Abstract
Iron is an essential nutrient that is often found in extremely limited available quantities within eukaryotic hosts. Because of this, many pathogenic bacteria have developed regulated networks of genes important for iron uptake and storage. In addition, it has been shown that many bacteria use available iron concentrations as a signal to regulate virulence gene expression. We have utilized DNA microarray technology to identify genes of the human pathogen Helicobacter pylori that are differentially regulated on a growth-inhibiting shift to iron starvation conditions. In addition, the growth phase-dependent expression of these genes was investigated by examining both exponential and stationary growth phase cultures. We identified known iron-regulated genes, as well as a number of genes whose regulation by iron concentration was not previously appreciated. Included in the list of regulated factors were the known virulence genes cagA, vacA, and napA. We examined the effect of iron starvation on the motility of H. pylori and found that exponential- and stationary-phase cultures responded differently to the stress. We further found that while growing cells are rapidly killed by iron starvation, stationary-phase cells show a remarkable ability to survive iron depletion. Finally, bioinformatic analysis of the predicted promoter regions of the differentially regulated genes led to identification of several putative Fur boxes, suggesting a direct role for Fur in iron-dependent regulation of these genes.
The essential nature of iron for virtually all organisms is a double-edged sword that requires a careful balancing act to ensure survival. On the one hand, iron is a required cofactor of many enzymes involved in the maintenance of basic metabolic functions such as electron transport. On the other hand, interaction of free oxygen with iron leads to the Fenton reaction, which results in the production of oxygen radicals that can cause severe damage to the majority of cellular biomolecules (65). Due to these two opposing phenomena, it is perhaps no surprise that organisms have developed tightly regulated systems for both uptake and sequestration of iron. Within the context of mammalian systems this is accomplished in part through the action of iron binding molecules such as transferrin, heme and lactoferrin (42, 44). In addition to protecting cells from the harmful effects of oxygen radicals, the ability to sequester iron also limits bacterial growth within the mammalian host. This is evidenced by extensive epidemiological data suggesting that there is a strong link between the iron status of the host and susceptibility to a number of bacterial pathogens (12, 70).
In light of the requirement for iron and the stiff competition for this nutrient, it is no surprise that successful pathogenic microorganisms have developed intricate systems that allow them to acquire and store iron within the bacterial cell. In fact, genes that encode factors involved in these processes are often considered to be virulence factors and have been the subject of intense study by numerous groups (9). Intrinsic to the ability to acquire iron is the fact that microbes must also be able to sense and respond to changing iron concentrations within the environment. This is crucial to help maintain the homeostatic relationship between having enough iron to grow and avoiding iron toxicity. In a number of organisms, this regulation is accomplished by the ferric uptake regulator (Fur) protein (15). Found in both gram-negative and gram-positive bacteria, Fur functions by binding to promoter regulatory elements, called Fur boxes, in an iron-dependent manner. Typically, Fur-regulated promoters are repressed under iron-replete conditions and derepressed under iron-depleted conditions. A notable exception to this simple mode of regulation has recently been shown in Helicobacter pylori. The H. pylori Fur protein controls the transcription of both iron uptake and storage genes, frpB and pfr, respectively (18). This is accomplished by alteration of binding specificity based on the iron status of the Fur protein.
Other factors of H. pylori are known to show iron-dependent regulation, although thorough characterization of the roles of many of these is lacking. In addition to binding to the pfr and frpB promoters, Fur directly binds and autoregulates its own promoter (17). Expression of the genes fecA1, fecA2, feoB, and frpB2/3 is iron regulated in a Fur-dependent manner (16, 67), although it is unclear whether this dependency is direct or indirect. While all of these genes are suggested to play a role in iron uptake, a definitive role has been demonstrated only for feoB in H. pylori (68). Expression of a number of other proteins has also been shown to be regulated by iron. Among these, several outer membrane proteins (29, 31, 73), the vacuolating cytotoxin (vacA) (61), the riboflavin biosynthetic genes ribAB (72), and the fumarate reductase genes frdCAB have all been shown to exhibit changes in expression in response to varying concentrations of iron (20). Increased expression of the vacA toxin is in keeping with the finding that in addition to regulating genes that are important for iron uptake and homeostasis, many pathogenic organisms regulate virulence factors in response to iron concentration (9).
A thorough understanding of the genes involved in iron uptake and homeostasis is potentially of profound clinical importance because H. pylori colonization is associated with multiple forms of gastric disease. These range in severity from mild gastritis to duodenal and gastric ulcers and two forms of gastric malignancy, mucosa-associated lymphoid tissue lymphoma and adenocarcinoma (7, 22, 46). Additionally, H. pylori infection has been linked to iron deficiency anemia (IDA), particularly in children and premenopausal women (5, 53). Clinical evidence suggests that there are multiple mechanisms whereby H. pylori infection leads to IDA, either through the induction of impaired iron absorption by the host or through competition for the limited iron stores that are present (4). Even with approximately 50% of the world's population infected with H. pylori (37), the process of infection and survival of the bacterium within its gastric niche remains relatively poorly understood.
In a recent systematic global analysis of growth phase-dependent gene expression of H. pylori, the transcriptome of the bacterium was determined across the entire growth phase and revealed a dramatic switch in expression of many genes as bacteria made the transition from the exponential to the stationary phase of growth (63). Two major classes of genes affected at this transition were those known to encode virulence factors and those involved in iron homeostasis, suggesting the possibility that intracellular concentrations of iron may play a strong role in the modulation of both classes of genes. To better understand growth phase-modulated adaptive mechanisms utilized by H. pylori, we used spotted DNA microarrays to characterize the global response to iron starvation of both exponential- and stationary-phase cultures. Comparison of the transcription levels after growth-limiting iron chelation with those of the starting culture revealed three classes of genes: those affected specifically in the exponential phase or the stationary phase and those affected in both growth phases. Among these genes are many encoding proteins whose expression had previously been shown to be affected by iron concentration, as well as many whose iron-regulated gene expression had not previously been identified. Microscopic analysis of the effect of iron limitation on motility revealed that stationary-phase H. pylori cells show a marked ability to swim and survive for longer periods than do exponential-phase bacteria. Finally, analysis of the predicted promoter regions of the identified regulated genes led to the detection of several putative Fur boxes, thus suggesting a direct role for Fur in iron-dependent regulation of these genes.
MATERIALS AND METHODS
Bacterial strains and growth.
H. pylori strain SS1 (34) was maintained as a frozen stock at −80°C in brain heart infusion medium supplemented with 20% glycerol and 10% fetal bovine serum (FBS) (Gibco/BRL). Bacteria were grown on horse blood agar (HBA) plates containing 4% Columbia agar base (Oxoid), 5% defibrinated horse blood (HemoStat Labs, Dixon, Calif.), 0.2% B-cyclodextrin (Sigma), 10 μg of vancomycin (Sigma) per ml, 5 μg of cefsulodin (Sigma) per ml, 2.5 U of polymyxin B (Sigma) per ml, 50 μg of cycloheximide (Sigma) per ml, 5 μg of trimethoprim (Sigma) per ml, and 8 μg of amphotericin B (Sigma) per ml under microaerophilic conditions at 37°C. A microaerobic atmosphere was generated using a CampyGen sachet (Oxoid) in a gas pack jar. For liquid culture, H. pylori was grown in brucella broth (BB) (Difco) containing 10% FBS with shaking in a microaerobic environment. The MIC of 2,2′-dipyridyl (DPP) that inhibited bacterial growth but not bacterial viability for the duration of the time courses was determined by supplementation of exponential-phase H. pylori cultures with increasing concentrations of DPP and subsequent monitoring of changes in the optical density at 600 nm OD600 and viable CFU by plating on HBA plates.
Iron chelation and RNA isolation.
An overnight liquid culture of SS1 (OD600 of 0.5 to 1.0) was harvested by centrifugation and resuspended to an OD600 of 0.05 to 0.1 in BB supplemented with 10% FBS. Bacterial samples were collected after dilution and represent the start time point. The master culture was then split to yield either two independent 70-ml cultures (chelation experiments) or four independent 70-ml cultures (supplementation experiments). The cultures were maintained under microaerobic conditions with shaking overnight at 37°C. Next, DPP was added to a concentration of 200 μM for each exponential-phase culture (OD600 of 0.30 to 0.35) and stationary-phase culture (OD600 of 0.80 to 0.90). Bacterial samples were collected immediately prior to the addition of DPP and represent the t = 0 time point. All cultures were maintained under microaerobic conditions with shaking at 37°C, and samples were harvested 15, 30, 60, and 90 min (experiment 1) or 30, 60, and 120 min (experiment 2) after iron chelation. At each time point, aliquots were removed from the medium, plated to determine CFU, examined microscopically to determine the level of motility and harvested on a 0.45-μm-pore-size cellulose filter by vacuum filtration. Filters containing the H. pylori cells were immediately frozen in liquid nitrogen and subsequently used for isolation of bacterial RNA. H. pylori RNA was isolated using TRIzol reagent (Gibco/BRL) as previously described (39). The RNA concentration was quantitated by determination of the OD260, and RNA integrity was verified by visualization on a 1% agarose gel.
Iron add-back.
Growth of SS1 under iron-restricted conditions was conducted by first determining the concentration of DPP that would slow but not completely inhibit bacterial growth; 75 μM was determined to be a suitable concentration. An overnight liquid culture of SS1 was used to inoculate BB supplemented with 10% FBS and 75 μM DPP at a starting OD600 of 0.05. The cultures were maintained under microaerobic conditions with shaking overnight at 37°C. After approximately 19 h, the culture had reached an OD600 of 0.13. Iron was replenished in this culture by the addition of FeCl3 to a final concentration of 1 mM, a concentration that was shown in an independent experiment to relieve iron-dependent growth restriction. The cultures were maintained under microaerobic conditions with shaking at 37°C. Samples were taken immediately prior to the addition of FeCl3 (t = 0 time point) and 30, 60, and 120 min thereafter, as described above, and subsequently used for RNA isolation and microarray analysis as described below.
Microarray hybridization and analysis.
Equal concentration of each test RNA (t = 0, 15, 30, 60, 90, and 120 min from biologically independent experiments) and the reference RNA (t = 0 min) were used for cDNA synthesis in a standard reverse transcription reaction using Superscript II (Invitrogen) and Panorama H. pylori cDNA labeling primers (SigmaGenosys). Synthesized cDNA was purified using Qia-Quick PCR purification columns (Qiagen) as specified by the manufacturer and subsequently indirectly labeled with Cy5 (red) and Cy3 (green) fluorophores, respectively, as previously described (63). Individual Cy5 and Cy3 reactions were combined, and unincorporated dye was removed using a Qia-Quick PCR column as specified by the manufacturer. The eluates from the columns were concentrated by evaporation in a Speed Vac and resuspended in 11 μl of Tris-EDTA (TE). Then 1 μl of 25-mg/ml yeast tRNA, 2.55 μl of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and 0.45 μl of 10% sodium dodecyl sulfate were added to each of the labeled probes. These were heated to 99°C for 2 min, cooled briefly, and then added to the H. pylori microarray for hybridization and stringency washes as previously described (47). The hybridized slides were scanned and analyzed using 4000A scanner and the GENEPIX 3.0 software (both from Axon).
Data were collated using the Stanford Microarray Database (54). For the chelation experiments, spots showing obvious abnormalities were excluded from analysis. Spot quality was further filtered by requiring that the Cy3 mean spot intensity/Cy5 median background intensity ratio be greater than or equal to 1.5 and the Cy5 mean spot intensity/Cy3 median background intensity ratio be greater than or equal to 1.5. Genes were filtered by requiring that the log2 of the red/green normalized ratio (mean) have an absolute value greater than or equal to 1 in at least one array for each experimental set. Biologically independent experiments were analyzed separately, and the data were collapsed to reveal factors whose expression was consistently altered by iron starvation. For the iron add-back experiment, spot quality was filtered by requiring that the Cy3 net spot intensity be greater than or equal to 100. Genes were filtered by requiring that the log2 of the red/green normalized ratio (mean) have an absolute value greater than or equal to 0.59 in at least one array for each experimental set. Biologically independent experiments were analyzed separately, and the data were collapsed to reveal factors whose expression was consistently altered by iron addition. The list of factors was then cross-compared to the list generated for the iron chelation experiments, and the factors which overlap were obtained. All microarray data generated by this study are publicly available at http://genome-www.stanford.edu/microarray/.
Correlation coefficient determination.
To verify the growth status of cultures used in the chelation experiments, arrays where the t = 0 time point of the cultures was labeled in Cy5 and the start time point of the cultures was labeled in Cy3 for both the exponential- and stationary-phase time courses were compared to a previously published growth phase-dependent transcriptional profile of H. pylori (63). Correlation coefficients were determined by comparing the log2 red/green ratios from these two arrays with the data from each of the arrays in the described control time course (63) by using Microsoft Excel.
Motility analysis and supplementation.
For the motility and supplementation experiment, exponential- and stationary-phase cultures were grown and chelated as described above and maintained for extended periods. The motility of samples was monitored by live phase-contrast microscopy with glass slides and coverslips prewarmed to 37°C. A Hammamatsu C2400 video charge-coupled device camera was used to record movement in the field of view via an Argus-20 image processor (using the TRACE function) onto S-VHS video. Movement was traced over a 5-s period. Two sets of selected video frames for each time point were digitized for the generation of time-lapse films with the National Institutes of Health ObjectImage program. The percentage of motile bacteria at each time point was estimated using these films. The average percent motility data for each time point was plotted over time using Microsoft Excel.
Supplementation was performed at the 4-h time point postchelation and every 4 h thereafter by the removal of 5% of the culture and replacement with BB-FBS that had been chelated with 200 μM DPP. All cultures were maintained under microaerobic conditions with shaking at 37°C. CFU were determined by plating on HBA plates.
Identification of predicted Fur binding sites.
Each of the differentially regulated genes from the chelation experiment was visually examined to identify transcriptional organization in relation to the surrounding genes, and a list was generated that represented the predicted first gene in each putative operon identified. A Perl script was then written to extract the sequence of the 150 nucleotides immediately upstream of the predicted translational start site of the first gene in each predicted operon. A candidate Fur binding consensus sequence (AATAATNNTNA) for H. pylori was generated by aligning the high-affinity Fur binding sites of the frpB box (16) and fur box I (17) promoters, as well as the FURTA-Hp positive fecA2 promoter (26), using Clustal X (62). The 150-nucleotide regions were then searched for this consensus motif by using an additional Perl script, and sequences showing 100% identity were aligned using Clustal X.
RESULTS AND DISCUSSION
Microarray analysis of iron-regulated gene expression.
Determining a suitable DPP concentration that would not inhibit bacterial survival was a critical experimental parameter since decreased viability at later time points could greatly affect the transcriptional profiles obtained. Consequently, growth and survival curves were determined in BB supplemented with 10% FBS which had been chelated with increasing concentrations of DPP. The concentration determined to be suitable was 200 μM as it inhibited H. pylori growth and showed no effect on bacterial viability for time points up to several hours after chelation (data not shown).
Since previous microarray analysis had revealed that many of the genes that are subject to growth phase-dependent expression are known or predicted to play roles in iron uptake and homeostasis (63), we wished to assess the growth phase dependency of transcriptional changes induced by iron starvation. Since these previously observed differences were most evident between the exponential and stationary phases, we depleted iron from cultures of H. pylori in these two growth phases as described in Materials and Methods. Bacterial cells were harvested during a time course up to 2 h postchelation, and total bacterial RNA was prepared and subsequently used for microarray analysis. To ensure reproducibility, chelation experiments were replicated in a separate experiment and only the results for genes that consistently showed at least a twofold change in expression in both experiments are reported.
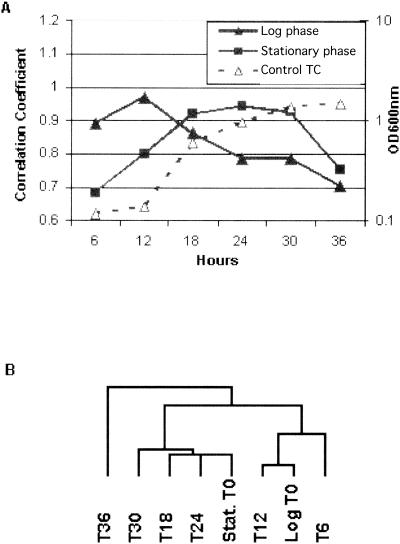
To ensure that the described iron-chelated cultures were in fact in the exponential and stationary phases, respectively, the genome-wide transcriptional profile of each of the starting cultures was compared to the previously published growth phase-dependent transcriptome of H. pylori (63). In this previous work, the complete transcriptional profile of H. pylori as it transits the growth curve from early exponential phase to late stationary phase was determined. Using this information, we were able to calculate a genome-wide correlation coefficient between the t = 0 time points of our exponential and stationary iron chelation cultures and each of the time points during the growth curve. These results are plotted in Fig. 1A. This plot shows that the predicted exponential-phase t = 0 iron chelation array correlates most highly with the 12-h time point of the growth curve, which represents the early exponential phase. On the other hand, the predicted stationary-phase t = 0 iron chelation array correlates most highly with the 24-h time point of the growth curve, which represents the early stationary phase. This result was also confirmed by performing an unsupervised hierarchical cluster of the iron chelation data sets with the growth curve data. The dendrogram depicted in Fig. 1B clearly shows that in each case, the exponential- and stationary-phase iron chelation arrays cluster with their respective counterparts across the growth curve. These data indicate that the respective chelation cultures were in the desired growth phase for determination of the growth phase-dependent response to iron starvation.
FIG. 1.
(A) Verification of the growth phase of the exponential- and stationary-phase H. pylori cultures used in the chelation experiment. Representative correlation coefficients for the t = 0 time point of the iron chelation cultures were calculated by comparison with published expression profiles of H. pylori across a standard growth curve time course (Control TC) (63). The OD600 of the Control TC is labeled on the right axis. Individual correlation coefficients between the exponential- and stationary-phase array data and the data for each of the arrays of the Control TC are indicated on the left-hand y axis. Time is indicated in hours on the x axis. The highest correlation coefficients for the indicated growth phase occur at t = 12 h and t = 24 h and represent the point at which the iron chelation experiments were initiated. (B) Dendrogram showing segregation of exponential-phase (denoted Log T0) and stationary-phase (denoted Stat. T0) t = 0 array data sets into respective branches generated by an unsupervised hierarchical cluster of the two t = 0 arrays from the chelation experiment with the array data from each of the time points of the Control TC. In each case, the chelation culture arrays cluster tightly with the concomitant growth phase arrays from the Control TC.
Classification of iron-regulated genes.
A total of 138 genes were shown to be differentially regulated by a minimum of twofold in the exponential-phase cultures (Table 1). The majority of these, 87 genes, were induced, while 51 genes showed a decrease in expression level. The transcriptional response of the stationary-phase cultures was much less dramatic, since only 75 genes displayed altered expression (Table 2). Of these, only 6 genes were repressed and the remaining 69 were induced. Direct comparison of the two tables revealed that only 30 genes overlapped between the exponential- and stationary-phase cultures (indicated by asterisks in Tables 1 and 2). Taken together, this indicates that a total of 183 individual genes showed altered expression on iron starvation of the two H. pylori cultures (Fig. 2).
TABLE 1.
Log-phase iron-regulated genes of H. pylori
| Gene designationa | Gene function and name | Maximum fold changeb in:
|
||
|---|---|---|---|---|
| TIGR | ASTRA | Expt 1 | Expt 2 | |
| Repressed genes | ||||
| Amino acid biosynthesis | ||||
| HP1229 | JHP1150 | Aspartokinase, lysC | 0.4 | 0.4 |
| Cell envelope/Surface structures | ||||
| HP0009 | JHP0007 | Putative OMP hopZ | 0.4 | 0.4 |
| HP0351 | JHP0325 | Flagellar basal-body M-ring protein, fliF | 0.3 | 0.3 |
| HP0655 | JHP0600 | Putative OMP | 0.4 | 0.4 |
| HP0858 | JHP0792 | ADP-heptose synthase, rfaE | 0.4 | 0.5 |
| HP0870 | JHP0804 | Flagellar hook, flgE | 0.4 | 0.5 |
| HP0912 | JHP0848 | OMP, hopC | 0.4 | 0.5 |
| #HP1119 | JHP1047 | Flagellar hook-associated protein 1 (HAP1), flgK | 0.3 | 0.4 |
| HP1275 | JHP1196 | Phosphomannomutase, algC | 0.3 | 0.4 |
| HP1450 | JHP1343 | 60-kDa inner membrane protein | 0.4 | 0.5 |
| Cellular processes | ||||
| *HP0103 | JHP0095 | Methyl-accepting chemotaxis protein, tlpB | 0.4 | 0.3 |
| HP0522 | JHP0471 | cag PAI protein, cag3 | 0.3 | 0.4 |
| HP1126 | JHP1055 | Colicin tolerance-like protein, tolB | 0.2 | 0.4 |
| HP1300 | JHP1220 | Preprotein translocase subunit, secY | 0.3 | 0.4 |
| HP1452 | JHP1345 | Thiophene and furan oxidizer, tdhF | 0.3 | 0.5 |
| DNA metabolism-restriction/modification | ||||
| HP0481 | JHP0433 | Adenine-specific DNA methyltransferase, MFOKI | 0.5 | 0.5 |
| Energy metabolism | ||||
| HP0509 | JHP0459 | Glycolate oxidase subunit, glcD | 0.4 | 0.4 |
| HP0574 | JHP0521 | Galactosidase acetyltransferase, lacA | 0.5 | 0.4 |
| HP1134 | JHP1062 | ATP synthase F1, subunit alpha energy metabolism, atpA | 0.3 | 0.5 |
| Hypothetical | ||||
| HP0248 | JHP0233 | Conserved hypothetical protein | 0.2 | 0.3 |
| HP0270 | JHP0255 | 0.5 | 0.4 | |
| HP0466 | JHP0418 | Conserved hypothetical protein | 0.3 | 0.4 |
| HP0554 | JHP0501 | 0.4 | 0.5 | |
| HP0659 | JHP0604 | 0.4 | 0.4 | |
| HP0783 | JHP0720 | 0.4 | 0.5 | |
| #HP0838 | JHP0776 | 0.4 | 0.3 | |
| HP0863 | JHP0797 | 0.3 | 0.5 | |
| HP1124 | JHP1053 | 0.3 | 0.4 | |
| HP1127 | JHP1056 | 0.5 | 0.4 | |
| HP1233 | JHP1154 | 0.3 | 0.4 | |
| Other categories, adaptations and atypical conditions | ||||
| HP1228 | JHP1149 | Invasion protein, putative dGTP pyrophosphohydrolase, invA | 0.3 | 0.3 |
| Purine ribonucleotide biosynthesis | ||||
| HP0255 | JHP0239 | Adenylosuccinate synthetase, purA | 0.5 | 0.4 |
| Transcription | ||||
| #HP0550 | JHP0497 | Transcription termination factor, rho | 0.2 | 0.3 |
| Translation | ||||
| HP0126 | JHP0116 | Ribosomal protein L20, rpl20 | 0.3 | 0.4 |
| HP0247 | JHP0232 | ATP-dependent RNA helicase, DEAD-box family, deaD | 0.2 | 0.4 |
| #HP0657 | JHP0602 | Putative processing protease | 0.4 | 0.4 |
| HP0658 | JHP0603 | Glu-tRNA amidotransferase, subunit b, gatB | 0.4 | 0.3 |
| HP0886 | JHP0818 | Cysteinyl-tRNA synthetase, cysS | 0.4 | 0.5 |
| HP1298 | JHP1218 | Translation initiation factor EF-1, infA | 0.5 | 0.5 |
| HP1299 | JHP1219 | Methionine aminopeptidase, map | 0.3 | 0.5 |
| HP1304 | JHP1224 | Ribosomal protein L6, rpl6 | 0.5 | 0.5 |
| HP1305 | JHP1225 | Ribosomal protein S8, rps8 | 0.5 | 0.5 |
| HP1306 | JHP1226 | Ribosomal protein S14, rps14 | 0.4 | 0.5 |
| HP1312 | JHP1232 | Ribosomal protein L16, rpl16 | 0.4 | 0.5 |
| *HP1313 | JHP1233 | Ribosomal protein S3, rps3 | 0.3 | 0.4 |
| HP1314 | JHP1234 | Ribosomal protein L22, rpl22 | 0.3 | 0.4 |
| *HP1315 | JHP1235 | Ribosomal protein S19, rps19 | 0.4 | 0.4 |
| HP1554 | JHP1445 | Ribosomal protein S2, rps2 | 0.4 | 0.4/PICK> |
| Transport and binding proteins | ||||
| HP0497 | JHP0449 | Putative sodium- and chloride-dependent transporter | 0.3 | 0.5 |
| HP1130 | JHP1058 | Biopolymer transport protein, exbB | 0.3 | 0.3 |
| *HP1400 | JHP1426 | Iron(III) dicitrate transport protein, fecA3 | 0.3 | 0.5 |
| Induced genes | ||||
| Amino acid biosynthesis | ||||
| HP0134 | JHP0122 | 3-Deoxy-d-arabino-heptulosonate-7-phosphate synthase, aroF | 3.7 | 3.7 |
| #HP0106 | JHP0098 | Cystathionine gamma-synthase, metB | 2.6 | 3.0 |
| #HP0107 | JHP0099 | Cysteine synthetase, cysK | 2.9 | 2.6 |
| HP0626 | JHP0570 | Tetrahydrodipicolinate N-succinyltransferase, dapD | 3.3 | 2.4 |
| *HP0649 | JHP0594 | Aspartate ammonia-lyase, aspA | 2.1 | 2.5 |
| *#HP0672 | JHP0615 | Solute binding signature and mitochondrial signature protein, aspB | 5.4 | 4.1 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | ||||
| HP0577 | JHP0524 | Methylene tetrahydrofolate dehydrogenase, folD | 2.7 | 2.4 |
| HP0665 | JHP0610 | Oxygen-independent coproporphyrinogen III oxidase, hemN | 2.3 | 2.4 |
| HP0844 | JHP0782 | Thiamine biosynthesis protein, thi | 2.6 | 2.2 |
| *#HP1582 | JHP1489 | Pyridoxal phosphate biosynthetic protein J, pdxJ | 2.4 | 4.8 |
| Cell envelope and surface structures | ||||
| HP0601 | JHP0548 | Flagellin A, flaA | 3.1 | 2.1 |
| HP0922 | JHP0856 | Toxin-like OMP, putative vacuolating cytotoxin (VacA) paralog | 3.6 | 3.0 |
| HP1052 | JHP0373 | UDP-3-O-acyl-N-acetylglcosamine deacetylase, envA | 3.2 | 2.5 |
| HP1177 | JHP1103 | OMP, omp27 | 2.4 | 2.2 |
| HP1192 | JHP1117 | Secreted protein involved in flagellar motility | 2.7 | 2.6 |
| HP1456 | JHP1349 | Membrane-associated lipoprotein, lpp20 | 5.5 | 3.5 |
| HP1469 | JHP1362 | OMP, omp31 | 2.0 | 2.1 |
| *HP1501 | JHP1394 | OMP, omp32 | 2.7 | 2.9 |
| Cellular processes | ||||
| HP0243 | JHP0228 | Neutrophil-activating protein (bacterioferritin), napA | 4.0 | 3.2 |
| HP0541 | JHP0489 | cag PAI protein, cag20 | 3.6 | 2.1 |
| HP0547 | JHP0495 | cag PAI protein, cag26 (cagA) | 3.5 | 2.5 |
| HP0875 | JHP0809 | Catalase, katA | 2.6 | 2.3 |
| HP0979 | JHP0913 | Cell divison protein, GTPase in circumferential ring formation, ftsZ | 2.0 | 2.3 |
| *HP1024 | JHP0400 | Cochaperone-curved DNA binding protein A, cbpA | 4.7 | 2.2 |
| HP1069 | JHP0356 | Cell division protein, ftsH | 4.6 | 3.0 |
| HP1431 | JHP1322 | 16S rRNA (adenosine-N6N6)-dimethyltransferase, ksgA | 2.4 | 2.7 |
| HP1563 | JHP1471 | Alkyl hydroperoxide reductase, tsaA | 2.8 | 2.3 |
| Central intermediary metabolism | ||||
| HP0020 | JHP0018 | Carboxynorspermidine decarboxylase, nspC | 2.2 | 2.3 |
| *HP0026 | JHP0022 | Citrate synthase, gltA | 2.3 | 2.2 |
| *#HP0294 | JHP0279 | Aliphatic amidase energy metabolism, amiE | 6.5 | 21.4 |
| HP0589 | JHP0537 | Ferredoxin oxidoreductase, alpha subunit, oorA | 2.2 | 2.0 |
| #HP0642 | JHP0586 | NAD(P)H-flavin oxidoreductase | 2.5 | 2.8 |
| HP0779 | JHP0716 | Aconitase B, acnB | 4.9 | 2.4 |
| *HP0900 | JHP0837 | Hydrogenase expression/formation protein, hypB | 3.2 | 4.1 |
| *HP0943 | JHP0878 | d-Amino acid dehydrogenase, dadA | 3.2 | 2.7 |
| *HP1186 | JHP1112 | Carbonic anhydrase | 2.3 | 2.2 |
| *HP1238 | JHP1159 | Aliphatic amidase, amiF | 2.3 | 2.9 |
| HP1398 | JHP1428 | Alanine dehydrogenase, ald | 3.9 | 2.8 |
| *HP1399 | JHP1427 | Arginase energy metabolism, rocF | 4.0 | 2.4 |
| #JHP0585 | Putative 3-hydroxyacid dehydrogenase | 2.2 | 3.3 | |
| Fatty acid and phospholipid metabolism | ||||
| *HP0690 | JHP0638 | Acetyl coenzyme A acetyltransferase (thiolase), fadA | 2.7 | 2.3 |
| *HP0871 | JHP0805 | CDP-diglyceride hydrolase, cdh | 8.2 | 6.9 |
| HP1045 | Acetyl-CoA synthetase, acoE | 4.5 | 3.6 | |
| Hypothetical | ||||
| HP0018 | JHP0016 | 2.5 | 3.0 | |
| HP0080 | JHP0074 | 3.5 | 3.3 | |
| HP0097 | JHP0089 | 5.1 | 2.4 | |
| HP0105 | JHP0097 | Conserved hypothetical protein | 3.2 | 2.9 |
| #HP0204 | JHP0190 | 2.2 | 2.0 | |
| HP0305 | JHP0290 | 2.3 | 2.3 | |
| *HP0309 | JHP0294 | Conserved hypothetical protein | 5.0 | 4.6 |
| HP0310 | JHP0295 | Conserved hypothetical protein | 6.7 | 4.1 |
| #HP0311 | JHP0296 | 4.6 | 2.3 | |
| *HP0318 | JHP0301 | Conserved hypothetical protein | 2.7 | 3.9 |
| HP0367 | JHP1014 | 2.1 | 2.5 | |
| *#HP0614 | JHP0557 | 2.1 | 2.7 | |
| HP0627 | JHP0571 | 2.3 | 2.3 | |
| HP0628 | 2.6 | 2.5 | ||
| HP0641 | JHP0584 | 2.0 | 2.3 | |
| *HP0710 | JHP0649 | Conserved hypothetical protein | 3.8 | 3.6 |
| *HP0746 | JHP0683 | 2.4 | 2.5 | |
| HP0920 | JHP0854 | Conserved hypothetical integral membrane protein | 3.0 | 2.7 |
| *HP0944 | JHP0879 | Conserved hypothetical protein | 3.0 | 3.7 |
| HP1391 | JHP1436 | 6.8 | 2.8 | |
| HP1394 | JHP1433 | Conserved hypothetical protein | 2.1 | 2.7 |
| HP1454 | JHP1347 | 2.8 | 2.7 | |
| HP1455 | JHP1348 | 4.1 | 3.5 | |
| HP1457 | JHP1350 | 3.9 | 3.3 | |
| HP1527 | JHP1416 | 3.4 | 2.1 | |
| #HP1588 | JHP1494 | Conserved hypothetical protein | 2.5 | 2.1 |
| Regulatory functions | ||||
| HP0224 | JHP0210 | Peptide methionine sulfoxide reductase, msrA | 3.2 | 2.6 |
| HP0714 | JHP0652 | RNA polymerase sigma-54 factor, rpoN | 2.3 | 2.1 |
| *#HP1027 | JHP0397 | Ferric uptake regulation protein, fur | 3.9 | 4.1 |
| Translation | ||||
| HP0403 | JHP0978 | Phenylalanyl-tRNA synthetase, alpha subunit, pheS | 2.0 | 2.5 |
| HP0570 | JHP0517 | Aminopeptidase a/I, pepA | 5.0 | 4.3 |
| *HP1012 | JHP0411 | Putative zinc protease, pqqE | 2.5 | 2.2 |
| Transport and binding proteins | ||||
| *#HP0686 | JHP0626 | Iron(III) dicitrate transport protein, fecAI | 2.1 | 3.3 |
| HP0715 | JHP0653 | ABC transporter, ATP-binding protein | 5.7 | 3.7 |
| *#HP0807 | JHP0743 | Iron(III) dicitrate transport protein, fecA2 | 4.1 | 4.3 |
| HP0940 | JHP0875 | Amino acid ABC transporter, periplasmic binding protein, yckK | 2.1 | 2.4 |
| *HP1017 | JHP0406 | Amino acid permease, rocE | 3.2 | 2.2 |
| HP1091 | JHP0334 | Alpha-ketoglutarate permease, kgtP | 2.8 | 2.5 |
| HP1174 | JHP1101 | Glucose/galactose transporter, gluP | 4.0 | 2.8 |
| HP1491 | JHP1384 | Phosphate permease | 3.6 | 2.4 |
| Unknown function | ||||
| HP0390 | JHP0991 | Adhesin-thiol peroxidase, tagD | 3.8 | 3.3 |
| HP0569 | JHP0516 | GTP binding protein, gtp1 | 2.1 | 2.6 |
| HP0625 | JHP0569 | Protein E, gcpE | 3.3 | 3.4 |
| *HP1104 | JHP1030 | Cinnamyl-alcohol dehydrogenase, cad | 3.1 | 3.1 |
*, these genes show differential regulation in both log- and stationary-phase cultures; #, these genes show differential regulation in the iron add-back experiment with the opposite change in expression.
Due to space limitations, the maximum fold change is given for only one of the time points analyzed per experiment.
TABLE 2.
Stationary-phase iron-regulated genes of H. pylori
| Gene designationa | Gene function and name | Maximum fold changeb in:
|
||
|---|---|---|---|---|
| TIGR | ASTRA | Expt 1 | Expt 2 | |
| Repressed | ||||
| Amino acid biosynthesis | ||||
| #HP0652 | JHP0597 | Phosphoserine phosphatase, serB | 0.2 | 0.2 |
| Cellular processes, chaperones | ||||
| HP0109 | JHP0101 | Chaperone and heat shock protein 70, dnaK | 0.4 | 0.5 |
| Energy metabolism | ||||
| #HP0632 | JHP0575 | Quinone-reactive Ni/Fe hydrogenase, large subunit, hyaB | 0.4 | 0.4 |
| #HP0633 | JHP0576 | Quinone-reactive Ni/Fe hydrogenase, cytochrome b subunit, hyaC | 0.5 | 0.5 |
| Hypothetical | ||||
| #HP0388 | JHP0993 | Conserved hypothetical protein | 0.3 | 0.4 |
| Transport and binding | ||||
| #HP0653 | JHP0598 | Nonheme iron-containing ferritin, pfr | 0.2 | 0.1 |
| Induced | ||||
| Amino acid biosynthesis | ||||
| *HP0649 | JHP0594 | Aspartate ammonia-lyase, aspA | 5.9 | 3.5 |
| *#HP0672 | JHP0615 | Solute binding signature and mitochondrial signature protein, aspB | 5.8 | 5.4 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | ||||
| HP0804 | JHP0740 | GTP cyclohydrolase II/3,4-dihydroxy-2-butanone 4-phosphate synthase, ribAB | 2.4 | 2.0 |
| HP1118 | JHP1046 | Gamma-glutamyltranspeptidase, ggt | 3.3 | 2.5 |
| *#HP1582 | JHP1489 | Pyridoxal phosphate biosynthetic protein J, pdxJ | 8.6 | 6.6 |
| #HP1583 | JHP1490 | Pyridoxal phosphate biosynthetic protein A, pdxA | 6.4 | 3.9 |
| Cell envelope and surface structures | ||||
| HP0115 | JHP0107 | Flagellin B, flaB | 4.0 | 2.1 |
| HP0175 | JHP0161 | Cell binding factor 2, putative peptidyl-prolyl cis-trans isomerase | 2.4 | 2.7 |
| *HP1501 | JHP1394 | OMP, omp32 | 2.0 | 2.1 |
| Cellular processes | ||||
| *HP0103 | JHP0095 | Methyl-accepting chemotaxis protein, tlpB | 2.0 | 4.9 |
| HP0887 | JHP0819 | Vacuolating cytotoxin, vacA | 2.6 | 2.3 |
| *HP1024 | JHP0400 | Cochaperone-curved DNA binding protein A, cbpA | 4.6 | 3.5 |
| Central intermediary metabolism | ||||
| HP0072 | JHP0067 | Urease beta subunit, ureB | 2.5 | 2.1 |
| HP0073 | JHP0068 | Urease alpha subunit, ureA | 2.7 | 2.3 |
| *HP0900 | JHP0837 | Hydrogenase expression/formation protein, hypB | 2.6 | 2.1 |
| *HP1186 | JHP1112 | Carbonic anhydrase | 3.4 | 2.5 |
| Energy metabolism | ||||
| *HP0026 | JHP0022 | Citrate synthase, gltA | 2.6 | 2.2 |
| HP0176 | JHP0162 | Fructose-bisphosphate aldolase energy metabolism, fba | 2.5 | 3.0 |
| HP0277 | JHP0262 | Ferredoxin energy metabolism | 2.9 | 3.7 |
| *#HP0294 | JHP0279 | Aliphatic amidase, amiE | 25.3 | 17.0 |
| HP0691 | JHP0637 | 3-Oxoadipate coenzyme A-transferase subunit A, scoA | 3.6 | 3.7 |
| HP0692 | JHP0636 | 3-Oxoadipate coenzyme A-transferase subunit B, scoB | 3.8 | 3.3 |
| *HP0943 | JHP0878 | d-Amino acid dehydrogenase, dadA | 3.8 | 2.5 |
| *HP1238 | JHP1159 | Aliphatic amidase, amiF | 6.3 | 4.1 |
| *HP1399 | JHP1427 | Arginase energy metabolism, rocF | 2.6 | 2.1 |
| HP1538 | JHP1461 | Ubiquinol cytochrome c oxidoreductase, cytochrome c1 subunit, petC | 2.2 | 2.1 |
| HP1539 | JHP1460 | Ubiquinol cytochrome c oxidoreductase, cytochrome b subunit, fbcH | 2.6 | 2.6 |
| Fatty acid and phospholipid metabolism | ||||
| HP0416 | JHP0968 | Cyclopropane fatty acid synthase, cfa | 3.9 | 3.3 |
| *HP0690 | JHP0638 | Acetyl coenzyme A acetyltransferase (thiolase), fadA | 3.7 | 4.0 |
| *HP0871 | JHP0805 | CDP-diglyceride hydrolase, cdh | 3.5 | 2.2/PICK> |
| Hypothetical | ||||
| *HP0309 | JHP0294 | Conserved hypothetical protein | 3.3 | 2.6 |
| *HP0318 | JHP0301 | Conserved hypothetical protein | 4.8 | 4.9 |
| HP0595 | JHP0542 | 2.6 | 2.0 | |
| *#HP0614 | JHP0557 | 2.9 | 2.2 | |
| HP0693 | JHP0635 | Conserved hypothetical integral membrane protein, atoE | 3.2 | 2.8 |
| *HP0710 | JHP0649 | Conserved hypothetical protein | 2.6 | 2.1 |
| HP0719 | JHP0657 | 2.1 | 3.5 | |
| HP0721 | JHP0658 | 2.3 | 2.8 | |
| *HP0746 | JHP0683 | 2.1 | 2.0 | |
| HP0773 | JHP0710 | 2.0 | 2.1 | |
| HP0914 | JHP0850 | Putative OMP | 2.1 | 2.6 |
| *HP0944 | JHP0879 | Conserved hypothetical protein | 3.5 | 3.0 |
| HP1173 | JHP1100 | 2.1 | 2.2 | |
| HP1182 | JHP1108 | Conserved hypothetical protein | 2.6 | 2.5 |
| HP1507 | JHP1400 | Conserved hypothetical ATP binding protein | 2.6 | 2.1 |
| Other categories, adaptations, and atypical conditions | ||||
| HP1496 | JHP1389 | General stress protein, putative ribosomal protein L25, ctc | 2.2 | 2.2 |
| Purines, pyrimidines, nucleosides, and nucleotides | ||||
| HP0680 | JHP0621 | Ribonucleoside-diphosphate reductase 1 alpha subunit, nrdA | 2.3 | 2.2 |
| Regulatory functions | ||||
| HP0166 | JHP0152 | Response regulator, ompR | 2.4 | 2.4 |
| HP1025 | JHP0399 | Putative heat shock protein, putative hspR | 3.1 | 2.5 |
| *#HP1027 | JHP0397 | Ferric uptake regulation protein, fur | 3.6 | 3.0 |
| Translation | ||||
| HP0177 | JHP0163 | Translation elongation factor EF-P, efp | 2.1 | 2.5 |
| *HP1012 | JHP0411 | Protease, putative zinc protease, pqqE | 2.1 | 2.5 |
| HP1019 | JHP0405 | Serine protease, htrA | 2.9 | 4.1 |
| *HP1313 | JHP1233 | Ribosomal protein S3, rps3 | 2.3 | 2.7 |
| *HP1315 | JHP1235 | Ribosomal protein S19, rps19 | 2.3 | 2.1 |
| Transport and binding proteins | ||||
| HP0140 | JHP0128 | l-Lactate permease, lctP | 2.7 | 2.0 |
| HP0301 | JHP0286 | Dipeptide ABC transporter, ATP-binding protein, dppD | 2.5 | 2.1 |
| *#HP0686 | JHP0626 | Iron(III) dicitrate transport protein, fecA1 | 6.4 | 4.0 |
| *#HP0807 | JHP0743 | Iron(III) dicitrate transport protein, fecA2 | 8.2 | 3.3 |
| #HP0876 | JHP0810 | Iron-regulated OMP, frpB1 | 22.0 | 10.8 |
| HP0942 | JHP0877 | d-Alanine glycine permease, dagA | 2.7 | 2.5 |
| *HP1017 | JHP0406 | Amino acid permease, rocE | 2.4 | 2.4 |
| HP1172 | JHP1099 | Glutamine ABC transporter, periplasmic glutamine binding protein, glnH | 2.6 | 3.1 |
| #HP1339 | JHP1258 | Biopolymer transport protein, exbB | 2.4 | 2.5 |
| *HP1400 | JHP1426 | Iron(III) dicitrate transport protein, fecA3 | 3.5 | 5.5 |
| #HP1432 | JHP1321 | Histidine- and glutamine-rich protein | 5.4 | 2.4 |
| #HP1561 | JHP1469 | Iron(III) ABC transporter, periplasmic iron-binding protein, ceuE1 | 3.7 | 3.2 |
| HP1562 | JHP1470 | Iron(III) ABC transporter, periplasmic iron-binding protein, ceuE2 | 3.6 | 2.6 |
| Unknown function | ||||
| *HP1104 | JHP1030 | cinnamyl-alcohol dehydrogenase EL13-2, cad | 3.1 | 3.2 |
*, these genes show differential regulation in both log- and stationary-phase cultures; #, these genes show differential regulation in the iron add-back experiment with the opposite change in expression.
Due to space limitations, the maximum fold change is given for only one of the time points analyzed per experiment.
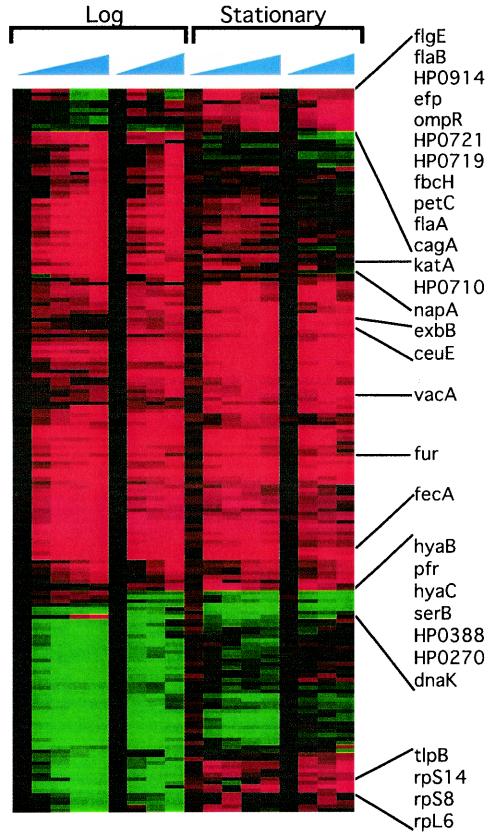
FIG. 2.
Microarray analysis of the iron starvation response of H. pylori. A cluster diagram shows the expression profile of all 183 genes meeting the filter criteria (explained in Materials and Methods) after addition of the iron chelator. Two independent experiments for each growth phase are depicted, and relative expression patterns are shown for each time point. Increasing time postchelation is indicated by the blue triangles above the array data. Red indicates an increase in expression, while green indicates reduced expression. Representative genes are listed, and their relative location is indicated by lines. A complete list of the regulated factors and the maximum relative changes in expression can be found in Tables 1 and 2.
Sorting of these genes into functional classes as defined by The Institute for Genomic Research revealed that a number of different biological functions are affected by the loss of iron. These include a large number of genes predicted to encode hypothetical or conserved proteins of unknown function, proteins involved in metabolism, components of the cell envelope or surface, and factors involved in transport and binding. For the sake of brevity, a few key genes are discussed below.
Iron starvation-repressed genes. (i) Iron sequestration factors.
The pfr locus was found to be strongly repressed in the iron-starved stationary-phase cultures of H. pylori, validating the ability of our experimental design to identify known iron-regulated genes. Pfr acts as the major H. pylori ferritin and is responsible for the storage of iron in subcellular granules within the bacterial cytoplasm (6). The activity of this protein plays an important role in the H. pylori life cycle because it is required for growth and survival under iron starvation conditions. Additionally, Pfr activity is important for maximal iron uptake, since mutation of pfr greatly decreases the rate of transport of 55FeCl2 and 55Fe3+ into the H. pylori cell (69). These activities are probably crucial within the tumultuous environment of the human stomach, since mutations in pfr result in an H. pylori strain that is unable to colonize the Mongolian gerbil (69).
Expression of pfr is tightly regulated by Fur. In keeping with this finding, we detected strong induction of fur expression in the iron-starved cultures (Tables 1 and 2). pfr was strongly regulated only in the stationary phase, suggesting that iron-regulated expression of this gene is connected to the growth phase (Fig. 2). This view is consistent with the recent findings that the expression levels of pfr increase dramatically during the late exponential phase in a non-iron-chelated rich medium while the level of fur remains fairly constant (63). This is presumably because during exponential growth, iron is rapidly being utilized and thus sufficient quantities are not present for storage. In contrast, as the cells reach stationary phase and are no longer actively multiplying, iron levels become high enough to require storage of intracellular iron for later utilization during periods of iron starvation and to prevent toxicity (1).
(ii) Genes involved in translation.
Analysis of the factors repressed by the removal of iron reveals that the majority of genes involved in translation were affected in exponential phase. Repression of many of these genes is most probably a direct result of the fact that the concentration of DPP used in this study resulted in cessation of growth in these cultures (data not shown). Since the bacteria are no longer dividing, the expression of factors that would be involved in growth was promptly down-regulated. This is evidenced by the fact that the largest class of repressed genes contains those predicted to play a role in translation. Included among these were genes encoding many ribosomal proteins and the translation initiation factor EF-1 (Table 1). It is perhaps for this reason that several genes involved in diverse forms of energy metabolism were repressed (Tables 1 and 2).
(iii) Genes encoding membrane proteins.
Several genes that encode membrane proteins were also repressed in the exponential-phase culture. These include an inner membrane protein encoded by HP1450 and three outer membrane proteins (OMPs). The 64 identified OMPs of the two sequenced H. pylori strains have been recently divided into five paralogous families based on sequence similarity and predicted secondary structure (3). The major OMP family is composed of two subfamilies: the Hop and the Hor proteins. The Hop subfamily contains the largest number of OMPs, including several factors that are predicted to function as porins (21, 25) on adhesins (43). Two of the OMPs (HP0009 and HP0912; encoded by hopZ and hopC) repressed in exponential phase belong to the Hop subfamily, while another Hop protein (HP1177; encoded by omp27) was induced on iron starvation (Table 1). This opposing regulation perhaps suggests a functional shift in porin activity. Little is known about the selectivity of most of the Hop porins. Alterations in expression of the Hop proteins, specifically those acting as porins, would presumably affect the permeability of the outer membrane and may play a previously unappreciated role in iron uptake and/or storage.
(iv) Miscellaneous.
Among the other five genes that were specifically repressed in the stationary phase are dnaK, which encodes the chaperone and heat shock protein 70 (Hsp70) and a conserved hypothetical protein, HP0388 (Table 2), that shows homology to predicted proteins in various pathogenic bacteria, such as Haemophilus influenzae, Campylobacter jejeuni, and Vibrio cholerae. Determination of the function of this hypothetical protein will be of interest in the future because its similar pattern of expression to pfr may suggest that it plays a role in iron sequestration. Extracellularly localized Hsp70 has been suggested to function as a sulfatide binding adhesion for H. pylori that facilitates attachment to sulfated glycolipids within the stomach mucus layer (28). Expression of dnaK is repressed by the HspR repressor protein (56), which we show here to be strongly induced by iron limitation, along with CbpA, encoded in the same operon (Table 2). We do not currently understand the significance of repression of dnaK on iron starvation; however, if surface-localized Hsp70 is in fact involved in mucosal adherence, decreased concentrations of iron may serve as a signal that promotes decreased adherence and also promotes relocalization of H. pylori to a more suitable environment within the gastric mucosa. In support of this, we observed effects on genes involved in chemotaxis (tlpB) and components of the flagellar apparatus. These are discussed in greater detail below.
Iron starvation-induced genes. (i) Transport and binding.
As expected, many genes that encode transport and binding proteins predicted to be involved in iron uptake were found to be regulated in our study. These include fecA1, fecA2, fecA3, and frpB1, which encode OMPs; exbB, which encodes a cytoplasmic membrane protein; and ceuE1 and ceuE2, which encode periplasmic binding proteins (Tables 1 and 2). frpB1 was one of the most strongly regulated genes in the study, showing greater than 10-fold induction in the stationary-phase culture. Iron starvation affected the expression of several of these genes in a growth phase-dependent manner, since while both fecA1 and fecA2 were strongly induced in both stationary-phase and exponential-phase cultures, transcription of fecA3 showed differential regulation depending on the growth status of the cells. In the stationary phase, fecA3 was induced in a similar manner to its counterparts; however, in the exponential phase, it was repressed by iron starvation. In addition, expression of ceuE1, ceuE2, and exbB is strongly regulated only in the stationary phase of growth. These data are in contrast with a recent report suggesting that expression of fecA3, ceuE1, and ceuE2 are unchanged by iron limitation (67). van Vliet et al. showed by spot blot hybridization that expression of these three genes remained virtually unchanged in H. pylori grown under iron-depleted conditions (67). Direct comparison of our results to theirs is confounded by the fact that while we analyzed both exponentially growing and stationary-phase cells, van Vliet et al. collected RNA from a single time point in the growth of their strain and do not report which stage of growth this point represents. This observed discrepancy between the two studies may reflect growth phase differences in expression of the genes in question.
(ii) Virulence factors.
Like other pathogenic bacteria, H. pylori seems to use iron limitation as a signal to regulate the expression of virulence factors. We observed induced expression of a number of genes that encode known virulence factors of H. pylori. These include cag20 and cag26 (cagA), both of which are located on the cag pathogenicity island (PAI). The PAI encodes a novel type IV secretion apparatus in some strains of H. pylori (2, 11), and it is well accepted that H. pylori strains carrying the PAI are more likely to be associated with serious manifestations of H. pylori infection (74). This is hypothesized to be because components of the island function in the delivery of CagA to the host cell, where it is inserted into the plasma membrane (52, 58) and induces striking morphological changes in the infected cell (51). Low concentrations of iron like those encountered within the host environment may serve as a signal that tells the bacterium that it is in a location where the production of virulence factors would be beneficial and may potentially induce the release of cellular components, such as iron, from damaged host cells.
Additionally, we observed increased expression of vacA, which encodes the vacuolating cytotoxin, VacA. VacA is a secreted protein that is internalized by host cells (38) and results in vacuolar degeneration that leads to the accumulation of large, nonfunctional endosomal-lysosomal hybrids (14, 35). The importance of VacA intoxication for the disease process remains elusive, but vacuolization of cells in human biopsy samples has been observed (10, 23). It was recently shown that vacA mutants have a 50% infectious dose that is 100-fold higher than that of an isogenic wild-type strain and show a significant colonization defect when coinfected with a wild-type strain (48). This suggests that VacA plays a role in establishing colonization, although the nature of this role is not immediately evident. Based on our and other (61) findings that vacA expression is up-regulated by iron starvation, one might propose that increased toxin production and the resulting increased cellular damage may lead to the release of host cell iron stores. Accordingly, it has been shown that administration of purified VacA to polarized T84 cell monolayers results in the increased permeability and release of Fe3+ and Ni2+ (45). Thus, toxin production and activity may facilitate the acquisition of iron by bacteria and promote bacterial survival in the host environment.
Induced expression of napA, which encodes the neutrophil-activating protein HP-NAP, was observed in the exponential-phase iron-depleted cultures. An independent study by Cooksley et al. recently showed that HP-NAP accumulation is influenced by Fur (13), once again confirming that our experimental design identified factors whose expression is affected by iron availability. HP-NAP was named for its ability to promote neutrophil adhesion to endothelial cells (24). It has been classified as a virulence factor and shown to be a major antigen in the human immune response to H. pylori (49). Additionally, is has been suggested to be a likely component for inclusion in an H. pylori vaccine due to its ability to provide colonization protection in mice orally immunized with purified HP-NAP (19, 49). Neutrophil activation and moderate levels of inflammation induced by H. pylori are likely to play critical roles in bacterial colonization because they promote cellular damage that results in the leakage of nutrients from host cells (8). Thus, as with VacA, increased expression of HP-NAP on iron starvation might serve as a means of releasing iron from host cells. Further support for this is evident in the fact that the HP-NAP has strong in vitro bacterioferritin activity, binding up to 500 atoms of iron (64). Due to the important nature of this virulence factor and the predicted Fur binding site found by the bioinformatic analyses performed by us and by Cooksley et al. (13) (discussed below), future studies will attempt to discern the importance of this iron-dependent regulation.
(iii) Nitrogen metabolism.
The most highly regulated gene in the stationary-phase cultures was amiE, which encodes an aliphatic amidase. It, along with amiF, which encodes a formamidase, were up-regulated to various degrees by iron starvation, regardless of the growth status of the culture (Tables 1 and 2). As a group, bacterial amidases catalyze the hydrolysis of short-chain aliphatic amides to ammonia and the corresponding organic acid and play a valuable role in nitrogen metabolism (55). This is particularly true in H. pylori since ammonia not only is the preferred source of nitrogen for the synthesis of amino acids, pyrimidines, and purines but also contributes to epithelial cell damage and chemotactic motility and is a crucial component of the acid resistance pathway for the bacterium (30, 40, 50, 71). Previous characterization of the AmiE and AmiF amidases has indicated that they show marked substrate specificity and that their activity is dependent on the activities of other enzymes of the nitrogen metabolism pathways of H. pylori, namely, those of urease and arginase (55). Expression of amiF and amiE strongly correlates with growth phase-dependent patterns of expression of fecA and frpB, perhaps suggesting that they have similar regulatory signals (63). Recently, van Vliet et al. reported that Fur regulates the transcription, expression, and activity of AmiE and indirectly regulates the activity of AmiF (66). Therefore, our finding of strong iron-dependent regulation is not surprising. Perhaps what is surprising, though, is the apparently strong level of coregulation of genes involved in nitrogen metabolism and iron starvation. We found that in addition to amiE and amiF, multiple genes that are involved in pathways of ammonia and urea production were iron regulated. These include genes encoding components of the urease enzyme, ureA and ureB; genes involved in arginine metabolism, rocE and rocF; and a component of the glutamine transporter, glnH (Tables 1 and 2). The reason for the coregulation of these two apparently very different metabolic pathways is currently unclear, but since ammonia causes epithelial cell damage, increased ammonia production may also serve as a mechanism by which H. pylori increases cellular damage and thus triggers the release of host cell iron stores. It is also likely that iron chelation affects iron-cofactored enzymes that play a role in nitrogen metabolism, and this might account to some degree for the apparent coregulation.
(iv) Flagellar genes and motility.
Observation of the growth phase-dependent expression of components of the flagellar apparatus included increased expression of flaA in exponential-phase cells but increased flaB expression in stationary-phase cells (Tables 1 and 2). Previous analysis of the H. pylori flagellar filament has shown that it consists of a copolymer of FlaA and FlaB (27, 36, 60), where FlaA is the predominant subtype and FlaB is present as a minor constituent (33). Both FlaB and FlaA are necessary for full motility (32). In comparison to most other bacteria, the existence of two distinct flagellin subunits present in different amounts is somewhat unusual. It has been suggested that H. pylori has the ability to alter the relative levels of FlaA and FlaB in response to different environmental stimuli (57, 59) and two recent independent studies have demonstrated that transcription of flaA and flaB was strongly regulated in a growth phase-dependent manner (41, 63). Interestingly, in those studies flaA showed maximal expression in the stationary phase while flaB expression peaked early in the exponential phase. This is opposite to the effect we observed when we starved the cells for iron, thus suggesting that in addition to growth phase dependency, further levels of flagellar regulation are governed by environmental signals. By altering relative concentrations of the two flagellar subunits, the bacterium could potentially adapt to microenvironments by alteration of the filament ultrastructure. This may alter flagellar stiffness or flexibility to better suit the particular environment being encountered.
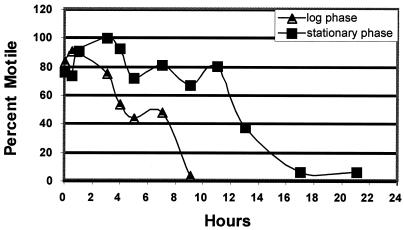
To determine whether iron starvation of H. pylori showed a differential effect on motility depending on the growth phase of the culture, we conducted video microscopy of H. pylori cells exposed to iron starvation conditions for extended periods as described in Materials and Methods. Analysis of these bacterial populations showed that prior to chelation, roughly equivalent numbers of bacteria (approximately 75%) were motile in the exponential and stationary phase cultures (Fig. 3). On iron starvation, the exponential phase decreased in the number of motile cells. By 9 h, the entire culture was nonmotile. This is in contrast to a normal growth curve, where motility decreases much more slowly, remaining at approximately 60% even after 12 h of growth (Table 3). Morphologically, at the later time points the iron-restricted bacteria in the exponenetial-phase culture appeared coccoid (data not shown). Analysis of the stationary-phase culture revealed that virtually the entire culture became motile by 3 h postchelation, and that overall levels of motility were maintained for much longer periods. This is similar to what is observed in a normal growth curve (Table 3). Of note, more than 60% of the iron-restricted culture was still highly motile after 9 h when all exponential-phase bacteria appeared coccoid. The motility of the stationary-phase cells remained relatively high for approximately 14 to 16 h after iron starvation; moreover, visually these cells appeared helical and noncoccoid for extended periods (data not shown). These data indicate that iron starvation shows growth phase-dependent effects on motility in H. pylori. The significance of this in the in vivo environment is not immediately clear, but the ability to sense the surrounding environment and alter motility probably serves as a critical component of successful colonization because it would direct H. pylori to a suitable environment for colonization. Of note, a single chemotaxis gene (tlpB) was shown to be differentially regulated by iron starvation, perhaps suggesting a role for this gene in responding to iron concentration.
FIG. 3.
Growth phase-dependent effect of iron starvation on H. pylori motility. Iron chelator was added to both exponential- and stationary-phase cultures of H. pylori, and the response of each culture was monitored by video microscopy as described in Materials and Methods. The percent motility of each culture over time is indicated.
TABLE 3.
Comparison of the progression of motility during iron chelation compared to normal growth at both exponential and stationary phases
| Exponential phase | Stationary phase
|
||||
|---|---|---|---|---|---|
| Condition | Growth time (h) | % Motile | Condition | Growth time (h) | % Motile |
| Control | Start | 82 | Control | Start | 66 |
| 12 | 66 | 12-13 | 34 | ||
| Chelation | Start | 87 | Chelation | Start | 76 |
| 9 | 4 | 12-13 | 38 | ||
Growth phase effects on iron starvation susceptibility.
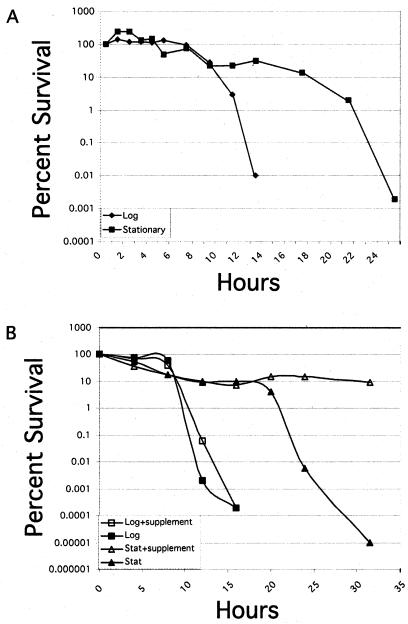
Based on our observation that iron-starved exponential-phase cells become nonmotile and coccoid considerably earlier than stationary-phase cells, we hypothesized that the growth phase might play a role in susceptibility of H. pylori to iron starvation. To investigate this, we conducted a chelation time course experiment and calculated the percent survival of iron-starved exponential- and stationary-phase cells. As shown in Fig. 4A, both exponential- and stationary-phase cells displayed similar survival profiles for the first 8-10 h of the experiment. After this point, survival rapidly decreased in the exponential-phase cells. However, a significant proportion of the stationary-phase cells remained viable for more than 20 h, suggesting that intracellular iron stores of the stationary-phase bacteria were sufficient to support extended periods of iron depletion. To ensure that decreased viability of the bacterial cultures was due to lack of iron and not due to deficiency of some other nutrient, we supplemented both exponential- and stationary-phase iron-starved cultures with freshly chelated medium as described in Materials and Methods. To our surprise, while this had no effect on the viability profile of the exponential-phase cells, it completely abrogated the decline in viability that we had originally observed in the later time points of the stationary-phase culture; we observed a 6-log-unit difference in the number of viable cells at the latest time point at which measurements were made (Fig. 4B). This suggests that stationary-phase cells do not die due to a lack of iron but die because of a lack of some other unidentified nutrient and indicates that stationary-phase H. pylori cultures have a previously unappreciated tremendous capacity for survival when faced with iron starvation, possibly due to the ability of Pfr to accumulate large intracellular iron stores. This ability to survive lengthy periods of iron starvation could be of significant clinical importance and probably plays a role in the environment of the human stomach, where available iron is tightly regulated through sequestration by the host.
FIG. 4.
(A) Iron starvation effects on cell viability. The viability of iron-depleted H. pylori cells was monitored by plating for CFU. Iron chelator was added to both exponential- and stationary-phase cultures, and titers were taken periodically thereafter. The percent survival was calculated based on initial titers taken at the t = 0 time point, and times indicated on the x axis indicate hours postchelation. (B) Effect of supplementation on cell viability. To determine whether decreased viability of iron-starved cells was due to expended stores of iron or of another nutrient, supplementation experiments were conducted. Both exponential- and stationary-phase cultures were maintained without supplementation as controls and showed kill curves similar to those depicted in panel A. For supplemented cultures, approximately 20% of the culture was removed every 4 h and replaced with previously chelated BB as described in Materials and Methods.
Bioinformatic analysis reveals putative Fur-regulated genes.
Since Fur regulation is a key component of iron acquisition and sequestration and since very few genes have actually been proven to be directly regulated by Fur binding in H. pylori, we performed a preliminary bioinformatic analysis of our differentially regulated genes to identify putative Fur binding sites in the predicted promoter regions. Using the sequence of the high-affinity Fur boxes from frpB, fur, and the FURTA-Hp-positive fecA2 promoter (16, 17, 26), a candidate Fur box consensus sequence (AATAATNNTNA) for H. pylori was generated (Fig. 5A). Homology searches of the predicted promoter regions of the 183 differential regulated genes identified 18 genes that contained a perfect match to the consensus sequence (Fig. 5B). Of these, three genes contained more than one putative Fur box: gluP, ribAB, and frpB1. Analysis of the list revealed a strong enrichment for genes that are predicted to play a role in iron acquisition or have previously been proposed to be Fur regulated, thus suggesting that our bioinformatic approach was able to identify at least a subset of Fur-regulated genes. Included among these are fecA1, fecA2, exbB, ribAB, napA, and amiE. Also included in this list are several genes whose putative regulation by Fur had not previously been hypothesized. Among these are several genes encoding hypothetical proteins, proteins of unknown function, and predicted membrane proteins. A goal of future research will be to directly investigate the Fur regulation of these genes.
FIG. 5.
(A) Identification of a Fur binding site consensus sequence. Sequence alignment of the high-affinity Fur binding sites of the frpB (designated box 1 in reference 16) and fur (designated box I in reference 18) promoters, as well as the FURTA-Hp positive fecA2 promoter (27). Conserved nucleotides are indicated in bold and denoted by asterisks. (B) Putative identification of Fur boxes in predicted promoter regions of iron-regulated genes. Sequence alignment of all genes shows a perfect match to the determined consensus sequence. Genes are listed in chromosomal order, and HP designations represent those given by TIGR. The “f” or “r” following the HP number indicates the forward and reverse strand, respectively, on which the consensus match was found. Conserved nucleotides are indicated in bold.
Iron add-back.
Finally, since the removal of iron from the exponentially growing culture of H. pylori results in growth cessation and thus has both direct and indirect effects on gene expression, we wished to conduct an additional approach to complement our growth phase dependence study. We therefore conducted microarray analysis of H. pylori grown in an iron-limited environment (see Materials and Methods) to examine the transcriptional effect of the addition of excess iron to these cells. We hypothesized that genes that showed altered expression might represent those that are the most strongly iron regulated. This iron add-back study revealed that 28 genes overlapped with the growth phase-dependent identified factors from the chelation experiments (denoted in Tables 1 and 2 by #). The 28 genes were equally distributed between the exponential- and stationary-phase lists of affected factors. Interestingly, multiple genes that have been previously shown to be iron regulated, such as vacA and napA (13, 61), were not identified by this method. This highlights the importance of using complementary conditions for comprehensive identification of regulons affected by a stimulus of interest.
Concluding remarks.
A formidable challenge in understanding how bacterial pathogens colonize and cause disease in their hosts is the development of a more thorough understanding of the adaptive mechanisms used by these pathogens within the host. The gastric pathogen H. pylori undoubtedly encounters a number of environmental stresses within the human stomach where it colonizes. One of these stresses is likely to involve periods of iron starvation because the human body tightly sequesters available iron to prevent oxidative damage and to inhibit bacterial growth. In an effort to more thoroughly define the response of H. pylori to iron starvation, we used DNA microarrays to investigate global temporal changes in gene expression of both exponentially growing and stationary-phase cultures. We chose to investigate both growth phases as a result of a recent study suggesting that many of the genes of H. pylori that are differentially regulated as the bacteria progress through the growth phase are predicted to be iron regulated or to play some role in iron acquisition (63). We therefore hypothesized that iron starvation might have drastically different effects on the transcriptome of H. pylori depending on the growth phase of the bacteria. This was indeed the case, since we noted that many regulated genes showed growth phase-dependent changes in transcription. This result highlights the importance of considering growth phase when interpreting the results of future studies directed at understanding the transcriptional responses of H. pylori.
Future experiments to identify the role of each of the identified factors in adaptation to iron starvation should provide valuable information concerning the mechanisms used by H. pylori during colonization within the gastric environment of the human host. Since the ability to acquire and store iron is a crucial part of the ability of H. pylori to survive in its host and cause disease, investigation of some of these factors may facilitate the elucidation of novel targets for vaccine development and antimicrobial therapy.
Acknowledgments
We are grateful to A. Camilli for critical review of the manuscript, M. Amieva and B. Neilan for invaluable discussion, and M. Goodrich and N. Salama for technical assistance.
D.S.M. is supported by NIH training grant AI07502-06, the Damon Runyon Cancer Research Fund, and the Stanford Digestive Disease Center DK56339. L.J.T. is supported by the National Health and Medical Research Council of Australia and a special grant provided by the Vice Chancellor of UNSW to complete this work at Stanford University. C.C.K. is supported by a predoctoral fellowship from the Howard Hughes Medical Institute and a Stanford Graduate Fellowship. Research in the laboratory of S.F. and L.S.T. is supported by grants CA92229 and AI38459 from the NIH.
D. Scott Merrell and Lucinda J. Thompson contributed equally to this work.
Editor: V. J. DiRita
REFERENCES
- 1.Abdul-Tehrani, H., A. J. Hudson, Y. S. Chang, A. R. Timms, C. Hawkins, J. M. Williams, P. M. Harrison, J. R. Guest, and S. C. Andrews. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 181:1415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol. 28:37-53. [DOI] [PubMed] [Google Scholar]
- 3.Alm, R. A., J. Bina, B. M. Andrews, P. Doig, R. E. Hancock, and T. J. Trust. 2000. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect. Immun. 68:4155-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barabino, A. 2002. Helicobacter pylori-related iron deficiency anemia: a review. Helicobacter 7:71-75. [DOI] [PubMed] [Google Scholar]
- 5.Barabino, A., C. Dufour, C. E. Marino, F. Claudiani, and A. De Alessandri. 1999. Unexplained refractory iron-deficiency anemia associated with Helicobacter pylori gastric infection in children: further clinical evidence. J. Pediatr. Gastroenterol. Nutr. 28:116-119. [DOI] [PubMed] [Google Scholar]
- 6.Bereswill, S., U. Waidner, S. Odenbreit, F. Lichte, F. Fassbinder, G. Bode, and M. Kist. 1998. Structural, functional and mutational analysis of the pfr gene encoding a ferritin from Helicobacter pylori. Microbiology 144:2505-2516. [DOI] [PubMed] [Google Scholar]
- 7.Blaser, M. J. 1998. Helicobacter pylori and gastric diseases. Br. Med. J. 316:1507-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blaser, M. J. 1993. Helicobacter pylori: microbiology of a ‘slow’ bacterial infection. Trends Microbiol. 1:255-260. [DOI] [PubMed] [Google Scholar]
- 9.Braun, V. 2001. Iron uptake mechanisms and their regulation in pathogenic bacteria. Int. J. Med. Microbiol. 291:67-79. [DOI] [PubMed] [Google Scholar]
- 10.Caselli, M., N. Figura, L. Trevisani, P. Pazzi, P. Guglielmetti, M. R. Bovolenta, and G. Stabellini. 1989. Patterns of physical modes of contact between Campylobacter pylori and gastric epithelium: implications about the bacterial pathogenicity. Am. J. Gastroenterol. 84:511-513. [PubMed] [Google Scholar]
- 11.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, H. L. 2003. The role of iron in infections with intracellular bacteria. Immunol. Lett. 85:193-195. [DOI] [PubMed] [Google Scholar]
- 13.Cooksley, C., P. J. Jenks, A. Green, A. Cockayne, R. P. Logan, and K. R. Hardie. 2003. NapA protects Helicobacter pylori from oxidative stress damage, and its production is influenced by the ferric uptake regulator. J. Med. Microbiol. 52:461-469. [DOI] [PubMed] [Google Scholar]
- 14.Cover, T. L., and M. J. Blaser. 1992. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J. Biol. Chem. 267:10570-10575. [PubMed] [Google Scholar]
- 15.Crosa, J. H. 1997. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol. Rev. 61:319-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delany, I., A. B. Pacheco, G. Spohn, R. Rappuoli, and V. Scarlato. 2001. Iron-dependent transcription of the frpB gene of Helicobacter pylori is controlled by the Fur repressor protein. J. Bacteriol. 183:4932-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delany, I., G. Spohn, A. B. Pacheco, R. Ieva, C. Alaimo, R. Rappuoli, and V. Scarlato. 2002. Autoregulation of Helicobacter pylori Fur revealed by functional analysis of the iron-binding site. Mol. Microbiol. 46:1107-1122. [DOI] [PubMed] [Google Scholar]
- 18.Delany, I., G. Spohn, R. Rappuoli, and V. Scarlato. 2001. The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol. Microbiol. 42:1297-1309. [DOI] [PubMed] [Google Scholar]
- 19.Del Giudice, G., A. Covacci, J. L. Telford, C. Montecucco, and R. Rappuoli. 2001. The design of vaccines against Helicobacter pylori and their development. Annu. Rev. Immunol. 19:523-563. [DOI] [PubMed] [Google Scholar]
- 20.de Vries, N., E. J. Kuipers, N. E. Kramer, A. H. van Vliet, J. J. Bijlsma, M. Kist, S. Bereswill, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2001. Identification of environmental stress-regulated genes in Helicobacter pylori by a lacZ reporter gene fusion system. Helicobacter 6:300-309. [DOI] [PubMed] [Google Scholar]
- 21.Doig, P., M. M. Exner, R. E. Hancock, and T. J. Trust. 1995. Isolation and characterization of a conserved porin protein from Helicobacter pylori. J. Bacteriol. 177:5447-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.el-Shoura, S. M. 1995. Helicobacter pylori. I. Ultrastructural sequences of adherence, attachment, and penetration into the gastric mucosa. Ultrastruct. Pathol. 19:323-333. [DOI] [PubMed] [Google Scholar]
- 24.Evans, D. J., Jr., D. G. Evans, T. Takemura, H. Nakano, H. C. Lampert, D. Y. Graham, D. N. Granger, and P. R. Kvietys. 1995. Characterization of a Helicobacter pylori neutrophil-activating protein. Infect. Immun. 63:2213-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Exner, M. M., P. Doig, T. J. Trust, and R. E. Hancock. 1995. Isolation and characterization of a family of porin proteins from Helicobacter pylori. Infect. Immun. 63:1567-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fassbinder, F., A. H. van Vliet, V. Gimmel, J. G. Kusters, M. Kist, and S. Bereswill. 2000. Identification of iron-regulated genes of Helicobacter pylori by a modified fur titration assay (FURTA-Hp). FEMS Microbiol. Lett. 184:225-229. [DOI] [PubMed] [Google Scholar]
- 27.Haas, R., T. F. Meyer, and J. P. van Putten. 1993. Aflagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using transposon shuttle mutagenesis. Mol. Microbiol. 8:753-760. [DOI] [PubMed] [Google Scholar]
- 28.Huesca, M., A. Goodwin, A. Bhagwansingh, P. Hoffman, and C. A. Lingwood. 1998. Characterization of an acidic-pH-inducible stress protein (hsp70), a putative sulfatide binding adhesin, from Helicobacter pylori. Infect. Immun. 66:4061-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Husson, M. O., D. Legrand, G. Spik, and H. Leclerc. 1993. Iron acquisition by Helicobacter pylori: importance of human lactoferrin. Infect. Immun. 61:2694-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Igarashi, M., Y. Kitada, H. Yoshiyama, A. Takagi, T. Miwa, and Y. Koga. 2001. Ammonia as an accelerator of tumor necrosis factor alpha-induced apoptosis of gastric epithelial cells in Helicobacter pylori infection. Infect. Immun. 69:816-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Illingworth, D. S., K. S. Walter, P. L. Griffiths, and R. Barclay. 1993. Siderophore production and iron-regulated envelope proteins of Helicobacter pylori. Zentbl. Bakteriol. 280:113-119. [PubMed] [Google Scholar]
- 32.Josenhans, C., A. Labigne, and S. Suerbaum. 1995. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J. Bacteriol. 177:3010-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kostrzynska, M., J. D. Betts, J. W. Austin, and T. J. Trust. 1991. Identification, characterization, and spatial localization of two flagellin species in Helicobacter pylori flagella. J. Bacteriol. 173:937-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, A., J. O'Rourke, M. C. De Ungria, B. Robertson, G. Daskalopoulos, and M. F. Dixon. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112:1386-1397. [DOI] [PubMed] [Google Scholar]
- 35.Leunk, R. D., P. T. Johnson, B. C. David, W. G. Kraft, and D. R. Morgan. 1988. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J. Med. Microbiol. 26:93-99. [DOI] [PubMed] [Google Scholar]
- 36.Leying, H., S. Suerbaum, G. Geis, and R. Haas. 1992. Cloning and genetic characterization of a Helicobacter pylori flagellin gene. Mol. Microbiol. 6:2863-2874. [DOI] [PubMed] [Google Scholar]
- 37.Matysiak-Budnik, T., and F. Megraud. 1997. Epidemiology of Helicobacter pylori infection with special reference to professional risk. J. Physiol. Pharmacol. 48(Suppl. 4):3-17. [PubMed] [Google Scholar]
- 38.McClain, M. S., W. Schraw, V. Ricci, P. Boquet, and T. L. Cover. 2000. Acid activation of Helicobacter pylori vacuolating cytotoxin (VacA) results in toxin internalization by eukaryotic cells. Mol. Microbiol. 37:433-442. [DOI] [PubMed] [Google Scholar]
- 39.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schoolnik, and A. Camilli. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura, H., H. Yoshiyama, H. Takeuchi, T. Mizote, K. Okita, and T. Nakazawa. 1998. Urease plays an important role in the chemotactic motility of Helicobacter pylori in a viscous environment. Infect. Immun. 66:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niehus, E., F. Ye, S. Suerbaum, and C. Josenhans. 2002. Growth phase-dependent and differential transcriptional control of flagellar genes in Helicobacter pylori. Microbiology 148:3827-3837. [DOI] [PubMed] [Google Scholar]
- 42.Nuijens, J. H., P. H. van Berkel, and F. L. Schanbacher. 1996. Structure and biological actions of lactoferrin. J. Mammary Gland Biol. Neoplasia. 1:285-295. [DOI] [PubMed] [Google Scholar]
- 43.Odenbreit, S., M. Till, D. Hofreuter, G. Faller, and R. Haas. 1999. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol. Microbiol. 31:1537-1548. [DOI] [PubMed] [Google Scholar]
- 44.Otto, B. R., A. M. Verweij-van Vught, and D. M. MacLaren. 1992. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit. Rev. Microbiol. 18:217-233. [DOI] [PubMed] [Google Scholar]
- 45.Papini, E., B. Satin, N. Norais, M. de Bernard, J. L. Telford, R. Rappuoli, and C. Montecucco. 1998. Selective increase of the permeability of polarized epithelial cell monolayers by Helicobacter pylori vacuolating toxin. J. Clin. Investig. 102:813-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parsonnet, J., G. D. Friedman, D. P. Vandersteen, Y. Chang, J. H. Vogelman, N. Orentreich, and R. K. Sibley. 1991. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325:1127-1131. [DOI] [PubMed] [Google Scholar]
- 47.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salama, N. R., G. Otto, L. Tompkins, and S. Falkow. 2001. Vacuolating cytotoxin of Helicobacter pylori plays a role during colonization in a mouse model of infection. Infect. Immun. 69:730-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Satin, B., G. Del Giudice, V. Della Bianca, S. Dusi, C. Laudanna, F. Tonello, D. Kelleher, R. Rappuoli, C. Montecucco, and F. Rossi. 2000. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a protective antigen and a major virulence factor. J. Exp. Med. 191:1467-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott, D. R., E. A. Marcus, D. L. Weeks, and G. Sachs. 2002. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology 123:187-195. [DOI] [PubMed] [Google Scholar]
- 51.Segal, E. D., J. Cha, J. Lo, S. Falkow, and L. S. Tompkins. 1999. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. USA 96:14559-14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Selbach, M., S. Moese, C. R. Hauck, T. F. Meyer, and S. Backert. 2002. Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo. J. Biol. Chem. 277:6775-6778. [DOI] [PubMed] [Google Scholar]
- 53.Seo, J. K., J. S. Ko, and K. D. Choi. 2002. Serum ferritin and Helicobacter pylori infection in children: a sero-epidemiologic study in Korea. J. Gastroenterol. Hepatol. 17:754-757. [DOI] [PubMed] [Google Scholar]
- 54.Sherlock, G., T. Hernandez-Boussard, A. Kasarskis, G. Binkley, J. C. Matese, S. S. Dwight, M. Kaloper, S. Weng, H. Jin, C. A. Ball, M. B. Eisen, P. T. Spellman, P. O. Brown, D. Botstein, and J. M. Cherry. 2001. The Stanford Microarray Database. Nucleic Acids Res. 29:152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skouloubris, S., A. Labigne, and H. De Reuse. 1997. Identification and characterization of an aliphatic amidase in Helicobacter pylori. Mol. Microbiol. 25:989-998. [DOI] [PubMed] [Google Scholar]
- 56.Spohn, G., and V. Searlato. 1999. The autoregulatory HspR repressor protein governs chaperone gene transcription in Helicobacter pylori. Mol. Microbiol. 34:663-674. [DOI] [PubMed] [Google Scholar]
- 57.Spohn, G., and V. Searlato. 2001. Motility, chemotaxis, and flagella, p. 239-248. In H. L. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 58.Stein, M., F. Bagnoli, R. Halenbeck, R. Rappuoli, W. J. Fantl, and A. Covacci. 2002. c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol. Microbiol. 43:971-980. [DOI] [PubMed] [Google Scholar]
- 59.Suerbaum, S. 1995. The complex flagella of gastric Helicobacter species. Trends Microbiol. 3:168-170; discussion, 170-171. [DOI] [PubMed] [Google Scholar]
- 60.Suerbaum, S., C. Josenhans, and A. Labigne. 1993. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J. Bacteriol. 175:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szczebara, F., L. Dhaenens, S. Armand, and M. O. Husson. 1999. Regulation of the transcription of genes encoding different virulence factors in Helicobacter pylori by free iron. FEMS Microbiol. Lett. 175:165-170. [DOI] [PubMed] [Google Scholar]
- 62.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson, L. J., D. S. Merrell, B. A. Neilan, H. Mitchell, A. Lee, and S. Falkow. 2003. Gene expression profiling of Helicobacter pylori reveals a growth phase dependent switch in virulence gene expression. Infect. Immun. 71:2643-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tonello, F., W. G. Dundon, B. Satin, M. Molinari, G. Tognon, G. Grandi, G. Del Giudice, R. Rappuoli, and C. Montecucco. 1999. The Helicobacter pylori neutrophil-activating protein is an iron-binding protein with dodecameric structure. Mol. Microbiol. 34:238-246. [DOI] [PubMed] [Google Scholar]
- 65.Touati, D. 2000. Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373:1-6. [DOI] [PubMed] [Google Scholar]
- 66.van Vliet, A. H., J. Stoof, S. W. Poppelaars, S. Bereswill, G. Homuth, M. Kist, E. J. Kuipers, and J. G. Kusters. 2002. Differential regulation of amidase- and formamidase-mediated ammonia production by the Helicobacter pylori Fur repressor. J. Biol. Chem. 278:9052-9057. [DOI] [PubMed] [Google Scholar]
- 67.van Vliet, A. H., J. Stoof, R. Vlasblom, S. A. Wainwright, N. J. Hughes, D. J. Kelly, S. Bereswill, J. J. Bijlsma, T. Hoogenboezem, C. M. Vandenbroucke-Grauls, M. Kist, E. J. Kuipers, and J. G. Kusters. 2002. The role of the ferric uptake regulator (Fur) in regulation of Helicobacter pylori iron uptake. Helicobacter 7:237-244. [DOI] [PubMed] [Google Scholar]
- 68.Velayudhan, J., N. J. Hughes, A. A. McColm, J. Bagshaw, C. L. Clayton, S. C. Andrews, and D. J. Kelly. 2000. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37:274-286. [DOI] [PubMed] [Google Scholar]
- 69.Waidner, B., S. Greiner, S. Odenbreit, H. Kavermann, J. Velayudhan, F. Stahler, J. Guhl, E. Bisse, A. H. van Vliet, S. C. Andrews, J. G. Kusters, D. J. Kelly, R. Haas, M. Kist, and S. Bereswill. 2002. Essential role of ferritin Pfr in Helicobacter pylori iron metabolism and gastric colonization. Infect. Immun. 70:3923-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wanachiwanawin, W. 2000. Infections in E-beta thalassemia. J. Pediatr. Hematol. Oncol. 22:581-587. [DOI] [PubMed] [Google Scholar]
- 71.Williams, C. L., T. Preston, M. Hossack, C. Slater, and K. E. McColl. 1996. Helicobacter pylori utilises urea for amino acid synthesis. FEMS Immunol. Med. Microbiol. 13:87-94. [DOI] [PubMed] [Google Scholar]
- 72.Worst, D. J., M. M. Gerrits, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 1998. Helicobacter pylori ribBA-mediated riboflavin production is involved in iron acquisition. J. Bacteriol. 180:1473-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Worst, D. J., B. R. Otto, and J. de Graaff. 1995. Iron-repressible outer membrane proteins of Helicobacter pylori involved in heme uptake. Infect. Immun. 63:4161-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiang, Z., S. Censini, P. F. Bayeli, J. L. Telford, N. Figura, R. Rappuoli, and A. Covacci. 1995. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect. Immun. 63:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]