Abstract
During meiosis, chromosomes undergo large-scale reorganization to allow pairing between homologues, which is necessary for recombination and segregation. In many organisms, pairing of homologous chromosomes is accompanied, and possibly facilitated, by the bouquet, the clustering of telomeres in a small region of the nuclear periphery. Taking advantage of the cytological accessibility of meiosis in maize, we have characterized the organization of centromeres and telomeres throughout meiotic prophase. Our results demonstrate that meiotic centromeres are polarized prior to the bouquet stage, but that this polarization does not contribute to bouquet formation. By examining telocentric and ring chromosomes, we have tested the cis-acting requirements for participation in the bouquet. We find that: (a) the healed ends of broken chromosomes, which contain telomere repeats, can enter the bouquet; (b) ring chromosomes enter the bouquet, indicating that terminal position on a chromosome is not necessary for telomere sequences to localize to the bouquet; and (c) beginning at zygotene, the behavior of telomeres is dominant over any centromere-mediated chromosome behavior. The results of this study indicate that specific chromosome regions are acted upon to determine the organization of meiotic chromosomes, enabling the bouquet to form despite large-scale changes in chromosome architecture.
Keywords: meiosis; bouquet; fluorescence in situ hybridization; chromosomes; maize
Introduction
Meiosis involves the precise segregation of homologous chromosomes to different daughter cells, reducing the diploid chromosome number by half. For this equal partitioning to occur, homologous chromosomes must first pair with each other and undergo recombination, ensuring their segregation to opposite spindle poles at the first meiotic division. The mechanisms of homologous chromosome pairing during meiotic prophase are not well understood. Pairing involves multiple discrete steps, including presynaptic alignment of chromosomes, in which homologous chromosomes roughly colocalize along their lengths, and synapsis, in which a protein scaffold, the synaptonemal complex, forms along the length of paired chromosomes. As synapsis begins at the leptotene–zygotene transition (for review see Zickler and Kleckner 1998), there is a striking rearrangement of chromosome ends to form the meiotic telomere cluster, or bouquet (Gelei, 1921). It has been proposed that the bouquet assists in presynaptic alignment (for review see Loidl, 1990; Scherthan, 2001). During the bouquet, all telomeres are localized to the same side of a surface during the bouquet, and the ends of all chromosomes (including homologues) are made codirectional and roughly colocalized. This organization has the effect of reducing the search space required for chromosomes to find their partners. The bouquet's evolutionary conservation among fungi, plants, and animals (for review see Dernburg et al., 1995), and its appearance coincident with homologue pairing, suggest that it may be important in mediating some aspects of homologue association, but the specific role of this telomere clustering has remained enigmatic.
Bouquet formation is thought to involve two distinct phases: the attachment of telomeres to the nuclear envelope (NE)* in leptotene (Zickler, 1977; Rasmussen and Holm, 1978), followed by their clustering to a small subregion (estimated at 5–10% in maize) of the NE (Bass et al., 1997). Electron microscopy has revealed large, electron-dense structures that extend from the ends of chromosome axial cores into the NE at the sites of telomere attachment (Esponda and Giménez-Martín, 1972); these attachment plaques are a conserved feature of meiosis (for review see Loidl, 1994). No specific proteins have yet been localized to these structures. Telomeres are known to associate with the NE in interphase by binding the nucleoporin Nup145 via the telomere-binding protein Ku and the nuclear pore complex–binding proteins Mlp1 and 2 (Strambio-de-Castillia et al., 1999; Galy et al., 2000). However, synaptonemal complex attachment sites in meiotic prophase seem to localize to regions of the NE devoid of nuclear pore complexes and enriched for the meiosis-specific lamin C2 (Alsheimer et al., 1999, 2000), implicating a different mechanism for telomere–NE association in meiosis that may involve lamin C2. Additionally, the loss of specific telomere-binding proteins (Taz1p in Schizosaccharomyces pombe [Cooper et al., 1997; Nimmo et al., 1998] and Ndj1p in Saccharomyces cerevisiae [Conrad et al., 1997; Trelles-Sticken et al., 2000]) can prevent telomere–NE association. Bouquet formation is also impaired in taz1 and ndj1 mutants. In ndj1 mutants, meiotic pairing is delayed by ∼2 h, underscoring the importance of the bouquet for the normal progression of meiosis (Trelles-Sticken et al., 2000).
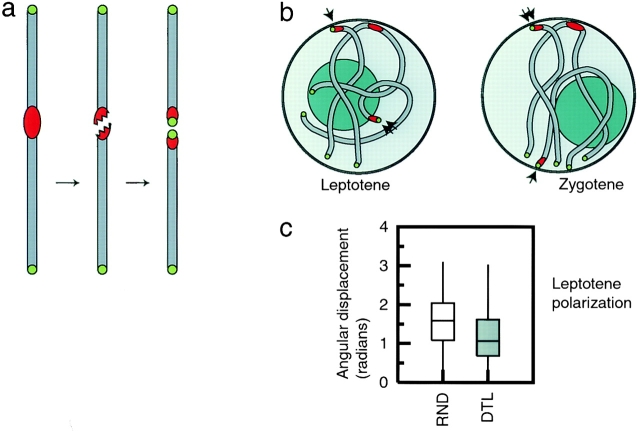
To understand the mechanism of bouquet formation, the state of the chromosomes before the bouquet must also be examined. A potential source of prebouquet chromosomal organization is the Rabl configuration, in which centromeres and telomeres occupy opposite sides of the nucleus due to the poleward movement of centromeres at the previous anaphase (Rabl, 1885). The bouquet superficially resembles the Rabl configuration, but is different in several respects (for review see Cowan et al., 2001). The Rabl configuration is ultimately caused by spindle forces at anaphase, in which centromeres move towards the spindle pole. However, in metazoan cells at the bouquet stage, it is the telomeres rather than the centromeres that are closely apposed to the centrosome. The extent of organization is also strikingly different; a comparison of images of the Rabl configuration and the bouquet in the same organism (Aragón-Alcaide et al., 1997a) shows the bouquet to be a very tight clustering of telomeres, whereas the Rabl configuration is a rather loose grouping of both centromeres and telomeres in opposite hemispheres. Hereafter we will refer to a close aggregation of chromosomal loci, as observed in the bouquet, as clustering, and refer to the looser grouping of chromosome regions into angularly confined domains as seen in the Rabl configuration as polarization. Both polarization and clustering are indicators of the order present in a nucleus, and can occur independently or concurrently.
Becasue both the Rabl configuration and the bouquet represent a polarized orientation of the chromosomes, it has been suggested that the Rabl configuration might contribute to the formation of the bouquet (Fussell, 1987). It has been proposed that in strong-Rabl organisms, the bouquet forms by a tightening of the existing Rabl organization (Aragón-Alcaide et al., 1997b). However, as has been demonstrated previously (Bass et al., 1997; Dong and Jiang, 1998), maize meiotic nuclei lack a strong, persistent Rabl configuration. In order to determine the extent to which preexisting and de novo factors contribute to bouquet formation, it is necessary to know whether the prebouquet nucleus retains any organization left over from the Rabl configuration. We address this question by quantitatively measuring both the polarization and the clustering of centromeres and telomeres over time, from the last cell division before meiosis until the completion of the bouquet.
Another outstanding question regarding bouquet formation is the basic nature of the forces involved; to what extent do forces acting on specific chromosome subregions (i.e., the telomeres), as opposed to forces acting equally on all chromosome regions, govern the behavior of entire chromosomes? Normal bouquet formation involving both chromosome ends occurs in many organisms that naturally possess acrocentric or telocentric chromosomes, e.g. mice (Scherthan et al., 1996) and grasshoppers (Suja and Rufas, 1994), and in which the Rabl orientation would not strictly oppose the centromeres and telomeres. Therefore, it is unlikely that large-scale processes acting on a preexisting Rabl organization are a universal mechanism for bouquet formation. It has also been shown (Bass et al., 2000) that maize chromosomes in an oat background participate in the bouquet normally, without starting in a Rabl configuration. However, prebouquet organization of chromosomes, generally involving pairing or clustering of centromeres, has been extensively noted in many species (mouse and man [Scherthan et al., 1996]; fission yeast [Chikashige et al., 1997]; lily [Suzuki et al., 1997]; budding yeast [Jin et al., 1998]; wheat [Martínez-Pérez et al., 1999]; rye [Mikhailova et al., 2001]; and cattle [Pfeifer et al., 2001]), indicating a widespread trend of centromere-based global chromosome organization. Chromosome derivatives provide a further way to investigate which type of process is acting, by placing chromosome subregions in an abnormal context. In fission yeast, a small (c. 390 kb) linear minichromosome derived from the centromeric region of chromosome III was found to separate from the rest of the centromeres and enter the bouquet normally (Chikashige et al., 1997), indicating a mechanism that may act directly on telomere sequences. We decided to examine two other types of chromosome derivatives readily available in maize: telocentric chromosomes and ring chromosomes. Telocentric chromosomes are formed in rare cases where univalent chromosomes undergo fission at the centromere at the first meiotic division (Weber et al., 1998). As telomeric sequence is added de novo to the broken centromere, telomeres and centromeres are juxtaposed. Ring chromosomes lack physical chromosome termini, but can leave telomeric repeats intact in an interstitial position. Ring chromosomes can be used to test the necessity of physical ends for bouquet formation, because they lack both DNA ends and axial element termini. Nevertheless, they can synapse normally (Haber et al., 1984; Rockmill and Roeder, 1998). The localization of these chromosome derivatives would not be greatly affected if positioning were determined by local telomere-based mechanisms, but would be altered if large-scale geometric properties of chromosomes were involved. In this study, we show that both telocentrics and rings display normal telomere clustering at the bouquet stage, indicating that neither centromere-distal location nor the presence of a physical end are required for bouquet participation.
Results
Detection of centromere and telomere position in nuclei
To establish the localization of centromeres and telomeres, we performed FISH on wild-type (line A344) maize anthers containing cells from sequential premeiotic and meiotic stages. Staging was carried out using criteria outlined in Dawe et al. (1994) and Bass et al. (1997). Briefly, meiotic cells were distinguished from nonmeiotic cells by cell size, cells in the interphase directly preceding meiotic prophase were distinguished from interphase cells of the preceding cell division by the size and overall shape of the nucleus, and prophase cells were distinguished from interphase cells by the condensation state of the chromosomes.
The earliest cells examined were in prophase of the final premeiotic mitosis (Fig. 1 a). Chromosomes in these early cells were both longer and straighter than those of somatic prophase cells. Centromere and telomere FISH signals were evident as doublets, one signal per sister chromatid. The centromere signals appeared to be restricted to roughly one half of the nucleus, whereas the telomeres were concentrated in the opposite half. However, a subset of telomere signals was found in the centromere hemisphere of the nucleus, indicating freedom of movement of the chromosome arms, and an incomplete or degenerate Rabl organization.
Figure 1.
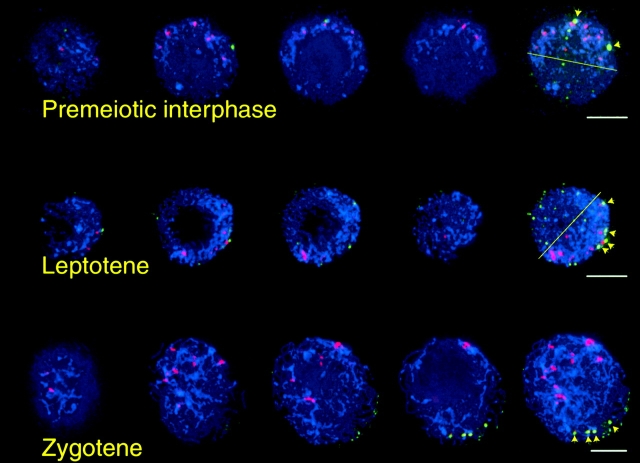
Centromere and telomere positions during the stages of maize meiosis used in this study are shown. Representative nuclei from each of the five stages observed are displayed as a series of semioverlapping quarter-volume projections. Cells were subjected to FISH using oligonucleotide probes to the telomere repeat (green) and the CentC repeat (red), and imaged in three dimensions. Chromosomes are counterstained with DAPI (blue). (a) The last prophase before meiosis. (b) Premeiotic interphase. (c) Leptotene. (d) Zygotene. (e) Tapetal cell prophase. Each stage is shown as a maximum-intensity projection of one quarter of the entire image stack. Arrowheads and insets in d indicate elongated centromere signals. Large fields of view containing several (5–20) cells were acquired and later cropped, resulting in three-dimensional datasets containing one nucleus each. The number of centromere signals detected ranged from 15 to 20 in nuclei before zygotene, and 9 to 16 in nuclei at zygotene. Bars, 5 μm.
Cells in premeiotic interphase (Fig. 1 b) showed subtle changes in nuclear organization. Chromatin was diffuse, and nuclear shape was less spherical compared with previous stages. In most cells examined, the centromeres were markedly confined to one hemisphere of the nuclear volume, and closely apposed to the NE. Telomeres were less visibly polarized than in the prophase preceding premeiotic interphase; they appeared randomly localized throughout the nuclear volume.
Further changes in nuclear organization were apparent in cells that had entered meiotic prophase. Leptotene nuclei (Fig. 1 c) were identified by the resolution of individual, unpaired chromosome fibers. In leptotene, centromeres remained visibly confined to half the nuclear volume, though outlying centromeres were observed more frequently than in earlier meiotic nuclei. Telomeres possessed no obvious polarization. A significant alteration of this pattern was evident in zygotene cells (Fig. 1 d), marked by the presence of paired chromosomes and formation of the telomere bouquet. Centromeres at this stage became randomized throughout the nuclear volume. The morphology of centromere signals also changed; in a given nucleus, six or seven of the signals became elongated (up to 2.5 μm in length, indicated by yellow arrowheads) and three or four of these possessed a nonstaining region separating two lobes of roughly equal size (Fig. 1 d, insets), though many signals remained small and punctate. This change coincides with the elongation previously observed for heterochromatic knobs (Dawe et al., 1994).
Next, we asked whether the centromere polarization we observed in prebouquet cells was a feature common to all maize cells or exclusive to meiotic cells and their immediate precursors. We examined the distribution of centromeres and telomeres in tapetal cells, somatic cells of the anther which surround meiocytes and are involved in subsequent pollen development. In tapetal cells, centromeres and telomeres were distributed throughout the nuclear volume, and did not display the persistent Rabl orientation found in prophase nuclei of the last premeiotic mitosis (Fig. 1 e). Thus, this polarization distinguished tapetal cells and meiocytes, which diverge in their lineage by several cell divisions (Chaubal et al., 2000).
In no stage or cell type was presynaptic centromere pairing seen. Prior to zygotene, we consistently detected between 16 and 20 centromere signals in each nucleus. As the haploid number of maize is 10, it appears that unlike polyploid wheat (Aragón-Alcaide et al., 1997a) and lily (Suzuki et al., 1997), maize does not undergo pairwise premeiotic centromere association.
Centromeres do not cluster at any stage of meiotic prophase
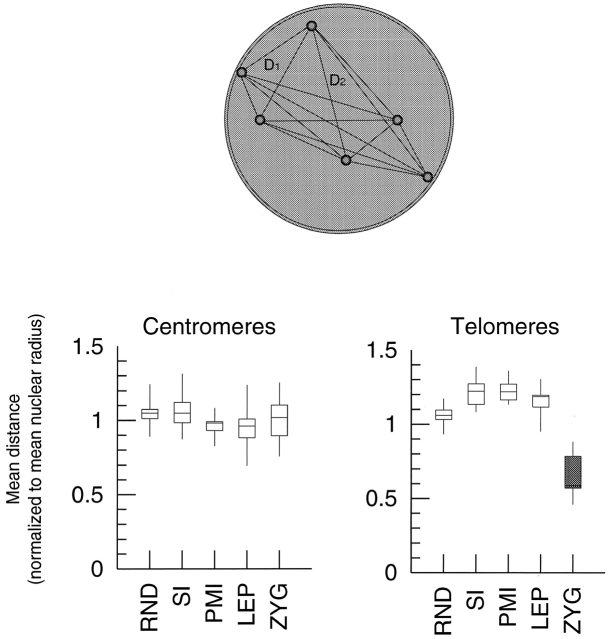
Our preliminary observations suggested a previously unreported organization of maize centromeres in premeiotic interphase and leptotene. In order to quantitatively confirm this organization, the three-dimensional images were submitted to spatial analysis and tested for significant deviations from random expectations. We first asked whether centromeres clustered in maize, as has been observed in some other organisms (Scherthan et al., 1996; Aragón-Alcaide et al., 1997b; Suzuki et al., 1997; Jin et al., 1998). We defined clustering of points as a shift in the distribution of distances between pairs of points to a significantly lower (closer) range than that of randomly placed points. Meiotic cells from premeiotic interphase through zygotene were obtained and FISH was performed against centromeres and telomeres. After converting deconvolved image stacks to three-dimensional models, the distance between signals was measured for all possible pairs of signals. The median pairwise distance values of each nucleus were pooled and compared with measurements from simulated nuclei containing randomly placed signals (see Materials and methods). No significant clustering of centromeres was observed at any stage, and no stages significantly differed from any other (Fig. 2; Table I). When examined individually, even nuclei with complete restriction of centromere signals to one hemisphere do not significantly differ from randomly generated nuclei by pairwise distance analysis (unpublished data). Thus, prebouquet en masse clustering of centromeres does not occur in maize. As a positive control, telomeres were submitted to an identical pairwise distance analysis. The clustering behavior of telomeres reiterates the findings of Bass et al. (1997), in that significant clustering occurs at zygotene but not at any other stage.
Figure 2.
Close clustering of signals is revealed by pairwise distance measurements. The diagram (above) illustrates the measurement taken: Euclidean distances are measured for each possible pair of signals, and all distances are normalized to the mean nuclear radius. The mean value of distances is taken from each nucleus. These mean values were grouped together by stage, and distributions for each stage are shown by a box-whisker diagram of centromere distances (left) and telomere distances (right). The only significantly clustered condition (shaded) is telomeres at zygotene; centromeres never display significant clustering. LEP, leptotene; PMI, premeiotic interphase; RND, random points placed in modeled nuclei; SI, somatic interphase; ZYG, zygotene. (See Table I for details).
Table I. Mean pairwise distance of signals normalized to nuclear radius.
| Random | Somatic | PMI | Leptotene | Zygotene | |
|---|---|---|---|---|---|
| Centromeres | 1.05 (0.07) | 1.06 (0.10) | 0.99 (0.10) | 0.94 (0.11) | 1.02 (0.15) |
| n = 1,000 | n = 31 | n = 10 | n = 53 | n = 15 | |
| Telomeres | 1.06 (0.05) | 1.22 (0.09) | 1.23 (0.09) | 1.16 (0.08) | 0.67 (0.14)a |
| n = 1,000 | n = 16 | n = 5 | n = 15 | n = 12 |
The mean pairwise distance (in nuclear radius units) of centromere and telomere signals shown by stage; SDs are in parentheses.
P < 0.01.
Centromeres, but not telomeres, are polarized within the nucleus before zygotene
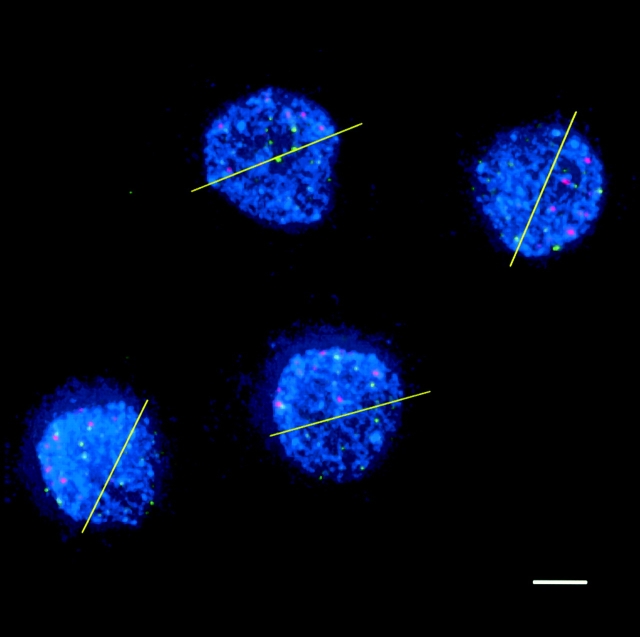
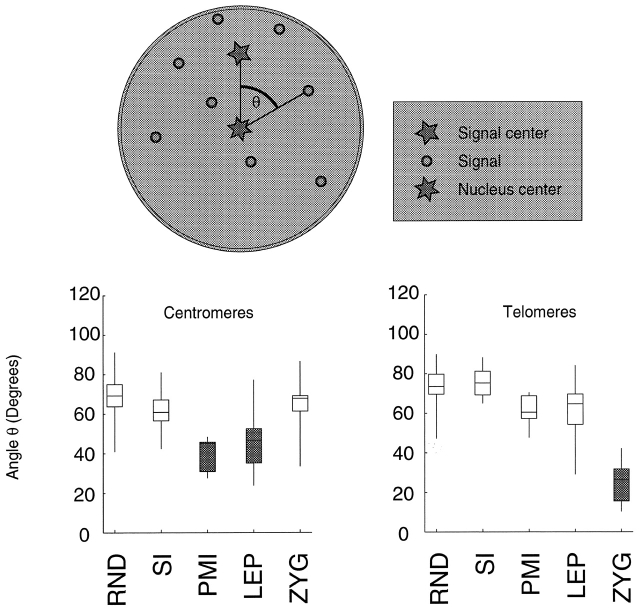
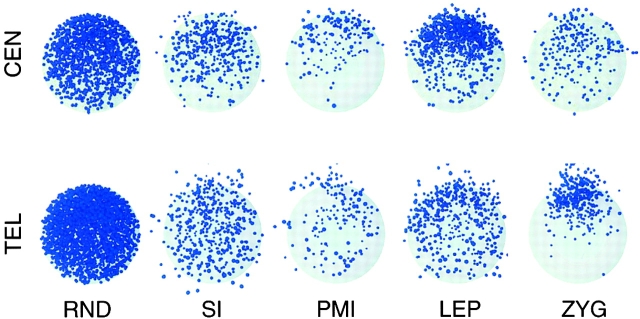
Although centromeres do not cluster, visual inspection suggested that centromeres were organized in a polar fashion, i.e., confined to one half of the nucleus. In Fig. 3, four leptotene nuclei are shown with lines drawn demarcating two halves: one containing all the centromeres, the other containing no centromeres. To confirm these observations quantitatively, we analyzed the angular distributions of centromere and telomere signals in a larger sample of nuclei. It is important to note that pairwise distance analysis provides no information about polarization. In the course of analyzing random distributions of signals, we found that signals confined to one hemisphere did not significantly differ in their pairwise distance distribution from signals distributed throughout an entire sphere. We decided to quantify polarization by using the more sensitive measure of the angular displacement of individual signals from the axis formed by the signal midpoint and the nuclear center. For each nucleus, an axis was defined by the line connecting the midpoint of the signals (centromeres or telomeres) and the nuclear center; the angular displacement of each individual signal from this axis, measured from the origin (nuclear center), could then be calculated. Equivalent measurements were performed for simulated nuclei (see Materials and methods). Significant polarization of centromeres is seen in both premeiotic interphase and leptotene (Fig. 4; Table II). Telomeres are unpolarized until the bouquet, indicating that the clustering of telomeres is coincident with their polarization. The overall polarization of centromeres at premeiotic interphase and leptotene, and telomeres at zygotene, can be visualized by superimposing all models of a given stage, rotated appropriately (Fig. 5).
Figure 3.
Four leptotene meiocytes with telomeres (green) and centromeres (red) labeled with FISH. Chromosomes (blue) are counterstained with DAPI. The yellow lines demarcate a hemisphere containing all the observed centromere signals in each nucleus. The telomeres cannot be divided at this stage. Bar, 5 μm.
Figure 4.
The polarization of signals is shown by measuring their angular separation. The diagram (above) shows the measurement taken. First, the nuclear center (six-pointed star) and signal midpoint (five-pointed star) are calculated. The angle θ formed by the axis connecting each individual signal with the nuclear midpoint with the axis connecting the signal midpoint to the nuclear midpoint is measured in three dimensions. The mean angular separation is calculated for each nucleus, and grouped by stage. These distributions are shown for each condition by box-whisker diagrams. Centromeres in premeiotic interphase and leptotene, and telomeres in zygotene, both display significant clustering (shaded boxes). LEP, leptotene; PMI, premeiotic interphase; RND, random points placed in a modeled nucleus; SI, somatic interphase; ZYG, zygotene. (See Table II for details).
Table II. Mean angular separation from nuclear center–signal center axis.
| Random | Somatic | PMI | Leptotene | Zygotene | |
|---|---|---|---|---|---|
| Centromeres | 68.47 (9.91) | 61.48 (8.82) | 42.4 (10.6)a | 44.86 (12.10) a | 63.00 (14.60) |
| n = 1,000 | n = 31 | n = 10 | n = 53 | n = 15 | |
| Telomeres | 74.14 (8.02) | 76.15 (7.33) | 60.90 (9.22) | 60.80 (14.84) | 25.27 (10.03)a |
| n = 1,000 | n = 16 | n = 5 | n = 15 | n = 12 | |
| Cen-Tel anglesb | 82.10 (36.27) | 97.46 (34.15) | 118.66 (40.62) | 109.55 (40.05) | 74.48 (30.37) |
The mean angular separation of signals (in degrees) from the signal midpoint is shown by stage; SDs are in parentheses.
a P < 0.001.
bSample sizes are the same as the row above.
Figure 5.
Ray-traced three-dimensional models showing superimposition of all nuclei used in this study. Small blue spheres are centromere (a) or telomere (b) signals. Each nucleus is rotated so that the signal midpoint satisfies (x = 0, y = 0, z > 0). LEP, leptotene; PMI, premeiotic interphase; RND, random points placed in a modeled nucleus; SI, somatic interphase; ZYG, zygotene. The overall polarization of centromeres at premeiotic interphase and leptotene and clustering of telomeres at zygotene are readily apparent.
Telocentric ends switch from centromere- to telomere-like behavior at the leptotene–zygotene transition
The quantitatively measured stage-specific behavior of centromeres and telomeres in wild-type maize nuclei provided a baseline to ask further questions about chromosome organization, using chromosome derivatives. The mechanisms of Rabl maintenance and bouquet formation were addressed through the use of lines containing either one copy (heterozygous) or two copies (homozygous) of telocentric chromosome 3 (Fig. 6). Heterozygous lines contain one copy of an unbroken chromosome 3. In principle, the behavior of a centromeric terminus before or after the bouquet could be centromere- (Fig. 6 b, top, single arrows) or telomere-like (Fig. 6 b, bottom, double arrows). To determine which mode of behavior was dominant, we performed FISH with telomere and centromere probes in both homozygous and heterozygous ditelocentric maize lines at all stages, and quantitatively analyzed their positions with respect to the normal centromeres and telomeres. The intensity and size of telomere signals was 2–5× greater at the centromeric ends of the telocentric chromosomes than at all other chromosome ends. Fortuitously, this larger size combined with the directly adjacent localization of CentC signal provided a convenient way of marking these ends without the need for a specific chromosome 3 centromere marker.
Figure 6.
The centromeric ends of telocentric chromosomes behave as centromeres before the bouquet, and as telomeres during the bouquet. (a) Steps in the formation of the telocentric chromosome. A normal chromosome 3 (left) undergoes rare centric misdivision, leading to a broken centromere (middle). Telomere sequence is then added to the centric ends by an unknown mechanism to form telocentric chromosomes (right). (b) A schematic of possible telocentric behaviors. In leptotene (left), the centromeric end can potentially either be polarized with the other centromeres (top, single arrow) or localize randomly like other telomeres (double arrow), whereas in zygotene, centromeric ends can either localize randomly like the other centromeres (top, double arrow) or participate in the bouquet with the other telomeres (bottom, single arrow). (c) Box-whisker plot of mean angular separation of the centromeric ends from the centromere midpoint at leptotene. The lower mean angle for the ditelocentric signals demonstrates that centromeric ends are significantly constrained compared with randomly placed points, and thus they are polarized with the other centromeres in the nucleus.
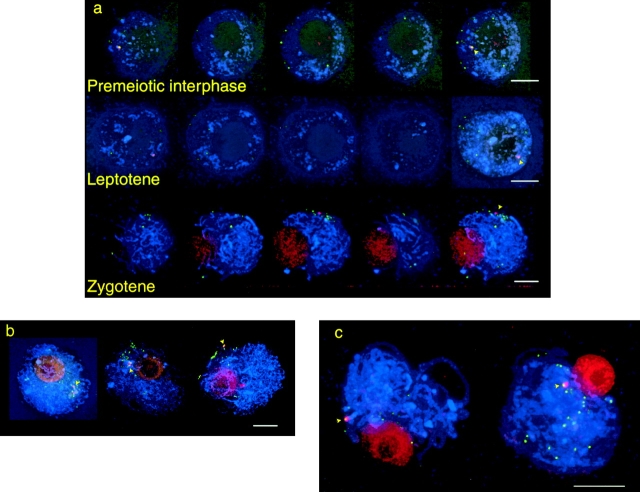
In premeiotic interphase (preceding leptotene but after the last premeiotic cell division) and leptotene nuclei, the centromeric ends of telocentric chromosomes were consistently found to lie in the same hemisphere with the majority of centromere signals (Fig. 7, top). Telocentric ends were analyzed for their polarization relative to the midpoint of the centromeres at leptotene. Their angular displacement from the axis formed by the nuclear midpoint and the centromere signal midpoint was measured, and compared to normal centromeres and to randomly placed points. For random points, two points were placed at random in the models and their angular separation from the centromere polarization axis was measured. The centromeric ends were found to be significantly constrained to one end of the axis (Figs. 6 c and 7, middle; Table III). The centromeric ends show no difference in polarity from the other centromeres, whereas they do significantly differ from random points. Although normal chromosome ends are randomly localized at leptotene, centromeric ends were constrained to one pole.
Figure 7.
Localization of telocentric ends in meiotic prophase. In each row, the first four images are quarter projections of a single nucleus; the fifth image is the full projection. Telomeres are labeled in green; centromeres are labeled in red; chromosomes (counterstained with DAPI) are blue. Telocentric ends, characterized by brighter than normal telomere staining with adjacent centromere staining, are highlighted in the full projections with arrows. In premeiotic interphase and leptotene (top two rows), the telocentric ends localize in the same hemisphere as the bulk of the centromeres (yellow line) and appear close to the NE. In zygotene, all the telocentric ends localize to the bouquet. The premeiotic nucleus has one wild-type copy of chromosome 3, and one telocentric (long and short arms separate) copy yielding two signals; the leptotene and zygotene nuclei have two telocentric copies which yield four signals.
Table III. Measurements of telocentric and ring chromosome positions.
| Random | Telocentric |
|---|---|
| 90.30 (38.56) | 67.04 (35.29) a |
| n = 1,000 | n = 19 |
The polarization of telocentric chromosomes' centromeric ends along the centromere polarization axis at leptotene, compared with the polarization of two randomly placed points.
P < .001.
In zygotene, the telocentric ends take on the behavior of normal telomeres, participating in the bouquet; projections of a typical nucleus are shown in Fig. 7, bottom. In nuclei with complete bouquet formation, the centromeric ends were never observed outside the bouquet. Thus, centric telomeres switch to normal telomeric behavior at zygotene. These observations demonstrate that a telomere adjacent to a centromere does not interfere with normal centromere behavior, nor does the nearby presence of a centromere interfere with normal telomere behavior.
The interstitial telomeres of ring chromosomes can participate in the bouquet
We analyzed the behavior of ring chromosomes to assess whether participation in the meiotic bouquet requires chromosome termini. The ring chromosomes used in the study were derived from an A–B translocation of maize chromosome 9. The initial translocation chromosome spontaneously circularized after a centric misdivision to give rise to a ring chromosome (Fig. 8 a). The interstitial telomeric repeats of the ring chromosome are thus located near the centromere, which contains a highly repetitive DNA sequence of the B repeat family (Kaszás and Birchler, 1996). Because this block of telomeric repeats is the only occurence of this sequence in the genome that is closely linked to B repeats, we could determine its localization by using FISH probes against both repeats in the same samples. Because the ring chromosome contains wild-type alleles of the seed markers shrunken (Sh) and bronze (Bz), its segregation can be monitored phenotypically.
Figure 8.
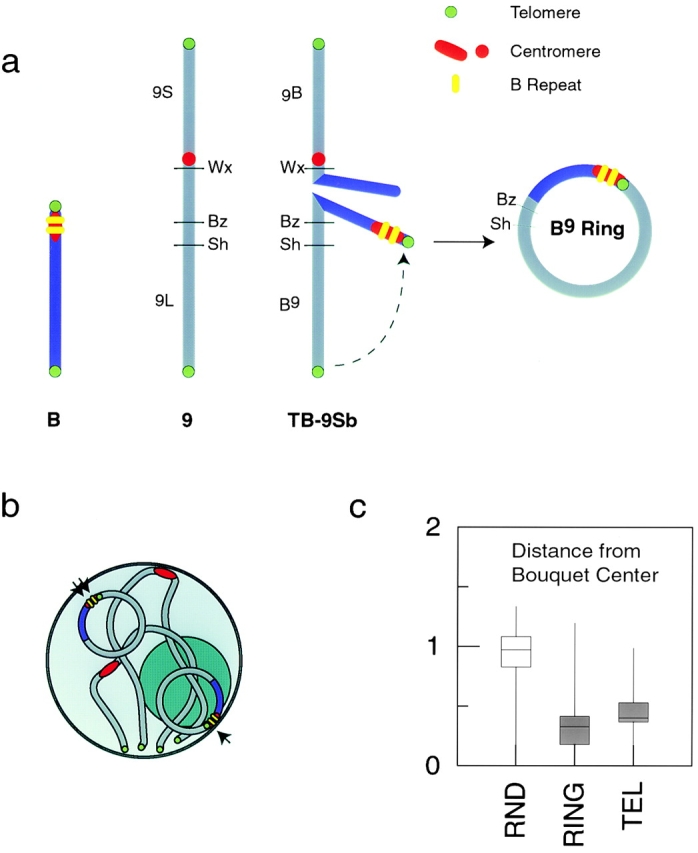
Meiotic behavior of a ring chromosome. (a) The origin of the ring chromosome used in this study. Firstly, a B chromosome underwent reciprocal translocation with chromosome 9 to give rise to translocation chromosome TB-9Sb. This chromosome then circularized to form the ring chromosome B9 ring. The resulting configuration places a B centromere (containing blocks of B repeat sequences) adjacent to a canonical telomere sequence, making possible its detection by FISH. (b) A schematic of possible ring chromosome behaviors during the bouquet. A ring chromosome may localize randomly within the cell (double arrows) or near the bouquet site (single arrow). (c) Box-whisker plots of distances from the bouquet center to the ring signals and to all telomere signals. RND, distances of randomly placed points in modeled nuclei. By this measure the ring signals do not significantly differ in their localization from telomere signals.
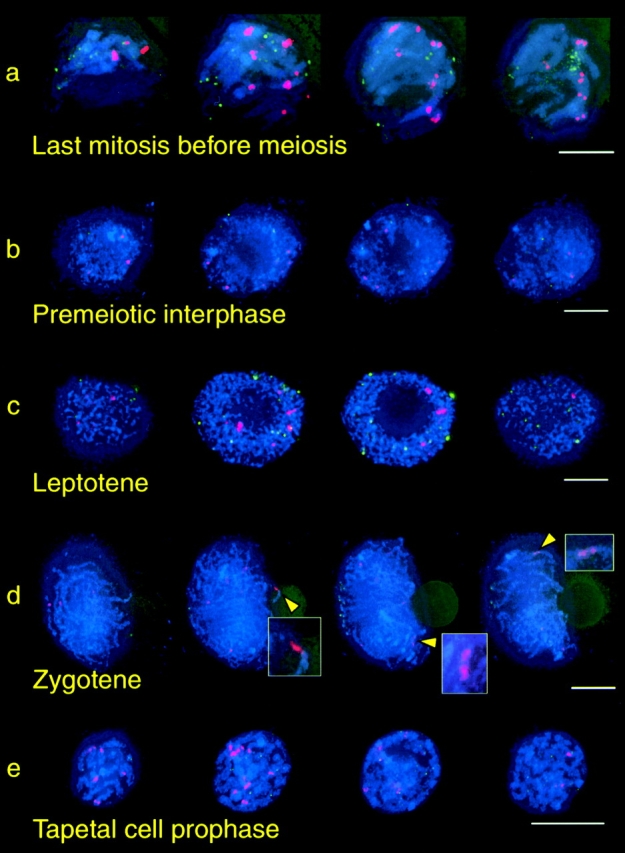
Cells at zygotene were probed simultaneously with telomere and B repeat oligonucleotides. In premeiotic interphase and leptotene (Fig. 9 a, top and middle), the B repeat sequence was localized at the nuclear periphery; however, the absence of centromere staining at this stage did not enable us to determine its polarization state. Inspection of later stages clearly indicated the presence of the B repeat sequence in the bouquet in almost all the bouquet cells examined (Fig. 9, a, bottom, and b). To determine that ring chromosome–bouquet associations were not random, a quantitative spatial analysis was carried out. Models were created of 25 nuclei, comprised of the nuclear (chromatin) boundary, nucleolus, telomere signals, and the ring signal. As before, the distance measurements in each model were normalized to the mean nuclear radius. The telomere midpoint was first calculated. The distance of ring signals from the telomere midpoint was compared with that of normal telomere signals and randomly placed points (Fig. 8 c). No differences between normal and ring telomeres were detected by a t test. The distribution of random points in simulated nuclei was significantly different from both telomeres and rings (Table IV). This indicates that ring chromosomes are localized significantly closer to the telomere midpoint than would be expected from random placement. Our observations demonstrate that ring chromosomes enter the bouquet with the same timing as normal chromosomes. Therefore, chromosome ends are not required to bring a chromosome region into the bouquet.
Figure 9.
Localization of ring chromosome before, during, and after the bouquet. (a) In each row, the first four images are quarter-projections of a single nucleus; the fifth image is the full projection. Telomeres are green; B repeat sequence (on the ring chromosome) is red; chromosomes (counterstained with DAPI) are blue. Ring chromosome locations is highlighted in the full projection with arrows. In premeiotic interphase and leptotene (top two rows) the ring chromosome are found (>80% of the time in >20 nuclei) at the outer edge of the DAPI-staining region, indicative of closeness to the NE. In zygotene (bottom row), the ring chromosome is in the telomere cluster. In this particular example the ring chromosome has entered the bouquet before at least four other telomeres, including those attached to the nucleolus on chromosome 6. (b) Three additional bouquet stage nuclei, demonstrating the presence of the B chromosome repeat in the immediate vicinity of the bouquet (arrows). (c) Two ring-containing nuclei at pachytene, after synapsis has completed and the bouquet has dispersed. The ring chromosome remains associated with the nuclear envelope (arrows) and is near the nucleolus. Bars, 5 μm.
Table IV. Mean signal–bouquet midpoint distance (normalized to nuclear radius).
The distance of ring chromosomes (as indicated by the B repeat signal) from the center of the bouquet at zygotene, in units of mean nuclear volume.
P < .001.
.01 < p < .02.
Discussion
The formation of the meiotic bouquet is a dramatic example of chromosome movement that occurs in the absence of a spindle. To form the bouquet, chromosome ends must be moved across large distances in the nucleus to a highly nonrandom position, and this movement must be coordinated such that all the ends arrive at the same place at nearly the same time. The mechanism of this drastic nuclear reorganization is unknown. In this work we have shown evidence that in maize, the mechanism does not act on global organization of the nucleus or of individual chromosomes, but rather acts locally on telomeres, whether or not these are located at a chromosome terminus.
A novel quantitative analysis of centromere position allowed us to determine that before the bouquet, centromeres are not randomly placed but are polarized, constrained to lie mostly within the same nuclear hemisphere. Thus, the bouquet is not the first sign of chromosome organization in maize meiotic prophase. This observation suggests that the meiotic nuclear organization of maize is similar to that of other model organisms (e.g., wheat, mice, humans) that display centromere-based organization prior to the bouquet. That this organization was not immediately obvious upon visual inspection highlights the fact that quantitative measurements are needed to exhaustively determine the organizational state of a cell or nucleus.
The leptotene–zygotene transition marks an abrupt shift in the behavior of centromeres and telomeres. Before zygotene, centromeres display significant angular constrainment, and afterwards they have no organization. Telomeres behave the opposite way: prior to zygotene there is no observable polarity, whereas afterwards the telomeres are both polarized and clustered. This indicates that meiotic nuclei in maize undergo a polarization switch, similar to the cluster switch described by Martínez-Pérez et al. (1999) in wheat. However, these authors reported that this switch occurs before the onset of meiotic prophase, earlier than we observe this transition in maize. In polyploid wheat, two types of centromere organization were observed: polarization (due to the Rabl configuration) and pairwise association. In diploid wheat, pairwise associations were not observed (Martínez-Pérez et al., 2000, 2001). The authors presented a model in which the polarization of centromeres promotes their pairwise association, which begins as nonhomologous pairs but are corrected to homologous pairs. Thus, in hexaploid wheat, the polarization of centromeres appears to directly contribute to the timely completion of the homology search. In contrast, although maize retains centromere polarity from the Rabl configuration, we observe no pairwise centromere associations until after the bouquet has formed. As maize is a diploid (though descended from a tetraploid ancestor [Gaut and Doebley, 1997]), the lack of centromere pairing in maize is not unexpected. Although we cannot rule out that maize centromere polarization contributes to the homology search in a subtle way, we conclude that it is largely nonfunctional and is not necessary for either bouquet formation or homologous pairing.
Our results are also reminiscent of observations of fission yeast at the onset of meiosis. Fission yeast centromeres are normally clustered near the spindle pole body in haploid cells, with telomeres loosely grouped at the other end of the nucleus; as cells and nuclei fuse and meiosis begins, the telomeres cluster at the spindle pole body and the centromeres are released (Chikashige et al., 1997). The polarization switch in maize is coincident with the transient elongation of both knob heterochromatin and CentC signals, consistent with the notion that the large-scale changes in nuclear organization may be mediated by changes at a lower-order chromatin level.
Somatic (tapetal) cells did not display any chromosomal organization during meiosis, in contrast to what has been reported for wheat (Martínez-Pérez et al., 1999). However, cells in the prophase immediately before meiosis did retain a degenerate Rabl organization, as Bass et al. (1997) also observed. Therefore, it is likely that a developmental decision takes place in the sporogenous lineage that results in the retention of larger degrees of chromosomal order. The cell divisions prior to meiosis in some organisms have been reported to require longer times to complete than divisions in somatic tissue (Bennett, 1977), indicating that preparation for meiosis could begin even before premeiotic S phase.
The reorganization of meiotic chromosomes in the nucleus was found to operate according to local, sequence-based rules, rather than large-scale geometric properties of chromosomes. The behavior of telocentric chromosomes, even when these are created de novo from metacentric configurations, suggests that the behavior of centromeres and telomeres occurs solely by virtue of their being centromeres and telomeres, and does not depend on their position along the chromosome or within the nucleus. The reasoning is as follows: in the all-metacentric progenitors of the chromosome 3 ditelocentric lines, chromosome 3 localized normally at zygotene, with both its telomeres participating in the bouquet, and its centromere randomly located with respect to the bouquet. The only regions of the chromosome brought near to the bouquet were those near the ends. However, after centromere misdivision and end healing, the regions adjacent to the centromere become competent to enter the bouquet. Therefore, the changes brought about to the chromosome ends as a result of the end-healing mechanism must be responsible for the new behavior. We detect telomere sequence at these ends by FISH, which suggests that telomere sequence was added during end-healing, and this sequence may be sufficient for bouquet entry. However, in the absence of a molecular understanding of the end-healing mechanism we cannot rule out the possibilities that nontelomere sequences are added to the ends, or an epigenetic property is conferred on the ends, either of which may play a functional role in telomere motility.
The ability of ring chromosomes to be transported to the bouquet site further supports the autonomy of chromosome regions for controlling behavior. Ring chromosomes do not require the geometric property of possessing a physical end for entry into the bouquet. Therefore, whatever machinery is involved in bringing telomeres to the bouquet site does not need to recognize a physical chromosome end, but rather a chromosome region which, at a minimum, contains telomere sequence. It must be noted that in both the telocentric and the ring cases, the telomeres are located next to large blocks of heterochromatin (centromeric heterochromatin in the case of telocentrics, and B repeat heterochromatin in the case of rings), and this may be an important determinant of their behavior, perhaps by providing an appropriate chromatin context for the telomeres to function.
The formation of the bouquet coincides with many other events, including the appearance of large numbers of Rad51 foci on meiotic chromosomes (Ashley et al., 1995; Terasawa et al., 1995; Franklin et al., 1999), and reorganization of microtubules and nuclear pores from a uniform to a clustered distribution (unpublished data; Scherthan et al., 2000). These latter extra-chromosomal reorganizations are of particular interest, in that they may suggest a physical link between cell and nuclear polarization. The strict coincidence of the loss of centromere polarization, the elongation of both knob and centromeric heterochromatin, and the clustering of telomeres strongly suggest that a specific event is occurring at the leptotene-zygotene transition whose role is to reorganize the nucleus, presumably so that the events of pairing and recombination may take place. The data in this paper suggest that as part of the leptotene–zygotene transition, telomere sequences associate with meiosis-specific machinery at the NE, which then actively transports the telomeres, along with the rest of the chromosome, to a bouquet site. The elucidation of this transport machinery will be a crucial step toward a complete understanding of the ways in which the meiotic nucleus can be organized.
Materials and methods
Cytological material
Maize (Zea mays) inbred line A344, ditelocentric lines, and ring chromosome lines were grown in greenhouses under standard conditions. A line containing the B9 ring chromosome was provided by Étienne Kaszás and James Birchler (University of Missouri, Columbia, MI) (Kaszás and Birchler, 1996, 1998). This line contains one normal chromosome 9 carrying recessive mutant alleles of shrunken and bronze, whereas wild-type copies of these genes are present on a ring chromosome derived from a TB translocation of chromosome 9. Lines with ditelocentric chromosome 3, originally developed by Katherine Rose and Rick Staub (Carleton College, Carlton, Victoria, Australia), were provided by Lisa Harper (University of California, Berkeley, CA). Tassels were harvested during daylight hours at various stages of development, generally at 6–8 wk postplanting. Anthers were dissected from florets and fixed with 4% paraformaldehyde in Buffer A (15 mM Pipes-NaOH, pH 6.8, 80 mM KCl, 20 mM NaCl, 0.5 mM EGTA, 2 mM EDTA, 0.15 mM spermine tetra HCl, 0.05 mM spermidine, 1 mM DTT, 0.32 M sorbitol) (Belmont et al., 1987; Dernburg et al., 1996) for 2 h at room temperature. After fixation, anthers were washed in Buffer A three times, and then stored at 4°C for up to 1 yr with no significant degradation in structural integrity.
Probes
CentC, a short (155 base pairs) tandem repeat localizing to all 10 maize centromeres (Ananiev et al., 1998) was provided by Dr. Evgueni Ananiev (Pioneer Hi-Bred International, Inc., Johnston, IA). Dot-plot analysis identified the most conserved region in the repeat: a 27-mer, 5′-CCTAAAGTAGTGGATTGGGCATGTTCG-3′. Direct 5′ labeled Cy5 or Texas red oligonucleotide probes with this sequence were obtained from Genset, Inc. The specific localization of this oligonucleotide to centromeres was confirmed by observing FISH signals at the pole-facing tips of meiotic metaphase I chromosomes and at the primary constrictions of mitotic prometaphase chromosomes (unpublished data). Oligonucleotides complementary to the maize telomere repeat (5′-3′ [CCCTAAA]4) were obtained from Genset Inc. with direct 5′ conjugation of fluorescein or Texas red. A Texas red–conjugated oligonucleotide complementary to the B repeat was provided by Étienne Kaszás (Novartis Agricultural Biotechnology Research institute, Research Triangle Park, NC).
FISH
Anthers were removed from florets and fixed in 4% paraformaldehyde (EM Sciences) in Buffer A. Meiocytes were extruded from fixed anthers into Buffer A and embedded in a 5% polyacrylamide pad. Hybridization was performed as described in Bass et al. (1997); briefly, oligonucleotide probes were added at a final concentration of 2–4 μg/ml to the hybridization mixture (2× SSC, 50% formamide); probes were preincubated on slides for 3–4 h at 37°; target DNA was denatured at 95° for 6 min; and postdenaturation hybridization was done at 30° for 8–12 h. DNA was counterstained with DAPI for < 10 min and slides were mounted in glycerol containing 1% n-propyl gallate.
Microscopy
Images were acquired on a DeltaVision (Applied Precision, Inc.) imaging station: an Olympus IX70 inverted microscope with 40×, 1.3 NA and 100×, 1.35 NA oil-immersion lenses and a Photometrics (Roper Scientific, Inc.) cooled CCD. The 100× lens was used for qualitative inspection; the 40× lens was used for acquisition of larger sample sizes for quantitative analysis. All images were taken with a Z step size of 0.2 microns, saved as three-dimensional stacks, and subjected to constrained iterative deconvolution (Chen et al., 1996).
Data analysis
The DeltaVision/softWoRx package (Applied Precision, Inc.) was used for three-dimensional model creation. DeltaVision 3-D model and 3-D object files were processed into text files containing raw (x, y, z) coordinates. Perl programs were written and used to analyze the three-dimensional data for clustering and polarization, and to generate random distributions for comparison.
Random point distributions
To determine whether any observations could be the result of chance, a random population of nuclei was generated for statistical comparison. Nuclei were represented by unit spheres (radius = 1). Points within, representing signal positions of centromeres and telomeres, were generated by randomly sampling spherical coordinates from uniform distributions on the azimuth (0° to 360°), the sine of the elevation (−1–1), and the cube of the distance from the center (0–1). The center of a void, representing the nucleolus, was picked in the same way. The nucleolar volume was set to 10% of the nuclear volume, in accordance with the mean volume of the nucleolus in modeled nuclei. Random points that fell within the void were excluded from the final list of points. 1,000 nuclei were thus generated, each containing from 15–20 centromere points, and from 30–40 telomere points.
Quantitative analysis of centromere and telomere positions
The original three-dimensional images were simplified into geometric models. The nucleolar and nuclear volumes were modeled by hand based on DAPI staining; FISH signals were automatically segmented based on an initial threshhold of the mean image intensity value plus one standard deviation, and visually confirmed for each nucleus. The error in automatic feature detection, compared to manual segmentation, was <5% and not significant (10 nuclei were modeled 10 times each both manually and automatically and volumes were compared; unpublished data). The three-dimensional coordinates defining each feature were saved as lists of x, y, and z coordinates for further analysis. Models were translated such that the nuclear midpoint was placed at the origin (at x = 0, y = 0, z = 0), and were individually scaled to set the mean nuclear radius to an arbitrary distance unit of 1. Thus, all distances in each model are expressed in units of their mean nuclear radius. For statistical comparisons, an unequal variance difference of means t test was used. In the figures which show quantitative measurements, distributions of values are shown by box-whisker diagrams. The horizontal line through the box marks the median value. The lines above and below the boxes extend to the entire range of observed measurements. The box regions above and below the median line contain the measurements 25% above and below the median, respectively. The conditions that differ significantly from the randomly generated measurements are indicated in the diagrams by solid boxes.
Acknowledgments
We thank E.V. Ananiev for providing CentC DNA, É. Kaszás and L.C. Harper for providing maize stocks, H.W. Bass for advice on FISH methodology; I. Golubovskaya for assistance with staging; and C.R. Cowan, A.F. Dernburg, L.C. Harper, and J.L. Paluh for critical reading of the manuscript.
This work was supported by the National Institutes of Health grant GM R01 46547.
P.M. Carlton's current address is Lawrence Berkeley National Laboratory, 1 Cyclotron Rd., Mailstop 84-171, Berkeley, CA 94720.
Footnotes
Abbreviation used in this paper: NE, nuclear envelope.
References
- Alsheimer, M., E. von Glasenapp, R. Hock, and R. Benavente. 1999. Architecture of the nuclear periphery of rat pachytene spermatocytes: distribution of nuclear envelope proteins in relation to synaptonemal complex attachment sites. Mol. Biol. Cell. 10:1235–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsheimer, M., E. von Glasenapp, M. Schnoelzer, H. Heid, and R. Benavente. 2000. Meiotic lamin C2: the unique amino-terminal hexapeptide GNAEGR is essential for nuclear envelope association. Proc. Natl. Acad. Sci. USA. 97:13120–13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev, E., R. Phillips, and H. Rines. 1998. Chromosome-specific molecular organization of maize. (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA.. 95:13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragón-Alcaide, L., S. Reader, A. Beven, P. Shaw, T. Miller, and G. Moore. 1997. a. Association of homologous chromosomes during floral development. Curr. Biol. 7:905–908. [DOI] [PubMed] [Google Scholar]
- Aragón-Alcaide, L., S. Reader, T. Miller, and G. Moore. 1997. b. Centromeric behaviour in wheat with high and low homologous chromosomal pairing. Chromosoma. 106:327–333. [DOI] [PubMed] [Google Scholar]
- Ashley, T., A.W. Plug, J. Xu, A.J. Solari, G. Reddy, E.I. Golub, and D.C. Ward. 1995. Dynamic changes in Rad51 distribution on chromatin during meiosis in male and female vertebrates. Chromosoma. 104:19–28. [DOI] [PubMed] [Google Scholar]
- Bass, H.W., W.F. Marshall, J.W. Sedat, D.A. Agard, and W.Z. Cande. 1997. Telomeres cluster de novo before the initiation of synapsis: a three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J. Cell Biol. 137:5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, H.W., O. Riera-Lizarazu, E.V. Ananiev, S.J. Bordoli, H.W. Rines, R.L. Phillips, J.W. Sedat, D.A. Agard, and W.Z. Cande. 2000. Evidence for the coincident initiation of homolog pairing and synapsis during the telomere-clustering (bouquet) stage of meiotic prophase. J. Cell Sci. 113:1033–1042. [DOI] [PubMed] [Google Scholar]
- Belmont, A.S., J.W. Sedat, and D.A. Agard. 1987. A three-dimensional approach to mitotic chromosome structure: evidence for a complex hierarchical organization. J. Cell Biol. 105:77–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M.D. 1977. The time and duration of meiosis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 277:201–226. [DOI] [PubMed] [Google Scholar]
- Chaubal, R., C. Zanella, M. Trimnell, T.W. Fox, M.C. Albertsen, and P. Bedinger. 2000. Two male-sterile mutants of Zea mays (Poaceae) with an extra cell division in the anther wall. Am. J. Bot. 87:1193–1202. [PubMed] [Google Scholar]
- Chen, H., D.D. Hughes, T.-A. Chan, J.W. Sedat, and D.A. Agard. 1996. IVE (image visualization environment): a software platform for all three-dimensional microscopy applications. J. Struct. Biol. 116:56–60. [DOI] [PubMed] [Google Scholar]
- Chikashige, Y., D.Q. Ding, Y. Imai, M. Yamamoto, T. Haraguchi, and Y. Hiraoka. 1997. Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J. 16:193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, M.N., A.M. Dominguez, and M.E. Dresser. 1997. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science. 276:1252–1255. [DOI] [PubMed] [Google Scholar]
- Cooper, J.P., E.R. Nimmo, R.C. Allshire, and T.R. Cech. 1997. Regulation of telomere length and function by a Myb—domain protein in fission yeast. Nature. 385:744–747. [DOI] [PubMed] [Google Scholar]
- Cowan, C.R., P.M. Carlton, and W.Z. Cande. 2001. The polar arrangement of telomeres in interphase and meiosis: Rabl organization and the bouquet. Plant Physiol. 125:532–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, R.K., J.W. Sedat, D.A. Agard, and W.Z. Cande. 1994. Meiotic chromosome pairing in maize is associated with a novel chromatin organization. Cell. 76:901–912. [DOI] [PubMed] [Google Scholar]
- Dernburg, A.F., J.W. Sedat, W.Z. Cande, and H.W. Bass. 1995. Cytology of telomeres. Telomeres. E.H. Blackburn and C.W. Greider, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 295–338.
- Dernburg, A.F., K.W. Broman, J.C. Fung, W.F. Marshall, J. Philips, D.A. Agard, and J.W. Sedat. 1996. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell. 85:745–759. [DOI] [PubMed] [Google Scholar]
- Dong, F., and J. Jiang. 1998. Non-Rabl patterns of centromere and telomere distribution in the interphase nuclei of plant cells. Chromosome Res. 6:551–558. [DOI] [PubMed] [Google Scholar]
- Esponda, P., and G. Giménez Martín. 1972. The attachment of the synaptonemal complex to the nuclear envelope. An ultrastructural and cytochemical analysis. Chromosoma. 38:405–417. [DOI] [PubMed] [Google Scholar]
- Franklin, A.E., J. McElver, I. Sunjevaric, R. Rothstein, B. Bowen, and W.Z. Cande. 1999. Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell. 11:809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussell, C.P. 1987. The Rabl orientation: a prelude to synapsis. Cell Biology: A Series of Monographs: Meiosis. P.B. Moens, editor. Academic Press, San Diego. 275–300.
- Galy, V., J.-C. Olivo-Marin, H. Scherthan, V. Doye, N. Rascalou, and U. Nehrbass. 2000. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 403:108–112. [DOI] [PubMed] [Google Scholar]
- Gaut, B.S., and J.F. Doebley. 1997. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA. 94:6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelei, J. 1921. Weitere Studien über die Oogenese des Dendrocoelum lacteum II. Die Längskonjugation der Chromosomen. Archiv für Zellforschung. 16:88–169. [Google Scholar]
- Haber, J.E., P.C. Thorburn, and D. Rogers. 1984. Meiotic and mitotic behavior of dicentric chromosomes in Saccharomyces cerevisiae. Genetics. 106:185–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Q.-W., E. Trelles-Sticken, H. Scherthan, and J. Loidl. 1998. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J. Cell Biol. 141:21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaszás, É., and J.A. Birchler. 1996. Misdivision analysis of centromere structure in maize. EMBO J. 15:5246–5255. [PMC free article] [PubMed] [Google Scholar]
- Kaszás, É., and J.A. Birchler. 1998. Meiotic transmission rates correlate with physical features of rearranged centromeres in maize. Genetics. 150:1683–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl, J. 1990. The initiation of meiotic chromosome pairing: the cytological view. Genome. 33:759–778. [DOI] [PubMed] [Google Scholar]
- Loidl, J. 1994. Cytological aspects of meiotic recombination. Experientia. 50:285–294. [DOI] [PubMed] [Google Scholar]
- Martínez-Pérez, E., P. Shaw, S. Reader, L. Aragón-Alcaide, T. Miller, and G. Moore. 1999. Homologous chromosome pairing in wheat. J. Cell Sci. 112:1761–1769. [DOI] [PubMed] [Google Scholar]
- Martínez-Pérez, E., P.J. Shaw, and G. Moore. 2000. Polyploidy induces centromere association. J. Cell Biol. 148:233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Pérez, E., P. Shaw, and G. Moore. 2001. The Ph1 locus is needed to ensure specific somatic and meiotic centromere association. Nature. 411:204–207. [DOI] [PubMed] [Google Scholar]
- Mikhailova, E., S. Sosnikhina, G. Kirillova, O. Tikholiz, V. Smirnov, R. Jones, and G. Jenkins. 2001. Nuclear dispositions of subtelomeric and pericentromeric chromosomal domains during meiosis in asynaptic mutants of rye (Secale cereale L.). J. Cell Sci. 114:1875–1882. [DOI] [PubMed] [Google Scholar]
- Nimmo, E.R., A.L. Pidoux, P.E. Perry, and R.C. Allshire. 1998. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature. 392:825–828. [DOI] [PubMed] [Google Scholar]
- Pfeifer, C., P.D. Thomsen, and H. Scherthan. 2001. Centromere and telomere redistribution precedes homologue pairing and terminal synapsis initiation during prophase I of cattle spermatogenesis. Cytogenet. Cell Genet. 93:304–314. [DOI] [PubMed] [Google Scholar]
- Rabl, C. 1885. Über Zellteilung. Morphol. Jahrb. 10:214–330. [Google Scholar]
- Rasmussen, S., and P. Holm. 1978. Human meiosis II: chromosome pairing and recombination nodules in human spermatocytes. Carlsberg Res. Commun. 42:275–327. [Google Scholar]
- Rockmill, B., and G.S. Roeder. 1998. Telomere-mediated chromosome pairing during meiosis in budding yeast. Genes Dev. 12:2574–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan, H. 2001. A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 8:621–627. [DOI] [PubMed] [Google Scholar]
- Scherthan, H., S. Weich, H. Schwegler, C. Heyting, M. Haerle, and T. Cremer. 1996. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J. Cell Biol. 134:1109–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan, H., M. Jerratsch, B. Li, S. Smith, M. Hulten, T. Lock, and T. de Lange. 2000. Mammalian meiotic telomeres: protein composition and redistribution in relation to nuclear pores. Mol. Biol. Cell. 11:4189–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia, C., G. Blobel, and M.P. Rout. 1999. Proteins connecting the nuclear pore complex with the nuclear interior. J. Cell Biol. 144:839–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suja, J.A., and J.S. Rufas. 1994. The telochore: a telomeric differentiation of the chromosome axis. Chromosome Res. 2:361–368. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., N. Ide, and I. Tanaka. 1997. Immunocytochemical visualization of the centromeres during male and female meiosis in Lilium longiflorum. Chromosoma. 106:435–445. [DOI] [PubMed] [Google Scholar]
- Terasawa, M., A. Shinohara, Y. Hotta, H. Ogawa, and T. Ogawa. 1995. Localization of RecA-like recombination proteins on chromosomes of the lily at various meiotic stages. Genes Dev. 9:925–934. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken, E., M.E. Dresser, and H. Scherthan. 2000. Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J. Cell Biol. 151:95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, D., M. Schneerman, W. Lee, and D. Muzumdar. 1998. Centromere mapping in Zea mays L. using telocentrics and isochromosomes. Cytogenet. Cell Genet. 81:146. [Google Scholar]
- Zickler, D. 1977. Development of the synaptonemal complex and the “recombination nodules” during meiotic prophase in the seven bivalents of the fungus Sordaria macrospora Auersw. Chromosoma. 61:289–316. [DOI] [PubMed] [Google Scholar]
- Zickler, D., and N. Kleckner. 1998. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32:619–697. [DOI] [PubMed] [Google Scholar]