Abstract
Recognition of human erythrocytes by Plasmodium species depends in part on Region II of the Duffy binding-like family of parasite ligands, which includes BA erythrocyte binding ligand (BAEBL) of P. falciparum. In previous studies of BAEBL from two clones, Dd2/Nm from Vietnam and E12 from Papua New Guinea (PNG), it was found that BAEBL bound different erythrocyte receptors. Because of variation in binding specificity, we studied the sequence and erythrocyte binding specificity of Region II of BAEBL in P. falciparum clones from different parts of the world. We observed five nucleotide substitutions leading to five amino acid changes and five polymorphisms in Region II of BAEBL in parasites from both PNG and other parts of the world. We expressed four of the polymorphisms on COS cells and determined their binding to enzyme-treated erythrocytes and to Gerbich-negative erythrocytes. We also performed erythrocyte-binding assay using the native protein from radiolabeled culture supernatant. Both assays demonstrated that each of the four polymorphisms in the parasite ligand, BAEBL, bound to a different receptor on erythrocytes. These results suggest that P. falciparum has evolved multiple invasion pathways dependent on polymorphisms in the BAEBL ligand.
Keywords: Plasmodium falciparum malaria, erythrocyte, parasite polymorphism, Gerbich negative, BAEBL
Introduction
The binding of merozoites from Plasmodium species to erythrocytes is dependent on two families of parasite ligands, the Duffy binding-like (DBL) and the reticulocyte binding-like (RBL) (1–3). Both are named for the erythrocyte receptors for Plasmodium vivax: the Duffy blood group proteins and a reticulocyte-specific receptor (4). Unlike P. vivax that is limited to Duffy positive reticulocytes, Plasmodium falciparum has greater flexibility in its invasion pathways (5). P. falciparum invades all aged erythrocytes; no human erythrocytes are known to be refractory to all P. falciparum clones. This flexibility is believed to result, in part, from multiple copies of DBL and RBL genes in P. falciparum (5). The erythrocyte specificities of the DBL and RBL genes are completely different for P. falciparum than for P. vivax. The DBL family in P. falciparum includes erythrocyte binding antigen-175 (EBA-175) and its para-logue, BA erythrocyte binding ligand (BAEBL; reference 1). Whereas EBA-175 bound only glycophorin A (6), BAEBL from two clones of P. falciparum had different erythrocyte receptors (7, 8). One clone, Dd2/Nm from Vietnam, failed to bind trypsin-treated erythrocytes and Gerbich-negative erythrocytes (7), a common human mutation of glycophorin C/D in Papua New Guinea (PNG; references 9 and 10). Another clone, E12 from PNG, bound to trypsin-treated erythrocytes, indicating that it used a receptor other than glycophorin C/D (8). One possibility was that the variation in receptors could result from mutations in BAEBL selected in PNG. Therefore we sequenced the erythrocyte binding domain of BAEBL (Region II) from multiple clones of P. falciparum isolated from PNG and from other parts of the world and determined the effect of polymorphisms in Region II of BAEBL on its erythrocyte binding specificity.
Materials and Methods
Parasite Clones Studied.
The isolates used in this study were obtained from Papua New Guinea: PNG 2, 3, 4, 13 (11), 5, 9–1, 9–3 and 10–1 (a gift from Russell Howard, Maxygen Corp., Redwood City, CA), 1917 and 1905 (a gift from Karen Day, Oxford University, Oxford, UK), E12 (12); from Africa: 3D7 isolated from the Amsterdam airport (13) but has microsatellites of an African parasite, M24, Fab9, Sc/D6; from South America: PC49, 7G8, DIV30, PC26 (11), HB3(14); and from Asia: Camp, T2/C6, MT/S-1, Dd2 (11), Dd2/Nm (15). Genotypes of the parasite lines were confirmed by microsatellite analysis (16). Parasite culture and DNA isolation were performed as described previously (17). Two P. falciparum clones, HB3 and Camp, were used for the erythrocyte binding assay described below.
Sequence Analysis of BAEBL Genes.
Open reading frame sequences of BAEBL were amplified from genomic DNA. Oligonucleotide primer sequences were 5′-TATCGTTTTTTTATGA GCAT-3′ and 5′-GTCAGAATAGGTACAATATT-3′. After treatment with SAP/ExoI (United States Biochemical Corp.), polymerase chain reaction products were directly sequenced using BigDye terminator chemistry on an ABI3100 sequencer (Applied Biosystems). DNA sequences were aligned using Sequencher 3.1 (Gene Codes Corp.) or AssemblyLIGN software (Oxford Molecular Ltd.).
Expression of Region II and its F1 and F2 Subregions on COS7 Cells.
Region II of BAEBL was cloned into the T8 vector (18). This vector contains a signal sequence and a glycosylphosphatidylinositol anchor that attaches the fusion protein to the surface of COS7 cells. Insertion of the variant sequences was accomplished by amplification with forward primer (5′-CGTGCGGCCGCCAATATACGTTTATACAGAAACGTACTCATTTGTTTGCT-3′) and reverse primer (5′-AGTGAATTCTATATCGTGTTTTGTTTTAGGATATTTA-3′) to generate the NotI and EcoRI sites, respectively. Insertion of the F1 subregion of BAEBL from P. falciparum clone Dd2/Nm was accomplished with forward primer (5′-CGTGCGGCCGCCAATATACGTTTATACAGAAACGTACTCATTTGTTTGCT-3′) and reverse primer (5′-AGTGAATTCGCAATCACATAAATCATCATATTCCTTTTCATTTTTG-3′). Insertion of the F2 subregion of BAEBL P. falciparum clone Dd2/Nm was accomplished with forward primer (5′-CGTGCGGCCGCAGATATACTGCTACTATTATTAAAAGTTTTCTAAATGGTC-3′) and reverse primer (5′-AGTGAATTCTATATCGTGTTTTGTTTTAGGATATTTA-3′). After a hot start at 94°C for 2 min, samples were cycled 30 times at 94°C for 30 s, 55°C for 50 s, and 72°C for 3 min, followed by a final 10-min incubation at 72°C. DBL2, a Duffy-like domain of P. falciparum erythrocyte membrane protein 1 (PfEMP-1), cloned in the T8 vector was used as a negative control in every experiment for the erythrocyte binding assay (19). This domain is expressed on the surface of COS7 cells and never binds erythrocytes.
Cell Culture and Transfection of COS7 Cells.
COS7 cells (American Type Culture Collection) were cultured in DMEM with 10% fetal calf serum (Invitrogen) in a humidified 5% CO2 incubator at 37°C. Cells (1.5 × 105) were seeded at 30% confluency onto 12-mm diameter glass coverslips of thickness between 0.13 and 0.17 mm (Fisher Scientific) in 3.5-cm diameter wells and transfected with 5 μg of plasmid DNA by the calcium phosphate precipitation method (20). The cells were washed three times with PBS 12–16 h after transfection. The transfected cells were used for immunofluorescence or erythrocyte binding assays 48 h after transfection.
Enzyme Treatment of Erythrocytes.
Whole blood was collected in 10% citrate phosphate dextrose and stored at 4°C. The erythrocytes were washed three times in RPMI 1640 (Invitrogen) containing 25 mM HEPES and 0.36 mM hypoxantine, pH 7.4 (Sigma-Aldrich) before being treated twice in 25 U of neuraminidase (Vibrio cholerae; Calbiochem) in RPMI 1640 for 2 h at 37°C. Erythrocytes were treated twice with 1 mg/ml of trypsin-TPCK (Sigma-Aldrich) for 2 h at 37°C. Trypsin activity was inhibited with 1 mg/ml soybean trypsin inhibitor (Sigma-Aldrich). Treated erythrocytes were resuspended to an hematocrit of 10% in DMEM with 10% fetal calf serum.
Erythrocyte Binding Assay to Transfected COS7 Cells.
A 10% erythrocyte suspension (40 μl) was added to transfected COS7 cells grown on coverslips (21). The erythrocytes were allowed to settle for 2 h at 37°C. The cells were washed by inversion of coverslips onto a glass support for 20 min in PBS. Transfected COS7 cells were scored using an inverted microscope. Erythrocyte binding assay was followed by an Immunofluorescence assay as described below.
Immunofluorescence Assay.
Transfected cells were assayed for surface expression 48 h after transfection. The cells were washed in PBS and fixed in 3.7% formaldehyde for 5 min at room temperature. This was followed by three washes in PBS for 5 min each at room temperature and staining with mouse mAb 179 for 1 h at room temperature at a final concentration of 4.1 ng/μl. The epitope is directed at an amino acid sequence in the T8 vector located at COOH terminus of Region II of BAEBL (18). Expression of Region II of BAEBL was also determined using the rat polyclonal antibody generated against Region II (7). The cells were washed three times for 5 min in PBS and visualized with fluorescein-conjugated goat anti–mouse Ab or goat anti–rat Alexa 488 (Jackson ImmunoResearch Laboratories/Molecular Probes) at a dilution of 1:100 in 10% normal goat serum (Jackson ImmunoResearch Laboratories). Excess secondary Ab was removed by washing three times in PBS for 5 min and mounted with Prolong Antifade (Molecular Probes).
Modified Erythrocyte Binding Assay.
Metabolically labeled supernatant from P. falciparum clones HB3 and Camp were used in absorption and elution assays described previously (7).
Results and Discussion
Initial characterization of erythrocyte receptors recognized by BAEBL from two different P. falciparum clones in two different laboratories suggested that each had a different erythrocyte receptor (7, 8). We sequenced the baebl gene from eight parasite clones and found polymorphisms restricted to Regions I and II of the molecules (GenBank/EMBL/DDBJ accession nos. AF605832, AF605833, AF605834, and AF424236). In total, we sequenced Regions I and II of BAEBL from 11 clones from PNG and 14 clones from other parts of the world where P. falciparum malaria is endemic. We observed five different sequence variants in Region II with polymorphisms in four amino acid positions (Table I). Four of the same sequence variants occurred in PNG as in the rest of the world (Table I). Because P. falciparum populations in PNG are isolated and there is no evidence of population admixture with parasites from other parts of the world (11), the BAEBL sequence variants found in PNG are likely to be from independent founder mutations.
Table I.
Position of Amino Acid Polymorphisms in BAEBL from Different Malaria-endemic Regions
| Region Ia
|
Region IIa
|
||||
|---|---|---|---|---|---|
| Clones | 112 | 185 | 239 | 261 | 285 |
| Papua New Guinea | |||||
| PNG2, PNG9-1 | L(CTT) | V(GTT) | S(AGT) | T(ACG) | K(AAA) |
| PNG4, PNG9-3, 1917, 1905 | L(CTT) | V(GTT) | S(AGT) | K(AAG) | K(AAG) |
| PNG13 | F(TTT) | V(GTT) | S(AGT) | K(AAG) | K(AAG) |
| PNG5, PNG10-1 | L(CTT) | I(ATT) | S(AGT) | K(AAG) | K(AAA) |
| E12 | F(TTT) | I(ATT) | S(AGT) | K(AAG) | K(AAA) |
| PNG3 | F(TTT) | I(ATT) | N(AAT) | R(AGG) | E(GAA) |
| Other areas of the world | |||||
| Dd2, Dd2/Nm (Vietnam) 7G8, DIV30 (Brazil) PC26 (Peru) |
L(CTT) | V(GTT) | S(AGT) | T(ACG) | K(AAA) |
| Sc/D6 (Sierra Leone) HB3 (Honduras) |
L(CTT) | V(GTT) | S(AGT) | K(AAG) | K(AAA) |
| MT/S-1 | F(TTT) | I(ATT) | S(AGT) | K(AAG) | K(AAA) |
| M24 (Kenya), 3D7 (Africa) PC49 (Peru), T2/C6 (Thailand) |
F(TTT) | I(ATT) | N(AAT) | K(AAG) | K(AAA) |
| Fab9 (Kwazulu) Camp (Malaysia) |
L(CTT) | I(ATT) | N(AAT) | R(AGG) | E(GAA) |
Polymorphisms in bases and amino acids are shown in bold.
For Regions I and II, numbers refer to the amino acids in the sequence of BAEBL from GenBank/EMBL/DDBJ accession no. AF33918.
Region II of P. falciparum DBL genes (1) is duplicated, forming the F1 and F2 domains. All base substitutions in Region II of BAEBL occurred in the F1 domain (Fig. 1) . EBA-175, another DBL gene of P. falciparum, differed from BAEBL in that mutations were scattered throughout both the F1 and F2 domains (22; Fig. 1). We expressed separately the F1 and the F2 region of BAEBL on the surface of COS7 cells and failed to observe erythrocyte binding to either region. As the sequence of F2 is conserved, it may be that both domains are required for erythrocyte binding.
Figure 1.
Location of polymorphisms in BAEBL and EBA-175. Erythrocyte-binding domain is duplicated into F1 (light gray) and F2 (dark gray). Positions of polymorphism in both BAEBL (this study) and EBA-175 (reference 21) are highlighted.
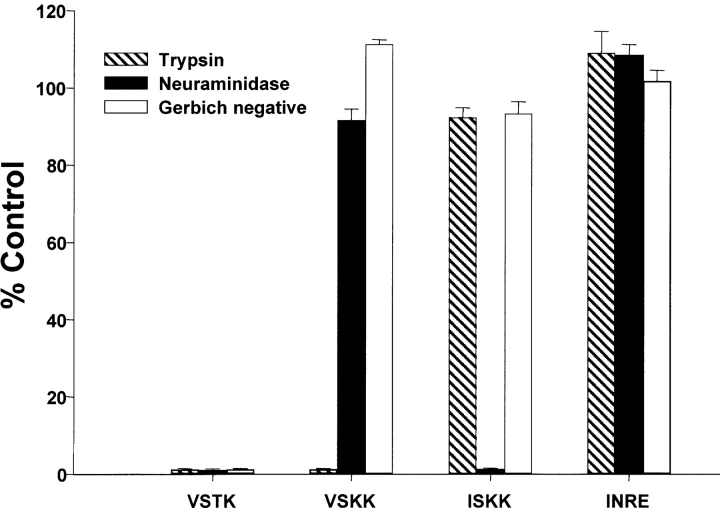
We investigated the functional significance of polymorphisms in the erythrocyte-binding domain (Region II) of BAEBL. We expressed transiently Region II of four polymorphic sequences on the surface of COS cells with the T8 vector. For all transfected constructs, positive surface expression was confirmed by immunofluorescence with two antibodies: one directed against the epitope on the COOH terminus of the heterologous protein and the other directed against Region II of BAEBL (7). Binding to the COS cells was performed with normal and enzyme-treated (trypsin and neuraminidase) erythrocytes and Gerbich-negative erythrocytes (exon 3 deletion of glycophorin C/D). Each of the four polymorphisms led to a different binding specificity as demonstrated by different binding patterns to enzyme-treated and Gerbich-negative erythrocytes (Fig. 2) . Region II from Dd2/Nm (VSTK) was sensitive to neuraminidase and trypsin and was the only sequence to bind poorly to Gerbich-negative erythrocytes; the other three sequences bound equally well to Gerbich-negative erythrocytes as to normal erythrocytes. At least three other patterns of binding specificity were observed: clone E12 (ISKK) bound trypsin-treated but not neuraminidase-treated erythrocytes; PNG3 (INRE) bound both neuraminidase- and trypsin-treated erythrocytes; and HB3 (VSKK) bound neuraminidase- but not trypsin-treated erythrocytes. We showed that a single base change leading to a single amino acid substitution in the erythrocyte-binding domain of BAEBL led to recognition of different molecules on human erythrocytes (VSTK to VSKK and VSKK to ISKK).
Figure 2.
Binding patterns of BAEBL variants to enzyme-treated and Gerbich-negative erythrocytes. Region II of BAEBL expressed in COS cells is from amino acid 143 to 606 (GenBank/EMBL/DDBJ accession no. AF332918) and contains the mutations shown at the positions delineated in Table I. COS cells with five or more attached RBCs were counted and the total per coverslip recorded. Data are shown as the mean of three independent experiments, and the error bar is the standard deviation. Data from enzyme-treated and Gerbich-negative erythrocytes are expressed as the percentage of binding to normal, untreated erythrocyte. The control normal untreated erythrocyte samples contained between 30 and 120 COS cells with bound erythrocytes. Two controls were included in each experiment; untransfected COS cells as well as COS cells transfected with DBL2, a domain in the adhesion molecule PfEMP1, expressed in the T8 vector did not bind either untreated or enzyme-treated erythrocytes.
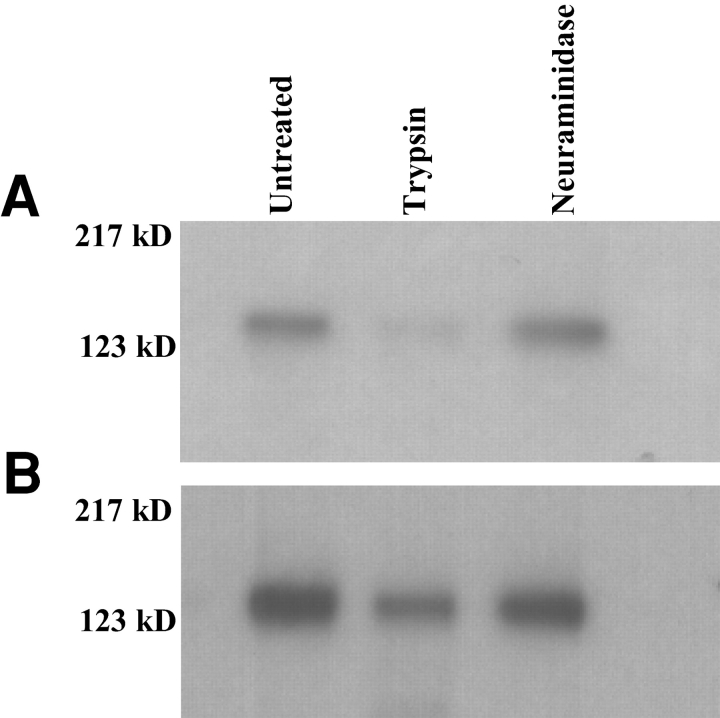
We next determined whether the specificity of binding of BAEBL observed on COS7 cells mirrors the binding of the native protein in culture supernatants. The E12 and Dd2/Nm sequences were derived from parasite clones that had been studied previously (7, 8). BAEBL immunoprecipitated from culture supernatants of E12 and Dd/Nm bound to erythrocytes with the specificity identical to that described for Region II expressed on COS cells, indicating that the two assays are measuring the same specificity (7, 8). Furthermore, BAEBL in culture supernatants from HB3 (VSKK) and Camp (INRE) clones also had identical binding specificities to enzyme treated erythrocytes as Region II of the corresponding sequences expressed on COS cells (Fig. 3) . The BAEBL variant from HB3 (VSKK) was not absorbed to neuraminidase-treated erythrocytes but was absorbed to trypsin-treated erythrocytes (unpublished data). Likewise, BAEBL variant from Camp (INRE) was not absorbed to either neuraminidase- or trypsin-treated erythrocytes (unpublished data). Thus, like Duffy of P. vivax (21) and EBA-175 of P. falciparum (6), Region II of BAEBL defines the erythrocyte binding specificity of the native protein.
Figure 3.
Immunoprecipitation of BAEBL eluted from untreated and enzyme treated erythrocytes. BAEBL immunoprecipitated from supernatant of P. falciparum clone HB3 (VSKK; A) and P. falciparum clone Camp (INRE; B) has similar binding characteristics as Region II of the corresponding BAEBL variants expressed on COS cells (see Fig. 2). The molecular weight standard is shown on the left.
Such polymorphism in sequence and receptors was described for influenza hemagglutinin selected for binding to sialic acid linked to galactose by α2,6 instead of α2,3 (23). The change in receptor specificity was caused by a point mutation that led to a single amino acid change in the receptor pocket. The amino acid changes that we have noted in BAEBL that led to changes in receptor specificity may be located in the receptor pocket, although the structure of Region II of BAEBL has yet to be solved. Mutations in Region II of EBA-175 do not affect its requirement for sialic acid (21). As EBA-175 only binds sialic acid in the context of glycophorin A (6), it remains to be determined whether mutations in EBA-175 affect binding to glycophorin A. Other highly polymorphic merozoite proteins such as apical membrane antigen 1 (AMA1) may encode variation in receptors, but these polymorphisms as for EBA-175 are more likely to play a role in immune evasion (21, 24, 25).
Members of multigene families (e.g., the DBL family of P. falciparum, the P. yoelii family of 230-kD proteins, and the var family of genes) have evolved new functions through gene duplication (6, 7, 26–30). Here, we describe the ability of a single P. falciparum gene to recognize various erythrocyte receptors depending on polymorphisms in the erythrocyte-binding domain.
Why has the parasite evolved to have such diversity? What is the advantage to the parasite to have multiple niches with different erythrocyte receptors? In PNG, an area of 50% allelic frequency of Gerbich-negative erythrocytes (9, 10), this diversity may allow the parasite to survive despite this human mutation in a BAEBL receptor. BAEBL polymorphisms occurred, however, in other regions besides PNG. Several possibilities may explain the diversity in receptors. First, Gerbich-negative cells may occur at high frequency in other parts of the world. This human polymorphism may have been missed because few reagents exist for detecting the Gerbich blood group, and they are all in private collections. In addition, there is little interest in this blood group system because variation does not cause erythroblastosis fetalis and is not responsible for transfusion reactions. With the availability of molecular tools to identify deletions in exon 3 that define the Gerbich-negative phenotype (10), it is now possible to determine whether this mutation has been missed in other malarious areas. Second, other mutations in glycophorin C/D may occur in other endemic areas.
A third possibility is that the polymorphism in parasite ligands has an advantage unrelated to Gerbich-negative human erythrocytes. Polymorphisms in erythrocyte-binding ligands may provide the parasite population versatility in the invasion pathways, thus increasing P. falciparum fitness. Our data suggest three other erythrocyte receptors for BAEBL besides the Gerbich blood group antigen. It is possible that polymorphisms also exist in these other BAEBL erythrocyte receptors. Polymorphisms in these may have driven the variation seen in BAEBL. This question awaits the identification of these erythrocyte receptors.
P. falciparum has evolved great flexibility in its invasion pathways. In part, this may result from the multiple copy DBL family of erythrocyte binding ligands, three of which have different erythrocyte receptor specificities (6, 7, and unpublished data). This is in contrast to P. vivax, which has a single copy of the DBL family that recognizes the Duffy blood group proteins, explaining the resistance to P. vivax infection in West Africa, where humans lack the Duffy blood group proteins on their erythrocytes. In addition to the multiple copies of DBL, we now describe another mechanism for recognition of different molecules on the erythrocytes by various forms of a single P. falciparum erythrocyte binding ligand.
References
- 1.Adams, J.H. O. Kaneko, P.L. Blair, and D.S. Peterson. 2001. An expanding ebl family of Plasmodium falciparum. Trends Parasitol. 17:297–299. [DOI] [PubMed] [Google Scholar]
- 2.Galinski, M.R., C. Corredor-Medina, P. Ingravello, and J.W. Barnwell. 1992. A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell. 69:1213–1226. [DOI] [PubMed] [Google Scholar]
- 3.Galinski, M.R., and J.W. Barnwell. 1996. Plasmodium vivax: Merozoites, invasion of reticulocytes and considerations for malaria vaccine development. Parasitol. Today. 12:20–29. [DOI] [PubMed] [Google Scholar]
- 4.Adams, J.H., B.H.K. Sim, S.A. Dolan, X. Fang, D.C. Kaslow, and L.H. Miller. 1992. A family of erythrocyte binding proteins of malaria parasites. Proc. Natl. Acad. Sci. USA. 89:7085–7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller, L.H., D.I. Baruch, K. Marsh, and O.K. Doumbo. 2002. The pathogenic basis of malaria. Nature. 415:673–679. [DOI] [PubMed] [Google Scholar]
- 6.Sim, B.K.L., C.E. Chitnis, K. Wasnioswska, T.J. Hadley, and L.H. Miller. 1994. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science. 264:1941–1944. [DOI] [PubMed] [Google Scholar]
- 7.Mayer, D.C.G., O. Kaneko, D.E. Hudson-Taylor, M.E. Reid, and L.H. Miller. 2001. Characterization of a Plasmodium falciparum erythrocyte-binding protein paralogous to EBA-175. Proc. Natl. Acad. Sci. USA. 98:5222–5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson, J.K., T. Triglia, M.B. Reed, and A.F. Cowman. 2001. A novel ligand from Plasmodium falciparum that binds to a sialic acid-containing receptor on the surface of human erythrocytes. Mol. Microbiol. 41:47–58. [DOI] [PubMed] [Google Scholar]
- 9.Booth, P.B., D. Tills, A. Warlow, A.C. Kopec, A.E. Mourant, P. Teesdale, and R.W. Hornabrook. 1982. Red cell antigen, serum protein and red cell enzyme polymorphisms in Karkar Islanders and inhabitants of the adjacent North Coast of New Guinea. Hum. Hered. 32:385–403. [DOI] [PubMed] [Google Scholar]
- 10.Patel, S., R.K. Mehlotra, W. Kastens, C.S. Mgone, J.W. Kazura, and P.A. Zimmerman. 2001. The association of the glycophorin C exon 3 deletion with ovalocytosis and malaria susceptibility in Wosera, Papua New Guinea. Blood. 98:3489–3491. [DOI] [PubMed] [Google Scholar]
- 11.Wootton, J.C., X. Feng, M.T. Ferdig, R.A. Cooper, J. Mu, D.I. Baruch, A.J. Magill and X.Z. Su. 2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 418:320–323. [DOI] [PubMed] [Google Scholar]
- 12.Anders, A.F., G.V. Brown, and A. Edwards. 1983. Characterization of an S antigen synthesized by several isolates of Plasmodium falciparum. Proc. Natl. Acad. Sci. USA. 80:6652–6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walliker, D., I.A. Quakyi, T.E. Wellems, T.F. McCutchan, A. Szarfman, W.T. London, L.M. Corcoran, T.R. Burkot, and R. Carter. 1987. Genetics analysis of the human malaria parasite Plasmodium falciparum. Science. 236:1661–1666. [DOI] [PubMed] [Google Scholar]
- 14.Bhasin, V.K., and W. Trager. 1984. Gametocyte-forming and non-gametocyte-forming clones of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 33:534–537. [DOI] [PubMed] [Google Scholar]
- 15.Dolan, S.A., L.H. Miller, and T.E. Wellems. 1990. Evidence for a switching mechanism in the invasion of erythrocytes by Plasmodium falciparum. J. Clin. Invest. 86:618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su, X.Z., D.J. Carucci, and T.E. Wellems. 1998. Plasmodium falciparum: parasite typing by using a multicopy microsatellite marker, PfRRM. J. Exp. Parasitol. 89:262–265. [DOI] [PubMed] [Google Scholar]
- 17.Su, X.Z., L.A. Kirkman, H. Fujioka, and T.E. Wellems. 1997. Complex polymorphisms in an approximately 330 kDa protein are linked to chloroquine-resistant falciparum in Southeast Asia and Africa. Cell. 91:593–603. [DOI] [PubMed] [Google Scholar]
- 18.Buffet, P.A., B. Gamain, C. Scheidig, D. Baruch, J.D. Smith, R. Hernandez-Rivas, B. Pouvelle, S. Oishi, N. Fujii, T. Fusai, et al. 1999. Plasmodium falciparum domain mediating adhesion to chondroitin sulfate A: a receptor for human placental infection. Proc. Natl. Acad. Sci. USA. 96:12743–12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su, X.Z., V.M. Heatwole, S.P. Wertheimer, F. Guinet, J.A. Herrfeldt, D.S. Peterson, J.A. Ravetch, and T.E. Wellems. 1995. The large diverse gene family var encodes protein that are involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 82:89–100. [DOI] [PubMed] [Google Scholar]
- 20.Graham, F.L., and A.J. van der Eb. 1973. A new assay of infectivity of human adenovirus 5 DNA. Virology. 52:456–467. [DOI] [PubMed] [Google Scholar]
- 21.Chitnis, C.E., and L.H. Miller. 1994. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J. Exp. Med. 180:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang, H., and B.K. Sim. 1997. Conservation of structure and function of the erythrocyte-binding domain of Plasmodium falciparum EBA-175. Mol. Biochem. Parasitol. 84:241–245. [DOI] [PubMed] [Google Scholar]
- 23.Rogers, G.N., J.C. Paulson, R.S. Daniels, J.J. Shekel, I.A. Wilson, and D.C. Wiley. 1983. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 304:76–78. [DOI] [PubMed] [Google Scholar]
- 24.Crewther, P.E., M.L. Matthew, R.H. Flegg, and R.F. Anders. 1996. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect. Immun. 64:3310–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall, V.M., L.-X. Zhang, R.F. Anders, and R.L. Coppel. 1996. Diversity of the vaccine candidate AMA-1 of Plasmodium falciparum. Mol. Biochem. Parasitol. 77:109–113. [DOI] [PubMed] [Google Scholar]
- 26.Ogun, S.A., and A.A. Holder. 1996. A highly molecular mass P. yoelii rhoptry protein binds to erythrocytes. Mol. Biochem. Parasitol. 76:321–324. [DOI] [PubMed] [Google Scholar]
- 27.Smith, J.D., C.E. Chitnis, A.G. Craig, D.J. Roberts, D.E. Hudson-Taylor, D.S. Peterson, and L.H. Miller. 1995. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 82:101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baruch, D.I., J.A. Gormley, C. Ma, R.J. Howard, and B.L. Paloske. 1996. Plasmodium falciparum erythrocyte membrane protein 1 is a parasitized erythrocyte receptor for adherence to CD36, thrombospondin, and intracellular adhesion molecule 1. Proc. Natl. Acad. Sci. USA. 93:3497–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rowe, A.J., J.M. Moulds, C.I. Newbold, and L.H. Miller. 1997. P. falciparum rosetting mediated by a parasite-variant erythrocyte membrane protein and complement receptor 1. Nature. 388:292–295. [DOI] [PubMed] [Google Scholar]
- 30.Preiser, P.R., S. Khan, F.T. Costa, W. Jarra, E. Belnoue, S. Ogun, A.A. Holder, T. Voza, I. Landau, G. Snounou, and L. Renia. 2002. Stage-specific transcription of different repertoires of a multigene family during Plasmodium life cycle. Science. 295:342–345. [DOI] [PubMed] [Google Scholar]