Abstract
Particulate air pollution is widespread, yet we have little understanding of the long-term health implications associated with exposure. We investigated DNA damage, mutation, and methylation in gametes of male mice exposed to particulate air pollution in an industrial/urban environment. C57BL/CBA mice were exposed in situ to ambient air near two integrated steel mills and a major highway, alongside control mice breathing high-efficiency air particulate (HEPA) filtered ambient air. PCR analysis of an expanded simple tandem repeat (ESTR) locus revealed a 1.6-fold increase in sperm mutation frequency in mice exposed to ambient air for 10 wks, followed by a 6-wk break, compared with HEPA-filtered air, indicating that mutations were induced in spermatogonial stem cells. DNA collected after 3 or 10 wks of exposure did not exhibit increased mutation frequency. Bulky DNA adducts were below the detection threshold in testes samples, suggesting that DNA reactive chemicals do not reach the germ line and cause ESTR mutation. In contrast, DNA strand breaks were elevated at 3 and 10 wks, possibly resulting from oxidative stress arising from exposure to particles and associated airborne pollutants. Sperm DNA was hypermethylated in mice breathing ambient relative to HEPA-filtered air and this change persisted following removal from the environmental exposure. Increased germ-line DNA mutation frequencies may cause population-level changes in genetic composition and disease. Changes in methylation can have widespread repercussions for chromatin structure, gene expression and genome stability. Potential health effects warrant extensive further investigation.
Keywords: DNA adducts, DNA strand breaks, tandem repeat mutation
Combustion of fossil fuels results in the production of complex mixtures of chemicals that are released into the environment and potentially affect millions of people globally. Previous work demonstrated that the offspring of wild birds breeding near integrated steel mills on the North American Great Lakes inherited increased numbers of tandem repeat DNA sequence mutations compared with those from areas without steel mills (1, 2). Subsequent studies investigated expanded simple tandem repeat (ESTR) mutation in outbred laboratory mice caged near two integrated steel mills and a major highway in Hamilton, Ontario, Canada, and at a rural reference site (3, 4). Using a pedigree approach (5), a significant increase in germ-line mutation rate was found in mice housed in the industrial environment compared with the reference site. The majority of mutations were transmitted through the paternal germ line. High-efficiency particulate-air (HEPA) filtration of the ambient air resulted in a significant reduction in mutation frequency, down to levels measured at the reference location (4). Therefore, the particulate fraction of air in this industrial location was largely responsible for the mutagenic hazard.
These findings show that chemical pollutants may cause heritable mutation. Further research is required to confirm these results, and to evaluate the potential risk to humans exposed to particulate air pollution. The present study was designed to provide insight into the mechanisms operating in tandem repeat mutation induced by exposure to particulate air pollution, relationships to other types of DNA modifications, and potential health consequences.
In this study, mature male C57BL/CBA F1 mice were exposed to HEPA-filtered or ambient air in Hamilton, Ontario, Canada, near two integrated steel mills and a major highway. Induced ESTR mutations arising in sperm DNA were measured by using single-molecule PCR (SM-PCR) (6). Earlier studies investigated outbred mice. Therefore, the possibility of genetic confounders could not be excluded. For example, size of the repeat locus (7–9), cis- and trans- elements (8, 10, 11) and genetic background (8, 12, 13) can all affect mutation frequency. The mean spontaneous ESTR mutation frequency was elevated in these outbred mice relative to other inbred strains [≈2X greater than typical (3)]. Analysis of inbred mice should confirm that genetic confounders did not contribute to differences in mutation frequencies, and provide a basis for comparison of air pollution to reference mutagens examined in the laboratory in the same strains.
In addition to measuring germ-line tandem repeat mutation, we characterized DNA lesions resulting from the exposure. It is unknown whether chemical agents in air pollution directly reach the gonads and cause DNA damage and mutation, or whether there is an indirect event elsewhere that results in physiological changes that destabilize the germ line. We measured levels of bulky DNA adducts as an indicator of the presence of DNA reactive chemicals in testes DNA, as well as DNA single and double strand breaks in sperm of mice breathing ambient or HEPA-filtered air.
In this study, we quantified induced ESTR mutations at three time points to determine the stages of spermatogenesis that are susceptible to mutation and DNA modifications resulting from air pollution exposure. Previous experiments with individual mutagens in the laboratory suggest that ESTR mutations arise in diploid premeiotic stages of spermatogenesis (14–16).
Previous studies sampled the progeny of exposed parents; therefore, these mutations may be the result of events occurring postfertilization as a consequence of premutational lesions in DNA affecting early cellular divisions (17–20). Indeed, high levels of somatic mosaicism were found in the outbred Swiss–Webster mice used, and ESTR loci are known to be highly susceptible to destabilization in early embryogenesis (21–23). In this study, we directly evaluated mutation arising in sperm, to confirm that mutations measured were germ line in origin.
Lastly, induction of ESTR mutation does not appear to result from the direct conversion of damaged or adducted DNA into mutation, but rather is thought to result from indirect events (24–28). Therefore, we hypothesize that epigenetic changes may provide a potential signal resulting in ESTR mutation induction. DNA methylation is the most stable epigenetic modification of mammalian genomes and is crucial for normal development, proliferation and maintenance of genome stability (29–33). DNA methylation patterns are disrupted in cancers (29, 33–35) and following exposure to genotoxic agents (36–38). Differential methylation has been observed in the descendents of animals exposed to various agents (39–41), suggesting a potential link to nontargeted DNA mutations and transgenerational effects. Therefore, global levels of DNA methylation were evaluated in sperm DNA.
Results
Air Quality.
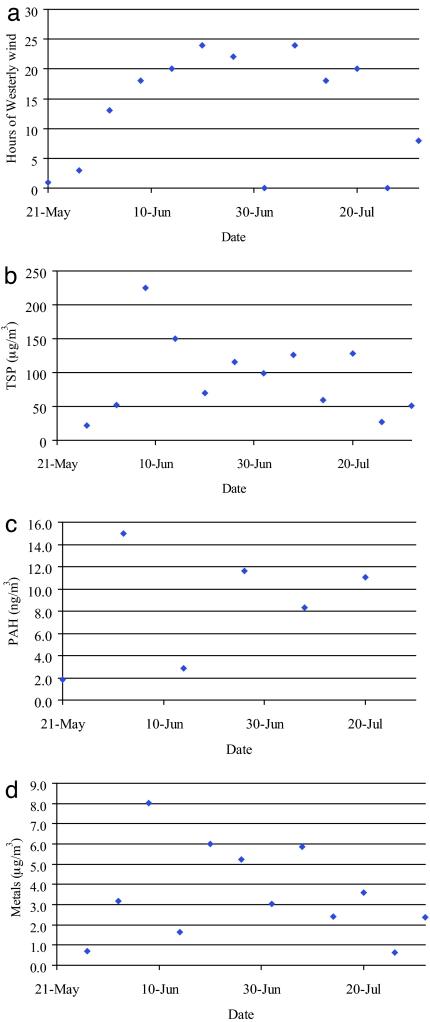
Air quality data were provided by the Ontario Ministry of the Environment (MOE). A westerly wind places the MOE samplers and animal enclosures downwind of the steel mills. Previous work demonstrated that levels of total suspended particles (TSP) and PAHs were positively correlated with westerly wind direction (4). Fig. 1 a–d shows the air quality parameters for the present study. The majority of the time the enclosures and air samplers were downwind of the steel mills. Exposures were lowest at the beginning and end of the experiment. Hours of westerly wind, TSP (mean 93.8 ± 17.0 μg/m3), and metal (mean 3.6 ± 0.7 μg/m3) concentrations, were highest at wk 4. Wk 3 had the highest PAH concentration (mean 8.3 ± 1.7 ng/m3). Benzo(b)-fluoranthene, benzo(k)fluoranthene, and indeno(123-cd)pyrene were the 3 most abundant PAHs. Iron, copper, and manganese were the 3 most abundant metals.
Fig. 1.
Air quality parameters. Air samplers were located ≈400 m north-east of the mouse sheds. Data were collected from the MOE database for May 14th to August 1st, 2004. PAH levels are based on measurement of benzo(a)anthracene, chrysene, benzo(b)fluoranthene, benzo(k)fluoranthene, benzo(a)pyrene, indeno(123cd)pyrene, dibenzo(a,h)anthracene and benzo(ghi)perylene. Metal levels are based on cadmium, chromium, iron, copper, lead, manganese, nickel, and vanadium. (a) Hours of westerly wind. (b) Total suspended particulate. (c) Polycyclic aromatic hydrocarbons. (d) Metals.
Mutations at Ms6-hm.
Mutation frequency at ESTR Ms6-hm locus was determined in sperm DNA sampled from mice exposed to HEPA-filtered or ambient air for 3 weeks (wks), 10 wks or 10 wks followed by 6 wks (16 wks) in the laboratory. A generalized score test showed a significant overall treatment effect (P = 0.036). Sperm sampled from mice exposed for 3 or 10 wks did not show elevated ESTR mutation frequencies (Table 1). Sperm samples collected at 16 wks developed from exposed spermatogonial stem cells. A significant increase in ESTR mutation frequency was found for spermatogonia exposed to whole air relative to HEPA filtered air (1.6-fold, generalized score test, P = 0.016).
Table 1.
Mutation data for the Ms6-hm locus for mice exposed to HEPA-filtered and whole air in Hamilton, Ontario
| Treatment | No. of mutants | No. of progenitors* | Mutation frequency, %* | Ratio relative to HEPA filtered† | P value‡ |
|---|---|---|---|---|---|
| HEPA (3 wks) | 24 | 638 (488–798) | 3.76 (3.01, 4.92) | ||
| Whole air (3 wks) | 26 | 719 (557–888) | 3.62 (2.93, 4.67) | 0.96 (0.71, 1.28) | 0.770 |
| HEPA (10 wks) | 33 | 772 (596–950) | 4.28 (3.47, 5.54) | ||
| Whole air (10 wks) | 31 | 716 (565–866) | 4.33 (3.58, 5.49) | 0.99 (0.88, 1.12) | 0.902 |
| HEPA (16 wks)§ | 35 | 879 (701–1057) | 3.98 (3.31, 4.99) | ||
| Whole Air (16 wks) | 50 | 796 (638–954) | 6.28 (5.24, 7.84) | 1.58 (1.28, 1.94) | 0.016 |
*Predicted, lower-upper range, mutation frequency and ratios are given with 95% confidence intervals (C.I.). The C.I. reflects uncertainties in estimating the number of template molecules.
†Ratios to HEPA filtered with 95% C.I. (estimated by inverting the Wald statistic).
‡Bold number indicates statistically significant value (generalized score test). This P value remains significant after application of a Bonferroni correction for three pair-wise comparisons.
§Ten wks in situ plus 6 wks in the laboratory.
DNA Adducts.
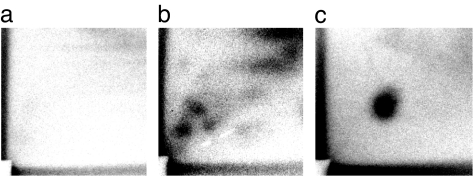
No detectable adducts were observed in testes samples at any of the time points (detection limit of 0.5 per 108 nucleotides). To ensure exposure in mice breathing whole air to DNA binding chemicals, whole lung samples were examined. Lungs of mice exposed to unfiltered air for 3 wks were positive for DNA adducts, showing a diagonal radioactive zone (DRZ) with distinct DNA adduct spots on chromatograms (Fig. 2b); HEPA mice showed only a faint DRZ (Fig. 2a). The total number of DNA adducts measure by 32P-postlabeling was significantly higher in mouse lung after 3 wks of exposure (1.0 ± 0.1 adducts per 108 total nucleotides) compared with mice from the HEPA filtered cages (0.7 ± 0.1 adducts per 108 total nucleotides) (1.4-fold, one-tailed Wilcoxon Rank test using a T approximation, P = 0.046). No detectable adducts were apparent at the other time points.
Fig. 2.
DNA adduct analysis. Examples of autoradiographs from two typical 3-wk lung samples and the positive control are shown. (a) HEPA-filtered air. (b) Whole air. (c) A BPDE-DNA adduct standard.
DNA Strand Breaks.
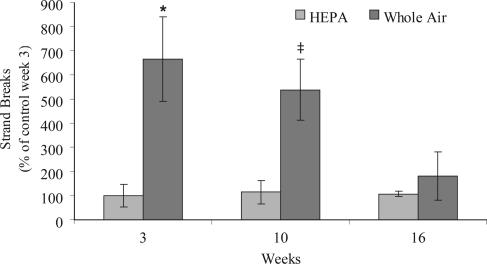
A significant treatment effect was observed (factorial analysis on ranks, P = 0.004); strand breaks were more abundant in sperm after exposure to particulate air pollution (Fig. 3). This increase was significant at 3 wks (P = 0.004) and marginally significant at 10 wks (P = 0.053). Strand breaks returned to HEPA-filtered levels after 6 wks in the laboratory (16 wks; P > 0.05).
Fig. 3.
DNA damage in mature sperm. Levels of DNA strand breaks are expressed as percent of the control at wk 3. Bars indicate standard error. *, P < 0.05; ‡, P = 0.053, one-tailed Wilcoxon rank test.
Global DNA Methylation.
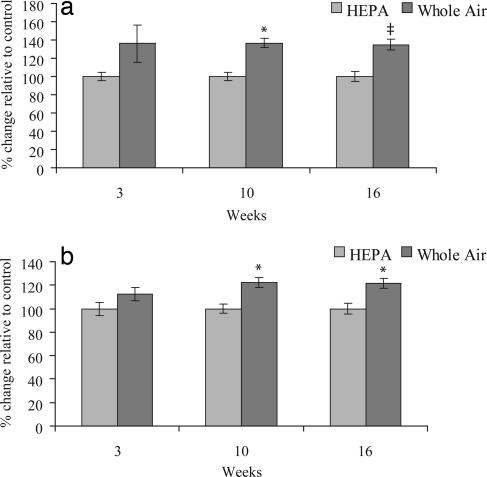
An increase in global methylation was found in sperm from mice exposed to whole air compared with HEPA filtered air using both the CEA and the MAA (Fig. 4 a and b). A factorial analysis revealed a significant treatment effect (P = 0.004 and P = 0.001 for the two assays respectively). There was no significant change in methylation at 3 wks (P = 0.126 and P = 0.162 for the two assays respectively). Hypermethylation arose in mice exposed continuously to particulate air pollution for 10 wks (P = 0.038 and P = 0.014 for the two assays respectively). Hypermethyation persisted in 16-wk samples (P = 0.067 and P = 0.021 for the two assays, respectively).
Fig. 4.
Global methylation of male germ-line DNA. (a) Percent global methylation change relative to age-matched controls using the cytosine extension assay. (b) Percent methylation change relative to age matched controls using the methyl-acceptance assay. Bars indicate standard error. *, P < 0.05; ‡, P = 0.067, two-tailed Wilcoxon rank test.
Discussion
We have demonstrated that exposure of inbred mice to particulate air pollution near two integrated steel mills and a major highway caused tandem repeat DNA mutation and hypermethylation in spermatogonial stem cells. Sperm sampled at 16 wks, but not at earlier time points, showed elevated ESTR mutation frequencies, indicating that exposure of spermatogonial stem cells to particulate air pollution for 10 wks caused germ-line mutations. Mutation induction in this study (1.6-fold) was similar to previous experiments examining the offspring of exposed outbred mice (1.5- to 2-fold) (4). Despite the presence of mutagenic PAHs in the particulate fraction of this air, DNA adducts were not detectable (<0.5 in 108 nucleotides) in the gonads of mice exposed to whole air, but were detected in lung after 3 wks of exposure. In contrast, increased DNA strand breaks were found in sperm collected at 3 and 10 wks. At 16 wks, strand breaks were similar in HEPA and whole air groups. Therefore, breaks that arose in spermatogonial stem cells during the 10-wk exposure period were repaired, or converted to gene or chromosomal mutations not detected in this study, in the subsequent stages of spermatogenesis following removal from the polluted environment. DNA hypermethylation was found in males sampled at 10 and 16 wks. These data suggest that epigenetic modification may have occurred in the early stages of spermatogenesis, but not the haploid stages (3 wks). Hypermethylation persisted through spermatogenesis and remained significantly elevated in mature sperm after removal from the environmental exposure.
Shorter exposures (3 and 10 wks) did not result in induced ESTR germ-line mutation. Despite the presence of DNA strand breaks in sperm sampled at 3 wks, lack of induced mutation was expected as spermatids are not generally susceptible to mutagen-induced ESTR mutation (14–16). The 10-wk exposure included 6 wks during which the cells were in diploid premeiotic stages of spermatogenesis, the most susceptible period for induced ESTR mutation (14–16). Because this was an ambient in situ exposure to ambient air pollution, day to day fluctuations may cause wide variations in exposure over the sampling period based on industrial emissions, traffic, wind direction, and other parameters. The mice were exposed to relatively high levels of particles and organic (PAHs) and inorganic (metals) pollutants over this period, and strand breaks were elevated at 3 and 10 wks. However, it is possible that spermatogonial stem cells were not exposed to a high enough cumulative dose to cause measurable mutation induction over this period. Alternatively, enough time may not have passed at the 10-wk time point to have accumulated a measurable effect in the germ line. It is possible that the exposure was not long enough to cause accumulation of mutations in the stem cells to obtain a significant increase in mutation frequency. For example, a power analysis revealed that the sample size of this study would need to be increased by ≈6-fold to obtain a statistically significant 1.2-fold increase based on the variability observed here.
The use of inbred strains eliminates genetic confounders such as cis- or trans-elements, genetic background or differences in ESTR allele sizes between groups. DNA mutation was measured directly in sperm, confirming that mutations arose in the germ line, rather than in early embryogenesis. Mutation frequency in spermatogonial stem cells was 1.6-fold higher in mice breathing whole air than HEPA-filtered air at 16 wks. Remarkably, mutation induction as a result of breathing particulate air pollution was similar to that observed in mice of the same genetic background after acute exposure to 0.5–1 Gy of X-rays (26, 42).
Bulky DNA adduct levels were elevated in the lungs of mice breathing whole air for 3 wks, confirming that they were exposed to DNA binding chemicals. Adduct levels were not elevated at 10 wks, likely resulting from variation in exposure through the season. DNA adduct levels were below detectable thresholds in testes at all points measured. DNA adducts predominantly reflect the presence of PAHs that are metabolized and bind covalently to DNA. PAHs are the principal mutagenic components of airborne particulates in Hamilton (43); thus, this finding suggests that PAHs that induce stable DNA adducts do not penetrate the male germ line and affect ESTR mutation rates. Alternatively, this finding supports the model that ESTR mutations arise through an indirect mechanism of mutation. DNA strand breaks were elevated in sperm samples collected at 3 and 10 wks. Strand breaks may reflect exposure to agents that generate reactive oxygen species (ROS), which can be produced by exposure to particulate matter, PAHs (reviewed in ref. 44), or metals (reviewed in refs. 45 and 46), and can damage DNA. Other factors that may contribute to oxidative stress include changes in metabolism or inflammation in mice exposed to particulate air pollution (44). It is unclear whether ROS generated elsewhere in the body leads to oxidative stress and DNA strand breaks in gametes. Future work should investigate the concentrations of metals, particles, and PAHs in gonadal tissue to address this hypothesis. In addition, it is possible that unstable DNA adducts that we did not measure may have caused oxidative damage and strand breaks, thus contributing to ESTR mutation induction. Some of such adducts would have been detected as strand breaks in our sperm assay. Further studies should consider the association of ESTR mutations and this class of DNA adduct. Previous work has focused almost exclusively on the mutagenic hazard of PAHs in this environment. Our findings suggest that other chemicals (e.g., metals), and particles in general, may play a more relevant role in germ-line mutagenesis than previously anticipated.
Previous work showed that the frequency of double and single strand breaks in mice exposed to radiation was too low to explain observed induced ESTR mutation frequency. An indirect, nontargeted mechanism was proposed whereby endogenous mutation processes are altered by radiation exposure (6, 15). Our study involved chronic exposure and did not estimate the frequency of DNA strand breaks, so a similar conclusion cannot be made from our data. However, several experiments have demonstrated that mutation spectra (i.e., patterns of gains and losses in repeat units) are identical in exposed and control mice, confirming the hypothesis that the same mechanism may operate in induced and spontaneous ESTR mutation (6, 47, 48). In our study, the ESTR mutation spectra of HEPA and whole air sperm samples were also identical (data not shown), suggesting that the same nontargeted mechanism may operate for chronic exposure to airborne particulates.
It is possible that the presence of DNA strand breaks in gametes elicits a signal that results indirectly in mutation at tandem repeat sequences. It has been proposed that alterations in cell cycle, potentially resulting from the introduction of DNA damage leading to delays, cause replication fork pausing, which in turn, may affect ESTR mutation rate (13). The formation of secondary structures across the ESTR allele during this pause may lead to the gain or loss of repeat units when replication is reinitiated. The nontargeted model for ESTR instability also suggests that other signals may be involved in mutation induction. Epigenetic changes can be transmitted through the germ line (49, 50) and therefore provide a potential mechanism through which environmental factors can exert heritable effects. In the germ line, methylation is strictly regulated and DNA methyl-transferases (DNMTs) are very active (51, 52). DNMTs are up-regulated during DNA damage and bind with high affinity to many DNA lesions (53). It has been hypothesized that DNMT1 (involved in methylation maintenance; ref. 54) is, in fact, an ancient DNA repair enzyme (55). Theoretically, with elevated DNA damage resulting from exposure to particulate air pollution, up-regulation of DNMTs may lead to hypermethylation over time. Correspondingly, we found that the sperm of mice exposed chronically to particulate air pollution was hypermethylated relative to age-matched HEPA-treated mice. DNA hypermethylation may be linked with structural changes in chromatin, decreased gene expression and decreased rates of transposon movement (56–58). Increased gamete DNA methylation may lead to potential problems for progeny, but it is unknown whether methylation changes inherited via mature sperm lead to altered epigenetic reprogramming in the fertilized egg. The global methylation measured here does not necessarily reflect the methylation status of active genes; therefore, the consequences of these changes are unknown.
The relationship between methylation and ESTR instability remains to be elucidated. Alterations in global methylation affecting gene expression, chromatin structure or genome stability could have an impact on spermatogonial stem cell function. It is possible that alterations in methylation are transmitted to subsequent generations, providing a persistent epigenetic signal. Future work should target localized regions of DNA and investigate the potential of urban air pollutants to modulate methylation of important genomic sites.
The overall implications of these findings for the health of humans are unclear. Heritable mutation, germ-line DNA damage and epigenetic modifications have the potential to affect disease incidence in the descendents of exposed individuals. In addition to its potential importance in the maintenance of genome stability, appropriate methylation of DNA is critical for imprinting (59), regulation of gene expression (60), mammalian development (61–63), and disease (33, 64).
Materials and Methods
Environmental Exposures.
C57BL/6 × CBA F1 hybrid mice (Jackson Laboratory, Bar Harbor, ME) were exposed as described in refs. 3 and 4. Thirty 7- to 9-wk-old males were housed in a modified utility shed 2 km from two integrated steel mills and 1 km from a major highway on Hamilton Harbor, Ontario, Canada from May 14, 2004, for 3 wks, 10 wks, or for 10 wks followed by 6 wks in the laboratory (16 wks). Each time point included five mice exposed to ambient air (Whole Air) and five mice exposed to air filtered through a HEPA filtration unit (True-HEPA, model 200–001, Bemis Manufacturing, Sheboygan Falls, WI). HEPA filtration removes at least 99.97% of particles 0.3 μm in diameter (65). Given the duration of spermatogenesis, sperm sampled from the caudal epididymis of mice held for 3 wks were postmeiotic spermatocytes and spermatids during the exposure period. Sperm sampled at 10 wks were exposed throughout the continuum of spermatogenesis from spermatogonial stages (for 6 wks) to spermatozoa during the experiment. In the last time point, a 6-wk break after the 10-wk exposure (16 wks) ensured that the collected sperm were at the spermatogonial stem stage for the entire exposure. Therefore, any measurable changes in these samples were the result of effects on stem cells. All animal procedures were carried out under the guidelines of the Canadian Council on Animal Care and procedures approved by the McMaster University Animal Research Ethics Board.
DNA Isolation.
For tandem repeat mutation, DNA adduct, DNA strand-break, and DNA methylation assays, all DNA extractions and downstream applications were performed on coded samples so that researchers were blind to their experimental treatment. In addition, control and exposed samples were assayed in mixed groups using the same batches of reagents. All manipulations before PCR were carried out in a laminar flow hood to minimize the risk of contamination. Genomic DNA was isolated from testes and caudal epididymis as described in ref. 6.
Tandem Repeat Mutations.
Size mutations at the Ms6-hm ESTR were measured by using SM-PCR (6). The ≈1.8-kbp CBA Mh6-hm allele was amplified from diluted aliquots of sperm DNA as described in refs. 6 and 66. The concentration of template DNA was adjusted for each sample until 40–60% of amplifications produced observable product. Approximately 100 reactions with product per animal were sized by using gel electrophoresis (40-cm agarose gels), Southern blotting and hybridization to radiolabeled Ms6-hm probe or internal size standard (1-Kb ladder, Invitrogen, Burlington, ON). The size of each product was measured against in-lane standards. Mutants were scored as bands with sizes differing by >4 repeat units (20 bp) from the progenitor by observers who were blind to experimental treatments. Mutation frequencies within animals were determined by dividing the number of mutant bands by the estimated total number of template molecules (66).
Bulky DNA Adducts.
The 32P-postlabeling assay was performed on lung and testes using existing methods (67). We focused this assay on the testes to determine whether DNA-reactive chemicals were transported from the site of exposure in the lungs, to the site of induced effects in the gonads. Briefly, DNA (10 μg) was digested by using micrococcal endonuclease and spleen phosphodiesterase. The modified nucleotides were radiolabeled (50 μCi/sample) by incubation with T4-polynucleotide kinase. Radiolabeled adduct nucleotide biphosphates were separated by chromatography on PEI-cellulose sheets. In each experiment, 3 standards of benzo(a)pyrene diol epoxide (BPDE) modified DNA with known modification levels (1 adduct per 107, 108 and 109 nucleotides) were run in parallel for quantification purposes using phosphor-imaging. Part of the digest was used to determine the final amount of DNA in the assay by HPLC-UV.
DNA Strand Breaks.
DNA strand breaks were detected by using a modification of the random oligonucleotide-primed synthesis (ROPS) (68) assay (69). Briefly, caudal epididymal sperm DNA was denatured and radiolabeled at the free 3′-OH termini using Klenow polymerase. The reactions were stopped, filtered and washed. Filters were analyzed by scintillation counting. Results are expressed relative to the week 3 HEPA animals.
DNA Methylation.
Global DNA methylation was analyzed by using two independent assays, the cytosine extension assay (CEA) and methyl-acceptance assay (MAA) (69–74). The CEA method uses methylation-sensitive restriction endonuclease HpaII, which has relatively frequent recognition sequences at CpG sites. A 5′ guanine overhang is left after DNA cleavage that can be used for subsequent single nucleotide extension with labeled (3H)dCTP; breaks without this overhang are not recognized. The extent of (3H)dCTP incorporation opposite the exposed guanine is directly proportional to the number of cleaved, unmethylated CpG sites, and inversely proportional to methylation levels. The higher the methylation, the lower the incorporation of (3H)dCTP″ (69–74). Briefly, for CEA, sperm genomic DNA was digested with HpaII. A second aliquot of DNA was incubated without restriction enzyme and served as the background control. The single-nucleotide extension reaction was performed by using TaqDNA polymerase to incorporate [3H]dCTP. Duplicate aliquots from each reaction were collected on ion-exchange filters and washed. Filters were dried and analyzed by scintillation counting. After background substraction, the results were expressed as percent change relative to the age-matched HEPA mice and are indicative of the level of methylation of cytosines at genomic 5′-(C)C(GG)-3′ sites.
In the MAA, the bacterial enzyme SssI was used to transfer radiolabeled methyl groups to unmethylated CpG sites in sperm DNA. Methylated DNA was isolated by filtering on Whatman DE81 ion exchange filters and washed. Dried filters were placed in scintillation liquid and measured. Blank values were obtained from duplicate incubations without the enzyme. The results are presented as percent change relative to the age-matched HEPA mice.
Statistical Analysis.
Statistical tests were carried out in SAS/STAT software, Version 8.2 (1999–2001 SAS Institute, Cary, NC). Statistical testing (ANOVA) on DNA adducts, strand breaks and methylation was performed on ranks. For Ms6-hm mutation frequencies, the number of PCR template molecules was determined from the number of positive PCR products using the Poisson distribution, estimated by taking the natural log of the observed number of PCR wells with no product to the total number of PCR wells run. The ratio of the exposed to control mutation frequencies was estimated by back-transforming from the Wald Statistic with the empirical covariance matrix using the GENMOD Procedure. A generalized score test was used to examine treatment effect.
ACKNOWLEDGMENTS.
We thank T. Singer, Y. Dubrova, and D. Desaulniers for comments on this manuscript, and J. Quinn for helpful discussions and use of exposure sheds. This research was supported by the Canadian Regulatory Systems for Biotechnology (G.R.D./C.Y. laboratory), National Sciences and Engineering Research Council, and the Alberta Cancer Board (O.K. laboratory).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Yauk CL, Quinn JS. Proc Natl Acad Sci USA. 1996;93:12137–12141. doi: 10.1073/pnas.93.22.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yauk CL, Fox GA, McCarry BE, Quinn JS. Mutat Res. 2000;452:211–218. doi: 10.1016/s0027-5107(00)00093-2. [DOI] [PubMed] [Google Scholar]
- 3.Somers CM, Yauk CL, White PA, Parfett CL, Quinn JS. Proc Natl Acad Sci USA. 2002;99:15904–15907. doi: 10.1073/pnas.252499499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Somers CM, McCarry BE, Malek F, Quinn JS. Science. 2004;304:1008–1010. doi: 10.1126/science.1095815. [DOI] [PubMed] [Google Scholar]
- 5.Singer TM, Lambert IB, Williams A, Douglas GR, Yauk CL. Mutat Res. 2006;598:164–193. doi: 10.1016/j.mrfmmm.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 6.Yauk CL, Dubrova YE, Grant GR, Jeffreys AJ. Mutat Res. 2002;500:147–156. doi: 10.1016/s0027-5107(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 7.Bois PR, Southgate L, Jeffreys AJ. Mamm Genome. 2001;12:104–111. doi: 10.1007/s003350010251. [DOI] [PubMed] [Google Scholar]
- 8.Gomes-Pereira M, Monckton DG. Mutat Res. 2006;598:15–34. doi: 10.1016/j.mrfmmm.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Martorell L, Gamez J, Cayuela ML, Gould FK, McAbney JP, Ashizawa T, Monckton DG, Baiget M. Neurology. 2004;62:269–274. doi: 10.1212/wnl.62.2.269. [DOI] [PubMed] [Google Scholar]
- 10.Brock GJ, Anderson NH, Monckton DG. Hum Mol Genet. 1999;8:1061–1067. doi: 10.1093/hmg/8.6.1061. [DOI] [PubMed] [Google Scholar]
- 11.Monckton DG, Neumann R, Guram T, Fretwell N, Tamaki K, MacLeod A, Jeffreys AJ. Nat Genet. 1994;8:162–170. doi: 10.1038/ng1094-162. [DOI] [PubMed] [Google Scholar]
- 12.Barber R, Plumb MA, Boulton E, Roux I, Dubrova YE. Proc Natl Acad Sci USA. 2002;99:6877–6882. doi: 10.1073/pnas.102015399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barber RC, Miccoli L, van Buul PP, Burr KL, van Duyn-Goedhart A, Angulo JF, Dubrova YE. Mutat Res. 2004;554:287–295. doi: 10.1016/j.mrfmmm.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Barber R, Plumb M, Smith AG, Cesar CE, Boulton E, Jeffreys AJ, Dubrova YE. Mut Res. 2000;457:79–91. doi: 10.1016/s0027-5107(00)00130-5. [DOI] [PubMed] [Google Scholar]
- 15.Dubrova YE, Plumb M, Brown J, Fennelly J, Bois P, Goodhead D, Jeffreys AJ. Proc Natl Acad Sci USA. 1998;95:6251–6255. doi: 10.1073/pnas.95.11.6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vilarino-Guell C, Smith AG, Dubrova YE. Mutat Res. 2003;526:63–73. doi: 10.1016/s0027-5107(03)00016-2. [DOI] [PubMed] [Google Scholar]
- 17.Niwa O, Kominami R. Proc Natl Acad Sci USA. 2001;98:1705–1710. doi: 10.1073/pnas.031439298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hales BF, Robaire B. J Androl. 2001;22:927–936. doi: 10.1002/j.1939-4640.2001.tb03431.x. [DOI] [PubMed] [Google Scholar]
- 19.Harrouk W, Codrington A, Vinson R, Robaire B, Hales BF. Mutat Res. 2000;461:229–241. doi: 10.1016/s0921-8777(00)00053-7. [DOI] [PubMed] [Google Scholar]
- 20.Marchetti F, Bishop JB, Cosentino L, Moore D, II, Wyrobek AJ. Biol Reprod. 2004;70:616–624. doi: 10.1095/biolreprod.103.023044. [DOI] [PubMed] [Google Scholar]
- 21.Kelly R, Gibbs M, Collick A, Jeffreys AJ. Proc Biol Sci. 1991;245:235–245. doi: 10.1098/rspb.1991.0115. [DOI] [PubMed] [Google Scholar]
- 22.Kelly R, Bulfield G, Collick A, Gibbs M, Jeffreys AJ. Genomics. 1989;5:844–856. doi: 10.1016/0888-7543(89)90126-2. [DOI] [PubMed] [Google Scholar]
- 23.Gibbs M, Collick A, Kelly RG, Jeffreys AJ. Genomics. 1993;17:121–128. doi: 10.1006/geno.1993.1292. [DOI] [PubMed] [Google Scholar]
- 24.Yauk CL. Mutat Res. 2004;566:169–182. doi: 10.1016/j.mrrev.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Dubrova YE, Plumb M, Brown J, Fennelly J, Bois P, Goodhead D, Jeffreys AJ. Proc Natl Acad Sci USA. 1998;95:6251–6255. doi: 10.1073/pnas.95.11.6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yauk CL, Dubrova YE, Grant GR, Jeffreys AJ. Mutat Res. 2002;500:147–156. doi: 10.1016/s0027-5107(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 27.Dubrova YE. Oncogene. 2003;22:7087–7093. doi: 10.1038/sj.onc.1206993. [DOI] [PubMed] [Google Scholar]
- 28.Niwa O. Mutat Res. 2006;598:61–72. doi: 10.1016/j.mrfmmm.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 29.Jones PA, Gonzalgo ML. Proc Natl Acad Sci USA. 1997;94:2103–2105. doi: 10.1073/pnas.94.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rizwana R, Hahn PJ. J Cell Sci. 1999;112(Pt 24):4513–4519. doi: 10.1242/jcs.112.24.4513. [DOI] [PubMed] [Google Scholar]
- 31.Swales AK, Spears N. Reproduction. 2005;130:389–399. doi: 10.1530/rep.1.00395. [DOI] [PubMed] [Google Scholar]
- 32.Doerfler W. Curr Top Microbiol Immunol. 2006;301:3–18. doi: 10.1007/3-540-31390-7_1. [DOI] [PubMed] [Google Scholar]
- 33.Jones PA, Baylin SB. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehrlich M. Curr Top Microbiol Immunol. 2006;310:251–274. doi: 10.1007/3-540-31181-5_12. [DOI] [PubMed] [Google Scholar]
- 35.Feinberg AP, Tycko B. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 36.Raiche J, Rodriguez-Juarez R, Pogribny I, Kovalchuk O. Biochem Biophys Res Commun. 2004;325:39–47. doi: 10.1016/j.bbrc.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Tryndyak VP, Kovalchuk O, Muskhelishvili L, Montgomery B, Rodriguez-Juarez R, Melnyk S, Ross SA, Beland FA, Pogribny IP. Mol Carcinog. 2007;46:187–197. doi: 10.1002/mc.20263. [DOI] [PubMed] [Google Scholar]
- 38.Minamoto T, Mai M, Ronai Z. Carcinogenesis. 1999;20:519–527. doi: 10.1093/carcin/20.4.519. [DOI] [PubMed] [Google Scholar]
- 39.Koturbash I, Baker M, Loree J, Kutanzi K, Hudson D, Pogribny I, Sedelnikova O, Bonner W, Kovalchuk O. Int J Radiat Oncol Biol Phys. 2006;66:327–330. doi: 10.1016/j.ijrobp.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 40.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skinner MK, Anway MD. Ann NY Acad Sci. 2005;1061:18–32. doi: 10.1196/annals.1336.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Somers CM. Mutat Res. 2006;598:35–49. doi: 10.1016/j.mrfmmm.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 43.Legzdin A, McCarry BE, Bryant DW. Polycyclic Aromatic Compouds. 1993;5:157–165. [Google Scholar]
- 44.Ma JY, Ma JK. J Environ Sci Health C. 2002;20:117–147. doi: 10.1081/GNC-120016202. [DOI] [PubMed] [Google Scholar]
- 45.Ercal N, Gurer-Orhan H, Aykin-Burns N. Curr Top Med Chem. 2001;1:529–539. doi: 10.2174/1568026013394831. [DOI] [PubMed] [Google Scholar]
- 46.Valko M, Morris H, Cronin MT. Curr Med Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- 47.Burr KL, van Duyn-Goedhart A, Hickenbotham P, Monger K, van Buul PP, Dubrova YE. Mutat Res. 2007 doi: 10.1016/j.mrfmmm.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 48.Dubrova YE. Rad Res. 2005;163:200–207. doi: 10.1667/rr3296. [DOI] [PubMed] [Google Scholar]
- 49.Chang HS, Anway MD, Rekow SS, Skinner MK. Endocrinology. 2006;147:5524–5541. doi: 10.1210/en.2006-0987. [DOI] [PubMed] [Google Scholar]
- 50.Anway MD, Leathers C, Skinner MK. Endocrinology. 2006;147:5515–5523. doi: 10.1210/en.2006-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trasler JM, Alcivar AA, Hake LE, Bestor T, Hecht NB. Nucleic Acids Res. 1992;20:2541–2545. doi: 10.1093/nar/20.10.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Biermann K, Steger K. J Androl. 2007;28:466–480. doi: 10.2164/jandrol.106.002048. [DOI] [PubMed] [Google Scholar]
- 53.James SJ, Pogribny IP, Pogribna M, Miller BJ, Jernigan S, Melnyk S. J Nutr. 2003;133:3740S–3747S. doi: 10.1093/jn/133.11.3740S. [DOI] [PubMed] [Google Scholar]
- 54.Bestor T, Laudano A, Mattaliano R, Ingram V. J Mol Biol. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- 55.Roberts RJ. Cell. 1995;82:9–12. doi: 10.1016/0092-8674(95)90046-2. [DOI] [PubMed] [Google Scholar]
- 56.Robertson KD. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 57.Slotkin RK, Martienssen R. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 58.Jirtle RL, Skinner MK. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pfeifer K. Am J Hum Genet. 2000;67:777–787. doi: 10.1086/303101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jaenisch R, Bird A. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 61.Monk M, Salpekar A. Mol Cell Endocrinol. 2001;183(Suppl 1):S35–S40. doi: 10.1016/s0303-7207(01)00575-5. [DOI] [PubMed] [Google Scholar]
- 62.Reik W, Dean W, Walter J. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 63.Reik W, Dean W. Nature. 2002;420:127. doi: 10.1038/420127a. [DOI] [PubMed] [Google Scholar]
- 64.Egger G, Liang G, Aparicio A, Jones PA. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 65.Ettinger HJ, DeField JD, Bevis DA, Mitchell RN. Am Ind Hyg Assoc J. 1969;30:20–26. doi: 10.1080/00028896909343074. [DOI] [PubMed] [Google Scholar]
- 66.Polyzos A, Parfett C, Healy C, Douglas G, Yauk CL. Mut Res. 2006;598:73–84. doi: 10.1016/j.mrfmmm.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 67.Godschalk RW, Maas LM, Van Zandwijk N, van 't Veer LJ, Breedijk A, Borm PJ, Verhaert J, Kleinjans JC, van Schooten FJ. Carcinogenesis. 1998;19:819–825. doi: 10.1093/carcin/19.5.819. [DOI] [PubMed] [Google Scholar]
- 68.Basnakian AG, James SJ. DNA Cell Biol. 1996;15:255–262. doi: 10.1089/dna.1996.15.255. [DOI] [PubMed] [Google Scholar]
- 69.Koturbash I, Rugo RE, Hendricks CA, Loree J, Thibault B, Kutanzi K, Progribny I, Yanch JC, Engelward BP, Kovalchuk O. Oncogene. 2006;25:4267–4275. doi: 10.1038/sj.onc.1209467. [DOI] [PubMed] [Google Scholar]
- 70.Pogribny I, Yi P, James SJ. Biochem Biophys Res Commun. 1999;262:624–628. doi: 10.1006/bbrc.1999.1187. [DOI] [PubMed] [Google Scholar]
- 71.Trasler J, Deng L, Melnyk S, Pogribny I, Hiou-Tim F, Sibani S, Oakes C, Li E, James SJ, Rozen R. Carcinogenesis. 2003;24:39–45. doi: 10.1093/carcin/24.1.39. [DOI] [PubMed] [Google Scholar]
- 72.Balaghi M, Wagner C. Biochem Biophys Res Commun. 1993;193:1184–1190. doi: 10.1006/bbrc.1993.1750. [DOI] [PubMed] [Google Scholar]
- 73.Tryndyak VP, Muskhelishvili L, Kovalchuk O, Rodriguez-Juarez R, Montgomery B, Churchwell MI, Ross SA, Beland FA, Pogribny IP. Carcinogenesis. 2006;27:1713–1720. doi: 10.1093/carcin/bgl050. [DOI] [PubMed] [Google Scholar]
- 74.Sibani S, Melnyk S, Pogribny IP, Wang W, Hiou-Tim F, Deng L, Trasler J, James SJ, Rozen R. Carcinogenesis. 2002;23:61–65. doi: 10.1093/carcin/23.1.61. [DOI] [PubMed] [Google Scholar]