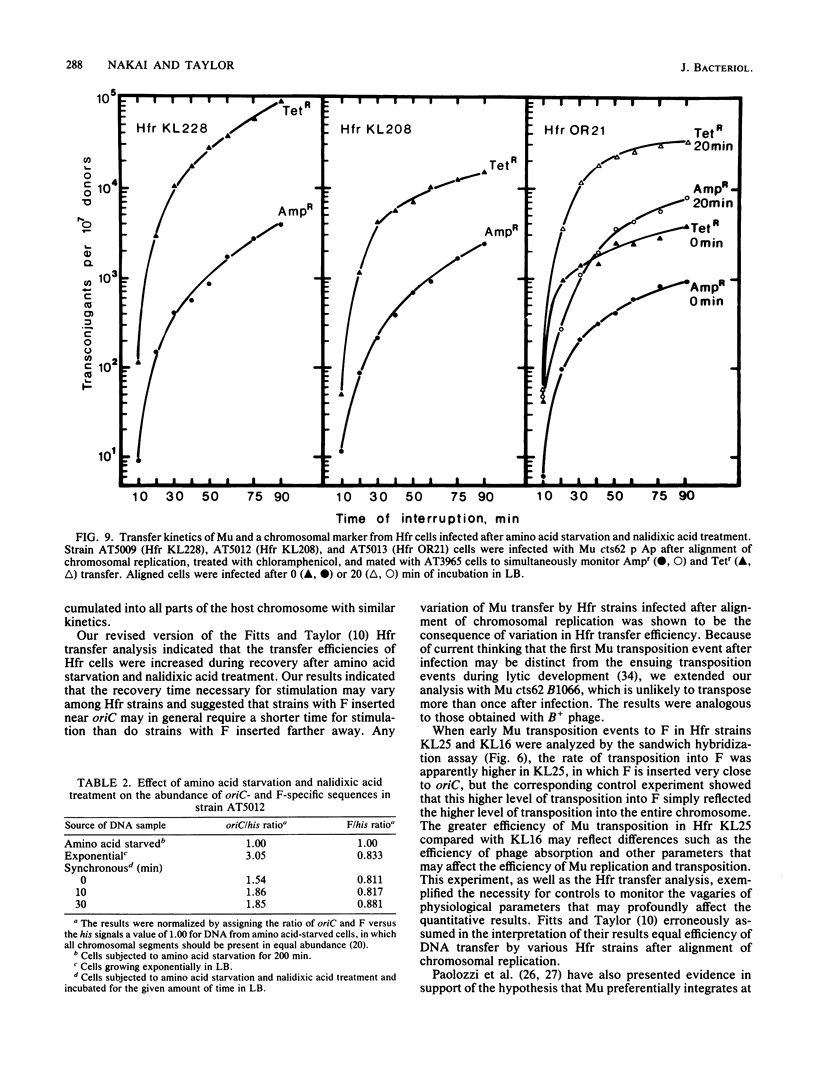
Bacteriophage Mu DNA integration in Escherichia coli strains infected after alignment of chromosomal replication was analyzed by a sandwich hybridization assay. The results indicated that Mu integrated into chromosomal segments at various distances from oriC with similar kinetics. In an extension of these studies, various Hfr strains were infected after alignment of chromosomal replication, and Mu transposition was shut down early after infection. The positions of integrated Mu copies were inferred from the transfer kinetics of Mu to an F- strain. Our analysis indicated that the location of Mu DNA in the host chromosome was not dependent on the positions of host replication forks at the time of infection. However, the procedure for aligning chromosomal replication affected DNA transfer by various Hfr strains differently, and this effect could account for prior results suggesting preferential integration of Mu at host replication forks.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C., Kemp D. J., Stark G. R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell E., Kohne D. E., Abelson J. Characterization of the inhomogeneous DNA in virions of bacteriophage Mu by DNA reannealing kinetics. J Virol. 1975 Apr;15(4):739–743. doi: 10.1128/jvi.15.4.739-743.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Fitts R. A., Taylor A. L. Integration of bacteriophage Mu at host chromosomal replication forks during lytic development. Proc Natl Acad Sci U S A. 1980 May;77(5):2801–2805. doi: 10.1073/pnas.77.5.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Kuempel P. L. Deoxyribonucleic acid-deoxyribonucleic acid hybridization assay for replication origin deoxyribonucleic acid of Escherichia coli. J Bacteriol. 1972 Jun;110(3):917–925. doi: 10.1128/jb.110.3.917-925.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach D., Symonds N. The isolation and characterisation of a plaque-forming derivative of bacteriophage Mu carrying a fragment of Tn3 conferring ampicillin resistance. Mol Gen Genet. 1979 May 4;172(2):179–184. doi: 10.1007/BF00268280. [DOI] [PubMed] [Google Scholar]
- Ljungquist E., Khatoon H., DuBow M., Ambrosio L., De Bruijn F., Bukhari A. I. Integration of bacteriophage mu DNA. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1151–1158. doi: 10.1101/sqb.1979.043.01.130. [DOI] [PubMed] [Google Scholar]
- Low B. Rapid mapping of conditional and auxotrophic mutations in Escherichia coli K-12. J Bacteriol. 1973 Feb;113(2):798–812. doi: 10.1128/jb.113.2.798-812.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low K. B. Escherichia coli K-12 F-prime factors, old and new. Bacteriol Rev. 1972 Dec;36(4):587–607. doi: 10.1128/br.36.4.587-607.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh R. C., Hepburn M. L. Inititation and termination of chromosome replication in Escherichia coli subjected to amino acid starvation. J Bacteriol. 1980 Apr;142(1):236–242. doi: 10.1128/jb.142.1.236-242.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBeth D. L., Taylor A. L. Growth of bacteriophage Mu in Escherichia coli dnaA mutants. J Virol. 1982 Nov;44(2):555–564. doi: 10.1128/jvi.44.2.555-564.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Neidhardt F. C., Vaughn V., Phillips T. A., Bloch P. L. Gene-protein index of Escherichia coli K-12. Microbiol Rev. 1983 Jun;47(2):231–284. doi: 10.1128/mr.47.2.231-284.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgard M. V., Emigholz K., Monahan J. J. Increased amplification of pBR322 plasmid deoxyribonucleic acid in Escherichia coli K-12 strains RR1 and chi1776 grown in the presence of high concentrations of nucleoside. J Bacteriol. 1979 Apr;138(1):270–272. doi: 10.1128/jb.138.1.270-272.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolozzi L., Ghelardini P., Kepes A., Marcovich H. The mechanism of integration of phage Mu in the chromosome of Escherichia coli. Biochem Biophys Res Commun. 1979 May 14;88(1):111–116. doi: 10.1016/0006-291x(79)91703-0. [DOI] [PubMed] [Google Scholar]
- Paolozzi L., Jucker R., Calef E. Mechanism of phage Mu-1 integration: nalidixic acid treatment causes clustering of Mu-1-induced mutations near replication origin. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4940–4943. doi: 10.1073/pnas.75.10.4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pato M. L., Reich C. Instability of transposase activity: evidence from bacteriophage mu DNA replication. Cell. 1982 May;29(1):219–225. doi: 10.1016/0092-8674(82)90106-4. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sadler J. R., Tecklenburg M., Betz J. L., Goeddel D. V., Yansura D. G., Caruthers M. H. Cloning of chemically synthesized lactose operators. Gene. 1977 Jul;1(5-6):305–321. doi: 10.1016/0378-1119(77)90036-1. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- TAYLOR A. L. BACTERIOPHAGE-INDUCED MUTATION IN ESCHERICHIA COLI. Proc Natl Acad Sci U S A. 1963 Dec;50:1043–1051. doi: 10.1073/pnas.50.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teifel J., Schmieger H. The influence of host DNA replication on the formation of infectious and transducing Mu-particles. Mol Gen Genet. 1981;184(2):308–311. doi: 10.1007/BF00272922. [DOI] [PubMed] [Google Scholar]
- Vasil M. L., Berka R. M., Gray G. L., Nakai H. Cloning of a phosphate-regulated hemolysin gene (phospholipase C) from Pseudomonas aeruginosa. J Bacteriol. 1982 Oct;152(1):431–440. doi: 10.1128/jb.152.1.431-440.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]