Abstract
The essential fatty acid eicosapentaenoic acid (EPA) present in fish oils displays beneficial effects in a range of human disorders associated with inflammation including cardiovascular disease. Resolvin E1 (RvE1), a new bioactive oxygenated product of EPA, was identified in human plasma and prepared by total organic synthesis. Results of bioaction and physical matching studies indicate that the complete structure of RvE1 is 5S,12R,18R-trihydroxy-6Z,8E,10E,14Z,16E-EPA. At nanomolar levels, RvE1 dramatically reduced dermal inflammation, peritonitis, dendritic cell (DC) migration, and interleukin (IL) 12 production. We screened receptors and identified one, denoted earlier as ChemR23, that mediates RvE1 signal to attenuate nuclear factor–κB. Specific binding of RvE1 to this receptor was confirmed using synthetic [3H]-labeled RvE1. Treatment of DCs with small interference RNA specific for ChemR23 sharply reduced RvE1 regulation of IL-12. These results demonstrate novel counterregulatory responses in inflammation initiated via RvE1 receptor activation that provide the first evidence for EPA-derived potent endogenous agonists of antiinflammation.
Clinical assessment of dietary supplementation with omega-3 polyunsaturated fatty acids (ω-3 PUFAs) indicate their beneficial impact in certain human diseases, particularly those in which inflammation is suspected as a key component in pathogenesis (1–3). Their molecular bases of action in reducing disease and local inflammation are important and of interest given the heightened awareness that inflammation and resolution are major mechanisms involved in many diseases, including cardiovascular disease, arthritis, Alzheimer's disease, asthma, and periodontitis (4, 5). ω-3 PUFAs are widely held to act via several possible mechanisms, such as preventing conversion of arachidonate to proinflammatory eicosanoids or serving as an alternative substrate producing less potent products (1). Of interest, fish leukocytes rich in ω-3 PUFA generate mediators from eicosapentaenoic acid (EPA) that play signaling roles (6). However, the pathophysiological role of enzymatic oxygenated compounds derived from EPA remains of interest in humans.
Recently, we uncovered a novel family of aspirin-triggered bioactive lipids biosynthesized during the spontaneous resolution phase of acute inflammation in vivo, coined the resolvins (resolution-phase interaction products), which are potent autacoids that now may provide molecular means that underlie ω-3 PUFA's protective actions (7, 8). At local sites, aspirin treatment enables EPA conversion to the novel 18R series of oxygenated products, which carry potent counterregulatory signals. One of the main bioactive compounds of this 18R series, namely 5,12,18R-trihydroxy-EPA (termed resolvin E1 [RvE1]) can arise via cell–cell interactions in murine inflammatory exudates, also exemplified with human vascular endothelial cells carrying aspirin-acetylated cyclooxygenase (COX)-2 and leukocytes possessing lipoxygenase (LO) activity (7). Because the GISSI paper reported a beneficial impact of ω-3 PUFA treated in >11,000 cardiovascular patients also taking aspirin (2), our results suggest that the novel antiinflammatory action of EPA-derived RvE1 may, in part, be responsible for the beneficial impact of EPA in cardiovascular diseases.
Here, the novel lipid mediator RvE1 was identified in humans and its complete stereochemical structure was established as 5S,12R,18R-trihydroxy-6Z,8E,10E,14Z,16E-EPA. RvE1 radiolabel was prepared and shown to interact with a G-protein–coupled receptor (GPCR) ChemR23 that mediates its potent protective and antiinflammatory actions.
Results
RvE1 generation in vivo
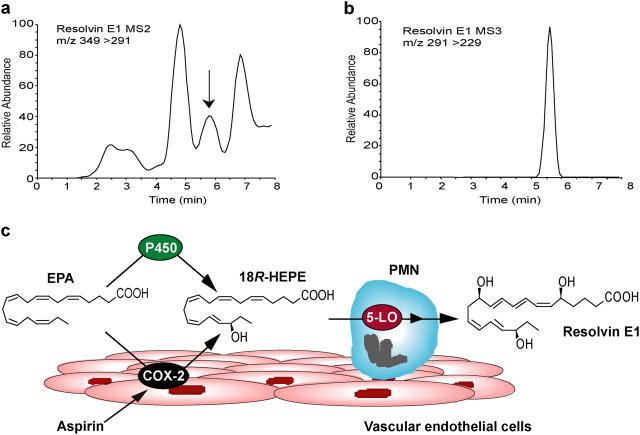
Originally RvE1 was found in vivo during resolution phase of inflammation in exudates from murine dorsal pouches treated with aspirin and EPA, and was generated during coincubation of human endothelial cells and neutrophils (7). RvE1 was monitored using liquid chromatography-tandem mass spectrometry (LC-MS/MS) in healthy human volunteers given EPA and aspirin (Fig. 1, a and b). The plasma values ranged from 0.1 to 0.4 ng/ml for six donors. The formation of RvE1 is consistent with the scheme that endothelial cells expressing COX-2 treated with aspirin transform vascular EPA to produce and release 18R-HEPE as we reported earlier (7). When human leukocytes and endothelial cells interact within the vasculature, 18R-HEPE is rapidly converted to RvE1 via transcellular biosynthesis (Fig. 1 c).
Figure 1.
Lipidomic analysis of RvE1 in human plasma. (a) Representative MS/MS selected ion chromatogram at m/z 291 and (b) MS3 selected ion chromatogram at m/z 229. (c) Scheme of RvE1 generation from EPA. Human endothelial cells expressing COX-2 treated with aspirin transform EPA by abstracting hydrogen at C16 to give R insertion of molecular oxygen to yield 18R-H(p)EPE. Alternatively, cytochrome P450 monooxygenase can convert EPA to 18R-HEPE (reference 28). They are further converted via sequential actions of a leukocyte 5-LO–like reaction, which leads to formation of RvE1.
Assignment of the complete stereochemistry of RvE1
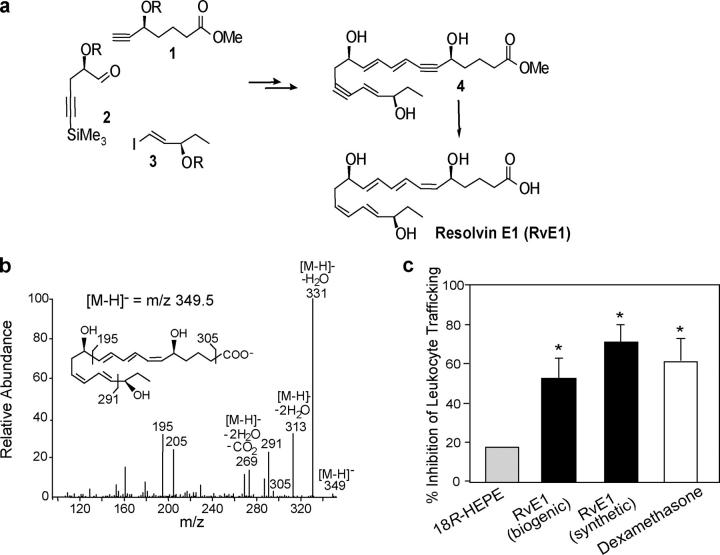
Although the basic structure of RvE1 and actions were initially reported (i.e., 5,12,18R-trihydroxy-EPA; reference 7), its complete stereochemical assignment remained to be established. To assign the complete stereochemistry of RvE1 and establish its biological activities, biogenic RvE1 was prepared (7) and matched with synthetic RvE1 (5S,12R,18R-trihydroxy-6Z,8E,10E,14Z,16E-EPA), having complete stereochemistry that was prepared by total organic synthesis from isomerically pure precursors (Fig. 2 a). Several geometric isomers were prepared by organic synthesis that carried different double bond geometries in native RvE1, namely 5S,12R,18R-6E,10E,14E,16E-triHEPE and 5S,12R,18R-6E,8Z,10E,14Z,16E-triHEPE, to establish their chromatographic and physical properties (Table S1, available at http://www.jem.org/cgi/content/full/jem.20042031/DC1). Because RvE1 is produced in subnanogram amounts in vivo, to assign its complete stereochemistry it was necessary to prepare both synthetic and biogenic materials for matching of their physical properties using UV spectroscopy, LC-MS/MS, gas chromatography mass spectrometry, and importantly compared biological activities. Of the isomers prepared and studied, the matching synthetic compound eluted beneath a single peak in HPLC with UV absorbance maxima 271 nm and 234 nm, indicative of conjugated triene and diene in the molecule. MS/MS fragmentation ions were essentially identical with the biogenic material, namely a parent ion at m/z 349 = [M-H]− and diagnostic product ions at m/z =291 and 195 (Fig. 2 b). Results of physical matching studies are summarized in Table S1. Administration of as little as 100 ng/mouse of synthetic RvE1 stopped leukocyte infiltration into inflammatory loci by 50∼70% in the TNF-α–induced dorsal air pouch, which proved to be as potent as the biogenic material (Fig. 2 c). For comparison in this model, local administration of dexamethasone (10 μg/mouse) gives 60% inhibition (Fig. 2 c) and aspirin (1.0 mg/mouse) gives 70% inhibition of leukocyte recruitment (9), indicating that RvE1 at nanomolar level is as potent as higher doses of dexamethasone or aspirin in stopping leukocyte infiltration. Also indomethacin (100 ng/mouse) gave 25% inhibition and RvE1 (100 ng/mouse) gave 50∼60% inhibition of leukocyte recruitment in zymosan-induced peritonitis (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20042031/DC1). The 18S isomer gave essentially equivalent activity as native RvE1 containing 18R, whereas the 6-trans,14-trans isomer showed reduced potency (∼70%) for reducing leukocyte infiltration in zymosan-induced peritonitis (unpublished data). Based on matching of physical and biological properties, the 18R series RvE1, a potent antiinflammatory lipid mediator, was assigned the complete structure 5S,12R,18R-trihydroxy-6Z,8E,10E,14Z,16E-EPA.
Figure 2.
Properties of RvE1. (a) Total organic synthesis of RvE1. Precursors 1–3 were prepared in isomerically pure form from starting materials with known stereochemistry and coupled sequentially to form acetylenic intermediate compound 4, which was selectively hydrogenated to form isomerically pure RvE1. (b) MS/MS spectrum of synthetic RvE1. (c) Inhibition of leukocyte trafficking in murine dorsal air pouch. 18R-HEPE and RvE1 (biogenic and synthetic) are at 100 ng/mouse, and dexamethasone is at 10 μg/mouse. Values represent mean ± SEM from five different mice. *, P < 0.05 (vs. vehicle control).
Identification of receptor candidate
The murine air pouch is widely used to assess dermal inflammation (Fig. 2 c); it is characterized by a cavity and a lining composed of both fibroblast-like and macrophage-like cells (10). Intrapouch application of TNF-α evokes leukocyte infiltration by stimulating local release of chemokines and chemoattractants that are often produced by fibroblasts and phagocytes via regulation of NF-κB transcription factors (11). Systemic administration of RvE1 dramatically attenuated leukocyte recruitment (Fig. 2 c), suggesting the receptor target for RvE1 was expressed in those cells, which counterregulate TNF-α–induced NF-κB activation. RvE1 and lipoxin (LX) A4 have different structures and are formed via different biosynthetic pathways and precursors (EPA vs. arachidonate), yet they appear to share redundant beneficial properties that dampen excessive leukocyte recruitment (12); therefore, we reasoned that RvE1 receptors might share similar structural features to LO-derived eicosanoid receptors such as LXA4 receptor (ALX) and leukotriene B4 receptor (13).
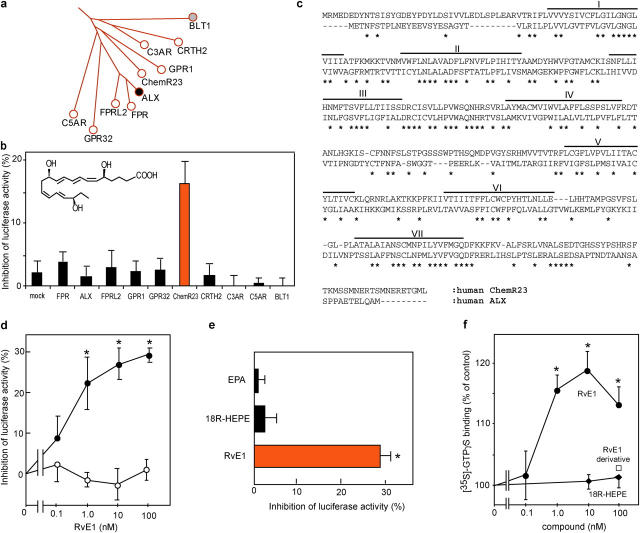
Fig. 3 a shows a branch of the phylogenetic tree of human ALX with closely related GPCRs. Expression plasmids of each GPCR were introduced into HEK293 cells, and the ability of RvE1 to inhibit TNF-α–stimulated NF-κB activation was monitored by cotransfection with NF-κB response element–luciferase reporter plasmid. This permitted an unbiased analysis of the activation of the relevant postligand receptor “stop” signaling for down-regulation of NF-κB activation as, for example, demonstrated with ALX-transfected cells and its ligands (14). Among those screened (Fig. 3 b), the orphan receptor denoted earlier as ChemR23 (15) was specifically activated by RvE1 and, at 10 nM, inhibited NF-κB activation (Fig. 3 b). ChemR23 shares 36.4% identity with ALX in deduced amino acid sequences and, of note, it contains a highly conserved domain within its second intracellular loop with 75% identity and within the seventh transmembrane region with 69.5% identity (Fig. 3 c).
Figure 3.
RvE1 receptor. (a) Phylogenetic tree representing amino acid sequence similarities between the human LXA4 receptor (ALX) and related GPCRs. (b) Functional screening for RvE1 receptors. HEK293 cells cotransfected with pNF-κB-luciferase and pcDNA3-GPCRs were exposed to RvE1 (10 nM) and TNF-α. (c) Amino acid sequence alignment of human ChemR23 with ALX. Asterisks indicate conserved amino acids. Putative transmembrane domains are lined and labeled I–VII. (d) RvE1 inhibits luciferase activity in a concentration-dependent manner on cells transfected with pcDNA3-ChemR23 (closed circles) but not pcDNA3 (open circles). (e) Ligand specificity for ChemR23. Cells transfected with pcDNA3-ChemR23 were exposed to 100 nM of each compound. Results are expressed as percent inhibition of luciferase activity and represent the mean ± SEM from n = 3 (b) or n = 4 (d and e); *, P < 0.05. (f) Actions of RvE1 (closed circles), RvE1 derivative (6,14-diacetylenic-RvE1; open rectangle), and 18R-HEPE (closed diamonds) on [35S]-GTP-γS binding to membrane expressing ChemR23. Results are expressed as a percentage of vehicle control with the mean ± SEM (n = 3). *, P < 0.05.
RvE1 gave concentration-dependent inhibition of TNF-α–induced NF-κB activation with an EC50 of ∼1.0 nM in ChemR23-transfected cells, but not in mock-transfected cells (Fig. 3 d). In this system, 1 mM aspirin, a known inhibitor of NF-κB at high concentrations (namely millimolar range; reference 16), gave nonreceptor-dependent inhibition of 26.2 ± 4.9% for the transfected cells. Neither EPA nor 18R-HEPE at 100 nM, both metabolic precursors of RvE1, inhibited NF-κB in ChemR23-transfected cells (Fig. 3 e). The double bond isomer of RvE1, 6-trans,14-trans-RvE1, at 100 nM showed reduced potency for NF-κB inhibition that was essentially the same magnitude reduction in vivo. We also examined functional interactions between ChemR23 and G-proteins using ligand-dependent binding of [35S]-GTPγS, a hydrolysis resistant GTP analogue. Specific [35S]-GTPγS binding in isolated membranes obtained from cells expressing ChemR23 increased selectively with RvE1 in a concentration-dependent manner (Fig. 3 f). These results indicate that RvE1 transmits signal as a selective agonist via ChemR23 and counterregulates TNF-α–stimulated NF-κB activation.
Radioligand binding
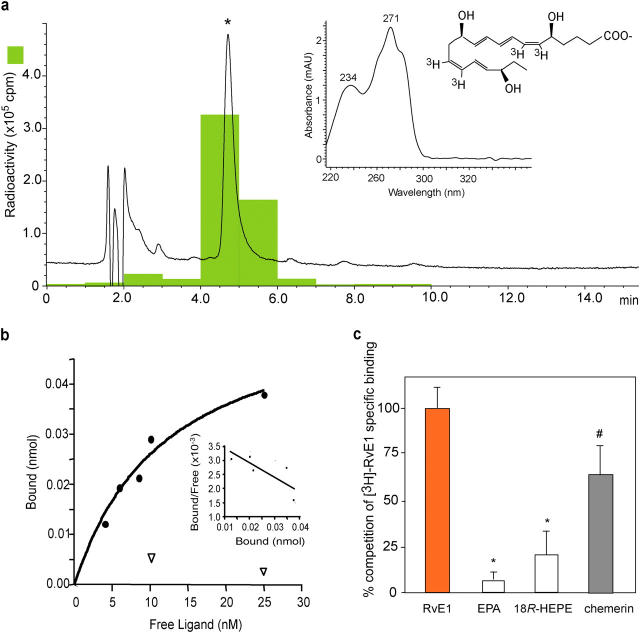
To determine RvE1 binding to ChemR23, tritium-labeled RvE1 was prepared by catalytic hydrogenation from synthetic diacetylenic RvE1 (Fig. 2 a, compound labeled 4). Integrity of the synthetic [3H]-RvE1 was confirmed by HPLC; radioactive compound coeluted with synthetic RvE1 beneath a single peak in HPLC. Its characteristic UV absorbance maxima at 271 nm and 234 nm were indicative of the conjugated triene and diene in RvE1 structure (Fig. 4 a). Analysis of [3H]-labeled RvE1-specific binding and Scatchard transformation is shown in Fig. 4 b. [3H]RvE1 bound to an apparent single site on ChemR23 transfectants with high affinity (Kd = 11.3 ± 5.4 nM, Bmax = 4,200 ± 1,050 binding sites per cell). RvE1 biosynthesis precursors, EPA and 18R-HEPE, did not compete for specific [3H]RvE1 binding (Fig. 4 c). We also tested the synthetic peptide fragment (YHSFFFPGQFAFS) derived from chemerin that was reported recently (while the present studies were in progress) to be a peptide ligand for this same receptor (17). Chemerin peptide at 10 μM competed for specific [3H]RvE1 binding by ∼70%, suggesting that lipid ligand RvE1 and peptide ligand chemerin share recognition sites on the same receptor ChemR23.
Figure 4.
Labeled RvE1 and specific binding. (a) Elution profile of the [3H]-labeled RvE1 on reverse phase HPLC. Tracing was recorded at 270 nm absorbance and chromatographic peak at the same retention time as standard RvE1 is indicated by the asterisk. The inset shows the online UV spectrum of the product. (b) [3H]-RvE1–specific binding to ChemR23. Human ChemR23–transfected (closed circles) or mock-transfected (open inverted triangles) CHO cells (106 cells) were incubated with indicated concentrations of [3H]-RvE1 in the presence or absence of 10 μM of unlabeled RvE1. Saturation curve and Scatchard plot (inset) are representative of n = 3. (c) Binding specificity. Human ChemR23-transfected CHO cells (106 cells) were incubated with 10 nM of [3H]-RvE1 in the presence of 10 μM of RvE1, EPA, 18R-HEPE, or chemerin peptide. Results are expressed as percent competition of [3H]-RvE1–specific binding. Results represent the mean ± SEM from duplicates of n = 3∼5. p-values were obtained by Student's t test comparing heteroligand (i.e., EPA, 18R-HEPE, or chemerin) with homoligand (i.e., RvE1). *, P < 0.01; #, P = 0.07.
Expression, regulation, and signaling properties of ChemR23
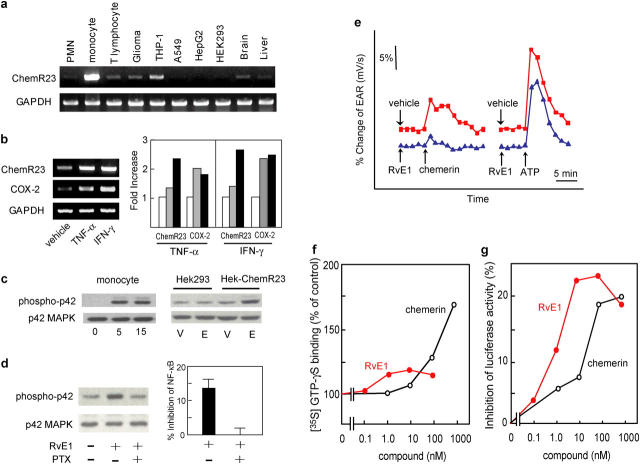
Tissue distribution of human ChemR23 was determined with dot blots containing mRNAs from human tissues that showed expression of ChemR23 in several tissues, such as cardiovascular system, brain, kidney, gastrointestinal tissues, and myeloid tissues (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20042031/DC1). Also, a murine receptor counterpart was found in developing bone using in situ hybridization (18). Among the human peripheral blood leukocytes, ChemR23 was abundantly expressed in monocytes, with lower amounts in neutrophils and T lymphocytes (Fig. 5 a), findings consistent with the observation that this receptor is expressed in APCs such as macrophages and DCs (15, 17). Both monocyte ChemR23 and COX-2 transcripts were highly up-regulated by treatment with inflammatory cytokines such as TNF-α and IFN-γ, and ChemR23 showed delayed induction to that of COX-2 (Fig. 5 b). RvE1 increased phosphorylation of extracellular signal-regulated kinase (ERK) mitogen-activated protein (MAP) kinase both in peripheral blood monocytes and HEK-ChemR23 cells, but not in mock-transfected HEK293 cells (Fig. 5 c). In addition, treatment of HEK-ChemR23 with pertussis toxin (PTX) abolished RvE1-dependent ERK activation and NF-κB inhibition, suggesting coupling to Gαi/o protein for the signal transduction (Fig. 5 d).
Figure 5.
Receptor expression. (a) RT-PCR analysis of human peripheral blood leukocytes and glioma (DBTRG-05MG), monocytic (THP-1), lung epithelial (A549), hepatoma (HepG2), embryonic kidney (HEK293) cell lines, and brain and liver. (b) RT-PCR analysis of human peripheral blood monocytes exposed to either buffer alone, 10 ng/ml TNF-α, or 25 ng/ml IFN-γ for 6 h (gray bars) and 24 h (black bars). Expression levels were quantified by a National Institutes of Health image, normalized by GAPDH levels, and expressed as fold increase over vehicle-treated cells. (c) MAP kinase activation in human peripheral blood monocytic cells were exposed to RvE1 for 5 or 15 min and HEK-ChemR23 cells were treated with 100 nM RvE1 (E) or vehicle (V). (d) Pertussis toxin (PTX) blocks RvE1-induced ERK activation and NF-κB inhibition in HEK-ChemR23 cells. (e) EAR with HEK-ChemR23 cells exposed to vehicle (red line), 100 nM RvE1 (blue line), 10 μM chemerin peptide, or 10 μM ATP using Cytosensor microphysiometer. These responses were observed from three separate experiments. (f) Actions of RvE1 (red) and chemerin peptide (black) on [35S]-GTP-γS binding to HEK293 cell membranes expressing human ChemR23. (g) Inhibition of TNF-α induced NF-κB luciferase activities by RvE1 (red) and chemerin peptide (black) on HEK293 cells expressing human ChemR23.
Because both RvE1 and chemerin peptide competed at the same receptor, it was of interest to determine whether there was a divergence between the activities of the two ligands. Chemerin-derived peptide activated ChemR23 as monitored by microphysiometry, evoking a response consistent with receptor-initiated increases in extracellular acidification rates (EARs; Fig. 5 e, red line). These results are in line with the reported properties of this peptide ligand (17). Of interest, the lipid RvE1 did not increase the EAR but selectively blocked the peptide-evoked responses (Fig. 5 e, blue line). Consistently, the extent of G-protein activation evoked by chemerin peptide was three times as much as RvE1 (Fig. 5 f). Alternately, RvE1 was an order of magnitude more potent than chemerin peptide for inhibiting NF-κB activation (Fig. 5 g). These results suggest that RvE1 interacts with ChemR23 and transmits signals via this receptor in a fashion different than that of the chemerin-derived peptide.
RvE1-ChemR23 interaction blocks splenic DC response to pathogen
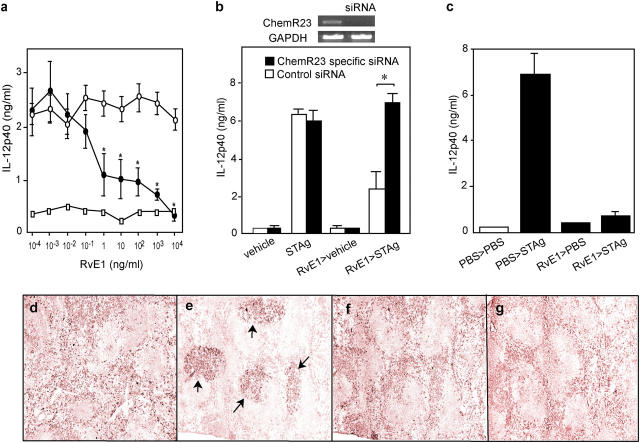
Given expression of ChemR23 in APCs and because APC function is influenced by dietary ω-3 PUFA supplementation (19), next we examined the activity of RvE1 on APC function using a microbial pathogen model. Injection of pathogen extract derived from Toxoplasma gondii soluble tachyzoite antigen (STAg) causes activation of splenic DCs to mobilize to T cell–enriched areas where they produce high amounts of IL-12 (20). Addition of increasing concentrations of RvE1 to isolated mouse splenic CD11c+ DCs markedly inhibited IL-12p40 production by STAg within the nanomolar range (Fig. 6 a). Next, we performed small interference RNA (siRNA) experiments to reduce ChemR23 in splenic DCs. Mouse receptor (18), which shares 80.3% identity with human ChemR23, was also present in splenic DCs. RvE1's action in regulating IL-12 production from DCs was eliminated by treatment with a siRNA specific for the mouse ChemR23 (Fig. 6 b). We confirmed that this siRNA treatment dramatically reduced ChemR23 mRNA expression in DCs (Fig. 6 b, inset) and cell surface expression of recombinant ChemR23 in HEK293 cells (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20042031/DC1). These results confer that RvE1's antiinflammatory action is mediated via ChemR23. In vivo treatment with RvE1 also blocked IL-12 production (Fig. 6 c) as well as DC migration into T cell areas of the spleen (Fig. 6, d–g).
Figure 6.
RvE1 regulates DCs. (a) RvE1 inhibits DC IL-12 production in vitro stimulated by pathogen extract (STAg). CD11c+ DCs incubated with vehicle (open circles) or RvE1 (closed circles) before STAg or no STAg (open squares). (b) Reduction of ChemR23 expression by siRNA eliminates RvE1 signaling. Expression of ChemR23 and GAPDH mRNA from DCs treated with either scramble or ChemR23-specific siRNAs (inset). Spleen cells transfected with siRNAs were treated with vehicle (ethanol, 0.1% vol/vol) or RvE1 (1.0 μg/ml). 8 h later, cells were stimulated with 10 μg/ml STAg and IL-12p40 was measured. Bars represent mean ± SD (n = 3), *, P < 0.05 (control vs. specific siRNA). (c) RvE1 blocks IL-12 production in vivo. Mice administered with either 100 ng RvE1 or vehicle were challenged intraperitoneally with PBS or STAg, and IL-12p40 secretion from splenic CD11c+ DCs was measured. (d–g) RvE1 blocks trafficking of CD11c+ DCs in spleen with pathogen extract challenge. Spleens from mice given 10 μg RvE1 or vehicle were stained for CD11c. (d) PBS plus vehicle (e) STAg plus vehicle (f) PBS plus RvE1 (g) STAg plus RvE1. Arrows indicate CD11c positive DCs accumulated in T cell–enriched area.
Discussion
Acute inflammation is a protective host response to challenge and/or tissue injury that can, if unopposed, lead to loss of tissue structure and function. In many chronic disorders, prolonged and unresolved inflammation is held to contribute to pathogenesis (4). The resolution of inflammation is an active process controlled in part by endogenous chemical mediators that counterregulate proinflammatory gene expression and cell trafficking as well as stimulate inflammatory cell clearance (11, 21). Also, our finding that cytokines up-regulated ChemR23 as well as COX-2 in monocytes suggests that, in scenarios where COX-2 is induced during inflammation, monocytes as well as endothelial cells treated with aspirin can also generate and respond to RvE1. Hence, RvE1 may serve an autocrine and/or paracrine signal during resolution to terminate further NF-κB activation and cytokine production in a temporal and spatially regulated fashion. RvE1 is generated in healthy human volunteers given EPA and aspirin (Fig. 1), results that are consistent with the notion that COX-2 is also constitutively expressed within the vasculature of healthy subjects (22, 23). Thus, the present results support the notion that aspirin, in addition to its well-appreciated action to inhibit prostanoid formation, can exert its beneficial actions, in part, via EPA catabolic synthesis of 18R series RvE1 that in turn interacts with receptors such as ChemR23 to dampen further proinflammatory processes.
Endogenous chemically redundant antiinflammatory lipid autacoids act with high affinities (nM range) and stereoselectivity on structurally related receptors, as does aspirin-triggered LXA4 generated from arachidonic acid (24) to enhance resolution by “stopping” PMN recruitment and IL-12 production from APCs (20). RvE1 also blocked PMN transendothelial migration in vitro (7) and inhibited migration and IL-12 production from APCs (Fig. 6). It is of note that this bioactive lipid mediator RvE1 and new peptide ligand from chemerin (17) can each serve as endogenous ligands at the same GPCR but transmit different ligand-dependent signals (Fig. 5). It appears that the receptor activation evoked by ligand binding and their intracellular signal transductions are different for peptide versus lipid ligands of ChemR23 and, hence, they can dictate different functional responses in vivo. Chemerin was described recently as a novel chemoattractant peptide that selectively promotes chemotaxis of APCs such as immature DCs and macrophages in vitro (17). Alternately, here we showed that RvE1 blocked DC migration and IL-12 production in response to pathogen in vivo (Fig. 6). Temporal and spatial coordination of the recruitment and activation of APCs are important in inflammation and host defense, and a GPCR ChemR23 selectively expressed on APCs seems to play a key role by interacting with either endogenous lipid (RvE1) or peptide (chemerin) ligands in regulating migration and activation of this particular cell type in vivo. This appears to be a similar scenario to LXA4 and its receptor ALX expressed on PMNs, which also interacts with both specific lipid and peptide ligands to transmit ligand-dependent signals (reference 25 and for a recent review see reference 26). Because ChemR23 and ALX share overall structural similarities and strongly conserved domains, the “dual ligand” as well as antiinflammatory and proresolving properties might be conserved in these structurally related GPCRs.
Together, our findings offer an endogenous agonist–driven molecular mechanism that can underlie some of the beneficial actions of ω-3 EPA observed in many clinical situations (1–3) as well as identify novel components in endogenous antiinflammation/resolution, namely RvE1 and its receptor that are of interest as new checkpoint regulators (21) that may be involved in the pathogenesis of a wide range of human diseases.
Materials and Methods
LC-MS/MS analysis of RvE1.
Human plasma samples were collected at 4 h after oral administration of fish oil supplement (fish oil concentrate; Walgreens) containing 1 g EPA and 0.7 g DHA followed by 160 mg aspirin at 3 h in six healthy volunteers. Plasma samples were extracted by C18 solid phase extraction with d4-LTB4 (Cascade Biologics, Inc.) as internal standard for LC-MS/MS analysis (7) using a Finnigan LCQ liquid chromatography ion trap tandem mass spectrometer equipped with a LUNA C18-2 (100 × 2 mm × 5 μm) column and UV diode array detector using mobile phase (methanol:water:acetate at 65:35:0.01) with a 0.2 ml/min flow rate.
Murine dorsal air pouch model.
Dorsal air pouches were raised on male FvB mice (6–8 wk) by injecting 3 ml of sterile air subcutaneously on days 0 and 3. On day 6, 100 ng/mouse of compounds were injected into the tail vein. Inflammation in the air pouch was induced by intrapouch injection of mouse recombinant TNF-α (100 ng/pouch), pouch lavages were collected at 4 h, and cells were enumerated.
Zymosan-induced peritonitis.
For peritonitis, 100 ng/mouse of RvE1 or related structures was injected into tail vein and followed by 1 ml zymosan A (1 mg/ml) into the peritoneum. Peritoneal lavages were collected at 2 h and cells were enumerated.
GPCR cDNAs and phylogenetic tree.
GPCR cDNAs were cloned by RT-PCR using specific primers designed according to the GenBank/EMBL/DDBJ database; human FPR (P21462), ALX (P25090), FPRL2 (P25089), GPR1 (A55733), GPR32 (O75388), ChemR23 (Q99788), CRTH2 (Q9Y5Y4), C3AR (Q16581), C5AR (P21730), BLT1 (Q15722), and mouse ChemR23 (U79525). The phylogenetic tree was constructed using the “All All Program” at the Computational Biochemistry Server at ETHZ (http://cbrg.inf.ethz.ch/Server/AllAll.html).
NF-κB reporter gene expression.
HEK293 cells (1.0 × 105 cells) were transiently transfected with 50 ng pNF-κB luciferase (Stratagene), 500 ng of either pcDNA3 or pcDNA3-GPCRs, and the internal standard pRL-TK (Promega) using Superfect transfection reagent (QIAGEN). After 24 h, cells were exposed to the test compounds for 30 min and stimulated with 1.0 ng/ml recombinant human TNF-α (BD Biosciences) for 5 h. Luciferase activity was measured by the Dual-Luciferase reporter assay system (Promega). Basal induction of luciferase activity by TNF-α was >150-fold in this system. For PTX treatment, HEK293 cells were treated with 200 ng/ml PTX for 24 h before stimulation.
[35S]-GTPγS binding assay.
HEK293 cells stably expressing human ChemR23 were homogenized in ice-cold TED buffer (20 mM Tris-HCl, pH 7.5, 1 mM EDTA/5 mM MgCl2/1 mM DTT). Membrane fraction (10 μg) was incubated in 400 μl of GTP-binding buffer (50 mM Hepes, pH 7.5, 100 mM NaCl/1 mM EDTA/5 mM MgCl2/1 mM DTT) containing 0.1 nM [35S]-GTPγS (>1,000 Ci/mmol; Amersham Biosciences) and 10 μM GDP for 30 min at 30°C. The bound and unbound [35S]-GTPγS was separated by rapid filtration through GF/C filters, and counted by liquid scintillation. Nonspecific binding was determined in the presence of 50 μM of unlabeled GTPγS. Basal [35S]-GTP-γS binding was 81.6 ± 1.5 cpm/μg protein.
Radioligand binding.
Tritiated RvE1 (6,7,14,15-[3H]RvE1; ∼100 Ci/mmol) was obtained by custom catalytic hydrogenation of synthetic 6,14-diacetylenic RvE1 methyl ester (Fig. 2 a, compound 4) that was supplied to and performed at American Radiolabeled Chemicals, and the 3H-labeled RvE1 was saponified and isolated by HPLC. [3H]-RvE1–specific binding was performed with CHO cells (American Type Culture Collection) transfected with human recombinant ChemR23. Cells were suspended in Dulbecco's phosphate buffered saline with CaCl2 and MgCl2 (DPBS2+). For saturation binding, aliquots (106 cells) were incubated with increasing concentrations of [3H]-RvE1 in the presence or absence of unlabeled homoligand (10 μM) for 1 h at 14°C. To determine ligand specificity by competition binding, aliquots (106 cells) were incubated with 10 nM of [3H]-RvE1 in the presence of various competitors (10 μM) for 1 h at 14°C. Chemerin COOH-terminal bioactive peptide (YHSFFFPGQFAFS) was synthesized by SynPep Co. as described previously (17). The bound and unbound radioligands were separated by filtration through Whatman GF/C glass microfiber filters (Fisher Scientific) and radioactivity was determined. Scatchard plot was obtained and Kd and Bmax values were calculated using Prism (Graphpad Software, Inc.).
Dot blot hybridization and RT-PCR.
Hybridization to MTE array (CLONTECH Laboratories, Inc.) was performed using 1.1 kb.p. fragment encoding open reading frame of ChemR23 following the manufacturer's protocol. Primers used in amplifications were as follows: human ChemR23, 5′-ATGAGAATGGAGGATGAAGA-3′ and 5′-TCAAAGCATGCCGGTCTCC-3′; mouse ChemR23, 5′-ATGGAGTACGACGCTTACAA-3′ and 5′-TCAGAGGGTACTGGTCTCCTTCT-3′; COX-2, 5′-GCTGACTATGGCTACAAAAGCTGG-3′ and 5′-ATGCTCAGGGACTTGAGGAGGGTA-3′; and GAPDH, 5′-GACCACAGTCCATGACATCACT-3′ and 5′-TCCACCACCCTGTTGCTGTAG-3′. Amplified products were confirmed by direct sequencing.
MAP kinase activation.
MAP kinase activation in monocytes and HEK293 cells after treatment with 100 nM of each compound was determined. After incubations, cells were lysed in cold lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 1.0% NP-40, 0.5% sodium deoxycolate, 10 mM NaF, 10 mM sodium pyrophosphate) containing protease inhibitor cocktail (Sigma-Aldrich). 40 μg of protein was separated on SDS-PAGE and immunoblot was performed using anti–phospho-p44/42 MAP kinase (Cell Signaling Technology) and anti-ERK (Santa Cruz Biotechnology, Inc.) antibodies. For PTX treatment, HEK-ChemR23 cells were incubated with or without 200 ng/ml PTX for 24 h at 37°C and ERK activation was monitored by addition of 100 nM RvE1 for 5 min.
Ligand-activated EAR.
Changes in HEK293 EARs were evaluated by use of a Cytosensor microphysiometer (Molecular Devices) as described previously (27). Cells were seeded into 12-mm Transwell inserts (3.0 μm pore size) at 5 × 104 cells and cultured for 2 d. Capsules were placed in sensor chambers and equilibrated for 30 min in MD-RPMI 1640 (1 mM phosphate). EARs were monitored at 30-s potentiometric rate measurements (μV/s; pump off cycle) after an 80-s pump cycle with a 10-s delay (120 s total cycle time). Cells were perfused with the indicated agonists for 30-s before the first rate measurements.
Activation of spleen DCs with pathogen extract (STAg).
Experiments were performed essentially as described previously (20). STAg was prepared from sonicated T. gondii (RH strain) tachyzoites. For isolated DC experiments, 70–85% CD11c+ DCs were isolated from spleen. CD11c+ DC suspensions (106 cells/ml) were spread into 96-well plates and incubated for 24 h with RvE1 before the addition of 5 μg/ml STAg. After overnight culture, supernatants were collected and IL-12p40 was measured with a sandwich ELISA. For in vivo treatments, C57BL/6 mice (n = 3 per group) were injected intravenously with 100 ng RvE1. After 18 h, the animals were challenged intraperitoneally with PBS (0.2 ml/mouse) and 5 μg/ml STAg and killed after an additional 6 h. CD11c+ DCs were isolated from spleen and IL12-p40 secretion was measured at 24 h. For DC migration, splenic frozen section from mice treated as described before (but given 10 μg of RvE1 or vehicle) were stained for CD11c and counterstained with hematoxylin.
RNA interference.
Synthetic siRNA for mouse ChemR23, 5′-AACACUGUGUGGUUUGUCAACdTdT-3′, and nonspecific control IX siRNA, 5′-AUUGUAUGCGAUCGCAGACUU-3′, were obtained from Dharmacon Research. Spleen cells (106 cells/ml) were transfected using Chariot (Active Motif) following the manufacturer's instructions. In brief, siRNA was mixed with Chariot transfection reagent and incubated at room temperature for 30 min. Spleen cells were plated in serum-free RPMI 1640 medium and 200 ng siRNA/Chariot solution was added and incubated for 2 h at 37°C, followed by the addition of 10% FCS RPMI 1640 to the cultures. To assure effective inhibition of gene expression, cells were further incubated for 30 h at 37°C before STAg stimulation.
Online supplemental material.
Table S1 summarizes the chromatographic, physical, and chemical properties of the biogenic RvE1, synthetic RvE1, and its geometric isomers for both HPLC-UV-MS/MS and gas chromatography mass spectrometry analyses. Fig. S1 demonstrates the biological property of synthetic RvE1 that inhibits leukocyte infiltration in murine zymosan-induced peritonitis. Fig. S2 shows the human tissue distribution of ChemR23 by dot blot hybridization. ChemR23 is expressed in several tissues including the cardiovascular system, brain, kidney, gastrointestinal tissues, and myeloid tissues. Fig. S3 demonstrates the selectivity of mouse ChemR23-specific siRNA using transiently transfected HEK293 cells affirming its validation for use with mouse DCs (Fig. 6 b). Studies reported here were performed using protocols approved by Harvard Medical Area Standing Committee on animals and human subjects in accordance with the Brigham and Women's Human Research Committee. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20042031/DC1.
Acknowledgments
We thank M.H. Small for manuscript preparation; Dr. A. Ariel for peripheral blood cell isolation; Dr. K. Gronert for carrying out Cytosensor analyses; and E. Tjonahen, K. Gotlinger, and R. Moussignac for expert technical assistance. The diacetylenic RvE1 (see Fig. 2, compound 4) was a generous gift from Taisho Pharmaceutical Co. Ltd.
This study was supported in part by Uehara Memorial Foundation (to M. Arita), and National Institutes of Health grant nos. GM38765 (C.N. Serhan) and P50- DE-016191 (to C.N. Serhan and N.A. Petasis).
The authors have no conflicting financial interests.
Abbreviations used: ω-3 PUFA, omega-3 polyunsaturated fatty acid; COX, cyclooxygenase; EAR, extracellular acidification rate; EPA, eicosapentaenoic acid; ERK, extracellular signal-regulated kinase; GPCR, G-protein–coupled receptor; HEPE, hydroxyeicosapentaenoic acid; LC-MS/MS, liquid chromatography tandem mass spectrometry; LO, lipoxygenase; LXA4, lipoxin A4; MAP, mitogen-activated protein; PTX, pertussis toxin; RvE1, resolvin E1; siRNA, small interference RNA; STAg, soluble tachyzoite antigen.
References
- 1.De Caterina, R., S. Endres, S.D. Kristensen, and E.B. Schmidt. 1993. N-3 Fatty Acids and Vascular Disease. Bi & Gi Publishers, Verona. 166 pp.
- 2.GISSI-Prevenzione Investigators. 1999. Dietary supplementation with n-3 polyunsaturated fatty acids and vitamin E after myocardial infarction: results of the GISSI-Prevenzione trial. Lancet. 354:447–455. [PubMed] [Google Scholar]
- 3.Albert, C.M., H. Campos, M.J. Stampfer, P.M. Ridker, J. Manson, W.C. Willett, and J. Ma. 2002. Blood levels of long-chain n-3 fatty acids and the risk of sudden death. N. Engl. J. Med. 346:1113–1118. [DOI] [PubMed] [Google Scholar]
- 4.Weiss, U. 2002. Insight: inflammation. Nature. 420:845–891. [Google Scholar]
- 5.Funk, C.D. 2001. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 294:1871–1875. [DOI] [PubMed] [Google Scholar]
- 6.Rowley, A.F., D.J. Hill, C.E. Ray, and R. Munro. 1997. Haemostasis in fish, an evolutionary perspective. Thromb. Haemost. 77:227–233. [PubMed] [Google Scholar]
- 7.Serhan, C.N., C.B. Clish, J. Brannon, S.P. Colgan, N. Chiang, and K. Gronert. 2000. Novel functional sets of lipid-derived mediators with antiinflammatory actions generated from omega-3 fatty acids via cyclooxygenase 2-nonsteroidal antiinflammatory drugs and transcellular processing. J. Exp. Med. 192:1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serhan, C.N., S. Hong, K. Gronert, C.P. Colgan, P.R. Devchand, G. Mirick, and R. Moussignac. 2002. Resolvins: a family of bioactive products of omega-3 fatty acid transformation circuits initiated by aspirin treatment that counter proinflammation signals. J. Exp. Med. 196:1025–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clish, C.B., J.A. O'Brien, K. Gronert, G.L. Stahl, N.A. Petasis, and C.N. Serhan. 1999. Local and systemic delivery of a stable aspirin-triggered lipoxin prevents neutrophil recruitment in vivo. Proc. Natl. Acad. Sci. USA. 96:8247–8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawrence, T., D.A. Willoughby, and D.W. Gilroy. 2002. Anti-inflammatory lipid mediators and insights into the resolution of inflammation. Nat. Rev. Immunol. 2:787–795. [DOI] [PubMed] [Google Scholar]
- 11.Tessier, P.A., P.H. Naccache, I. Clark-Lewis, R.P. Gladue, K.S. Neote, and S.R. McColl. 1997. Chemokine networks in vivo. Involvement of C-X-C and C-C chemokines in neutrophil extravasation in vivo in response to TNF-α. J. Immunol. 159:3595–3602. [PubMed] [Google Scholar]
- 12.McMahon, B., S. Mitchell, H.R. Brady, and C. Godson. 2001. Lipoxins: revelations on resolution. Trends Pharmacol. Sci. 22:391–395. [DOI] [PubMed] [Google Scholar]
- 13.Yokomizo, T., T. Izumi, K. Chang, Y. Takuwa, and T. Shimizu. 1997. A G-protein-coupled receptor for leukotriene B4 that mediates chemotaxis. Nature. 387:620–624. [DOI] [PubMed] [Google Scholar]
- 14.Gewirtz, A.T., L.S. Collier-Hyams, A.N. Young, T. Kucharzik, W.J. Guilford, J.F. Parkinson, I.R. Williams, A.S. Neish, and J.L. Madara. 2002. Lipoxin A4 analogs attenuate induction of intestinal epithelial proinflammatory gene expression and reduce the severity of dextran sodium sulfate-induced colitis. J. Immunol. 168:5260–5267. [DOI] [PubMed] [Google Scholar]
- 15.Samson, M., A.L. Edinger, P. Stordeur, J. Rucker, V. Verhasselt, M. Sharron, C. Govaerts, C. Mollereau, G. Vassart, R.W. Doms, and M. Parmentier. 1998. ChemR23, a putative chemoattractant receptor, is expressed in monocyte-derived dendritic cells and macrophages and is a coreceptor for SIV and some primary HIV-1 strains. Eur. J. Immunol. 28:1689–1700. [DOI] [PubMed] [Google Scholar]
- 16.Kopp, E., and S. Ghosh. 1994. Inhibition of NF-κB by sodium salicylate and aspirin. Science. 265:956–959. [DOI] [PubMed] [Google Scholar]
- 17.Wittamer, V., J. Franssen, M. Vulcano, J. Mirjolet, E. Le Poul, I. Migeotte, S. Brezillon, R. Tyldesley, C. Blanpain, M. Detheux, et al. 2003. Specific recruitment of antigen-presenting cells by chemerin, a novel processed ligand from human inflammatory fluids. J. Exp. Med. 198:977–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Methner, A., G. Hermey, B. Schinke, and I. Hermans-Borgmeyer. 1997. A novel G protein-coupled receptor with homology to neuropeptide and chemoattractant receptors expressed during bone development. Biochem. Biophys. Res. Commun. 233:336–342. [DOI] [PubMed] [Google Scholar]
- 19.Endres, S., R. Ghorbani, V.E. Kelley, K. Georgilis, G. Lonnemann, J.W. van der Meer, J.G. Cannon, T.S. Rogers, M.S. Klempner, and P.C. Weber. 1989. The effect of dietary supplementation with n-3 polyunsaturated fatty acids on the synthesis of interleukin-1 and tumor necrosis factor by mononuclear cells. N. Engl. J. Med. 320:265–271. [DOI] [PubMed] [Google Scholar]
- 20.Aliberti, J., S. Hieny, C. Reis e Sousa, C.N. Serhan, and A. Sher. 2002. Lipoxin-mediated inhibition of IL-12 production by DCs: a mechanism for regulation of microbial immunity. Nat. Immunol. 3:76–82. [DOI] [PubMed] [Google Scholar]
- 21.Nathan, C. 2002. Points of control in inflammation. Nature. 420:846–852. [DOI] [PubMed] [Google Scholar]
- 22.Topper, J.N., J. Cai, D. Falb, and M.A. Gimbrone Jr. 1996. Identification of vascular endothelial genes differentially responsive to fluid mechanical stimuli: Cyclooxygenase-2, manganase superoxide dismutase, and endothelial cell nitric oxide synthase are selectively up-regulated by steady laminar shear stress. Proc. Natl. Acad. Sci. USA. 93:10417–10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng, Y., S.C. Austin, B. Rocca, B.H. Koller, T.M. Coffman, T. Grosser, J.A. Lawson, and G.A. FitzGerald. 2002. Role of prostacyclin in the cardiovascular response to thromboxane A2. Science. 296:539–541. [DOI] [PubMed] [Google Scholar]
- 24.Serhan, C.N. 2002. Endogenous chemical mediators in anti-inflammation and pro-resolution. Curr. Med. Chem. 1:177–192. [Google Scholar]
- 25.Chiang, N., I.M. Fierro, K. Gronert, and C.N. Serhan. 2000. Activation of lipoxin A4 receptors by aspirin-triggered lipoxins and select peptides evokes ligand-specific responses in inflammation. J. Exp. Med. 191:1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perretti, M. 2003. The annexin 1 receptor(s): is the plot unravelling? Trends Pharmacol. Sci. 24:574–579. [DOI] [PubMed] [Google Scholar]
- 27.Gronert, K., S.P. Colgan, and C.N. Serhan. 1998. Characterization of human neutrophil and endothelial cell ligand-operated extracellular acidification rate by microphysiometry: impact of reoxygenation. J. Pharmacol. Exp. Ther. 285:252–261. [PubMed] [Google Scholar]
- 28.Capdevila, J.H., S. Wei, C. Helvig, J.R. Falck, Y. Belosludtsev, G. Truan, S.E. Graham-Lorence, and J.A. Peterson. 1996. The highly stereoselective oxidation of polyunsaturated fatty acids by cytochrome P450BM-3. J. Biol. Chem. 271:22663–22671. [DOI] [PubMed] [Google Scholar]