Abstract
Splicing mutations in the human GH (hGH) gene (GH-1) that cause skipping of exon 3 result in a form of GH deficiency termed isolated GH deficiency type II (IGHD II). The GH-1 gene contains five exons; constitutive splicing produces the wild-type 22-kDa hormone, whereas skipping of exon 3 results in transcripts encoding a 17.5-kDa isoform that acts as a dominant-negative to block secretion of the wild-type hormone. Common characteristics of IGHD II include short stature due to impaired bone elongation, growth, and, in severe cases, anterior pituitary hypoplasia. Typically, IGHD II is treated by sc delivery of hGH, which can rescue stature but, unfortunately, does not inhibit pituitary hypoplasia. Direct destruction of transcripts encoding the dominant-negative 17.5-kDa isoform should both rescue stature and prevent hypoplasia. Here, we have used delivery of short hairpin RNAs to rescue a murine model of IGHD II by specifically targeting transcripts encoding the 17.5-kDa isoform using RNA interference. To our knowledge, this is the first example where a short hairpin RNA has been expressed to specifically degrade an incorrectly spliced transcript and rescue a dominant-negative disease phenotype in vivo.
THE GREAT MAJORITY of mutations that lead to isolated GH deficiency type II (IGHD II) occur in and around exon 3 and lead to aberrant splicing (1). These mutations include splice site mutations as well as disruption of splicing enhancer elements that are necessary to promote constitutive splicing (Fig. 1A) (2). GH-1 comprises five exons that are constitutively spliced to encode the wild-type 22-kDa isoform. Transcripts lacking exon 3 encode a 17.5-kDa dominant-negative isoform that prevents secretion of wild-type protein from somatotrophs (3,4). Previously, we reported the creation of mouse lines expressing a human GH-1 transgene containing a G to A transition at the 5′ splice site of intron 3 (IVS3 + 1) that leads to exclusive production of the 17.5-kDa isoform (5). This transgene is expressed from a cosmid containing the entire GH-1 locus control region including upstream DNA elements required for somatotroph-specific expression. The IVS3 + 1 mutant exerts a dominant-negative effect on wild-type mouse GH (mGH) and generates a transgene dose-dependent IGHD II phenocopy. Even in the presence of two wild-type mGH alleles, high-copy transgenic mice exhibit IGHD II with concomitant reduced weight, severely reduced pituitary GH content, and progressive anterior pituitary hypoplasia (5,6). Overproduction of the 17.5-kDa isoform triggers not only somatotroph death but also destruction of neighboring cells by macrophage invasion, leading to severe hypoplasia and additional anterior pituitary hormone deficiencies (5). The 17.5-kDa isoform arises because the normal exon 3 splice sites are relatively weak with accurate splicing requiring the presence of at least three splicing enhancer elements (2). Even in the normal pituitary, a small number of GH-1 transcripts (<1–3%) are incorrectly spliced and encode the 17.5-kDa isoform (7). Variability in the age of onset and severity of IGHD II among patients is thought to be due to the amount of the 17.5-kDa isoform relative to the full-length, 22-kDa protein (3,5,8). These phenotypic differences may reflect a threshold and dose dependency of the amount of the 17.5-kDa isoform produced from mutated alleles, sufficient to cause somatotroph death and trigger pituitary defects (5). Currently, recombinant human GH (hGH) is used in replacement therapy for IGHD II to overcome short stature but is relatively expensive, can lead to unwanted side effects (9), and importantly, does not prevent anterior pituitary hypoplasia and other ensuing anterior pituitary deficits (10). Because IGHD II arises as a direct consequence of excessive production of a specific dominant-negative isoform, strategies designed to decrease levels of the 17.5-kDa isoform without affecting the normal 22-kDa product could be an effective form of therapy. Here we have used RNA interference (RNAi) to specifically target the mutant GH-1 transcript encoding the 17.5-kDa isoform in vivo, and we show rescue of IGHD II in a mouse model.
Figure 1.
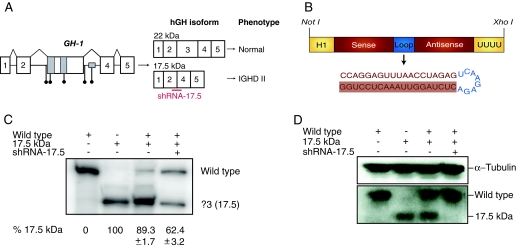
Allele-specific RNAi. A, The human GH-1 gene contains five exons as depicted. The region around exon 3 is enlarged to illustrate patient mutations (black circles) and splicing enhancer elements (gray boxes). Aberrant splicing of GH-1 produces a dominant-negative 17.5-kDa hGH isoform lacking exon 3 that prevents secretion of wild-type GH leading to IGHD II. An shRNA (shRNA-17.5) was designed complementary to the unique exon 2-exon 4 junction (red bar). B, Schematic of pSuper-sh17.5. A NotI-XhoI fragment of pSuper-sh17.5 containing an H1 promoter driving expression of shRNA-17.5 was used for pronuclear injections to generate transgenic mice. The guide strand is highlighted. C, AtT20 cells were transfected with hGH cDNA constructs and pSuper-sh17.5, as indicated. RNA was isolated and the different isoforms amplified by RT-PCR using primers in exon 2 and exon 5. Bands corresponding to the wild-type product (22-kDa isoform) and the exon 3 skipped product (Δ3; 17.5-kDa isoform) are as indicated. The percentage ± se of the Δ3 transcripts relative to wild-type is shown below based on at least three independent experiments. Quantitation is based on the ratio of products within a single lane and was calculated by phosphorimager densitometry. D, Western blots of cell lysates from the same transfections as in C were performed using antibodies against hGH or α-tubulin as a loading control.
Materials and Methods
Cell culture
Wild-type and Δ3 human GH cDNAs were cloned into pcDNA 3.1(+) (Invitrogen, Carlsbad, CA) as described (11) using identical primers. pSuper-sh17.5 was cloned as described (6). Mouse AtT20/D16v-F2 cells (European Collection of Cell Cultures, Salisbury, UK) were grown in DMEM with 10% fetal bovine serum and 1% penicillin/streptomycin with 4.5 g/liter glucose. Cells were transfected with hGH constructs and/or pSuper-sh17.5 using LT-1 (Mirus; Madison, WI) according to the manufacturer’s protocol.
RT-PCR
RNA was isolated 48 h after transfection using TRI-reagent (Molecular Research Center, Cincinnati, OH). For analysis of human GH-1 splicing patterns in AtT20 cells, RT-PCR was performed as described (6) with the exception that Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI) was used. For analysis of pituitary RNA, 500 ng total RNA was used for first-strand cDNA synthesis with the following primer that recognizes both mouse and human GH sequences 5′-CGGGGGCTGCCATCTTCCAGC-3′. The same primer was used for PCR amplification with a 32P-labeled forward primer, 5′-GCCTGCTCTGCCTGCYCTGGC-3′. PCR products were separated on 6% denaturing polyacrylamide gels and visualized on a phosphorimager. Amplifying both products with the same primers allows accurate quantitation within the same lane, obviating the need for a loading control. Results are thus shown as a ratio of the two products derived from a single lane.
To assay possible interferon responses, first-strand cDNA was synthesized using oligo d(T) with 2 μg total pituitary RNA. Subsequent RT-PCR amplification was done using OAS-1 forward 5′-CAGCCGTCAATGTCGTGTGTGATT-3′ and OAS-1 reverse 5′-TGTTAAGGAACACCACCAGGTCAG-3′.
Western blots
At-T20/D16v-F2 cells were lysed in 1× Laemmli loading buffer 48 h after transfection. Whole pituitaries were briefly sonicated in RIPA buffer [50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS] and mixed with an equal amount of 2× Laemmli loading buffer. All samples were denatured for 5 min at 95 C before loading onto 10% SDS gels. After transfer, polyvinylidene difluoride (PVDF) membranes were blocked with 5% milk and incubated with primary antibodies against hGH or α-tubulin (Abcam, Cambridge, MA). After incubation with horseradish peroxidase-conjugated secondary antibodies (GE Healthcare, Piscataway, NJ), proteins were visualized by enhanced chemiluminescence (PerkinElmer Life Sciences, Norwalk, CT).
Transgenic mice
A NotI-XhoI restriction fragment of pSuper-17.5 (Fig. 1B) was purified and used for pronuclear injection of C57BL/6 embryos. Injections and oviduct transfers were performed by the Vanderbilt Transgenic Core Facility using standard techniques in accordance with protocols approved by the Vanderbilt University Institutional Animal Care and Use Committee (VU IACUC). Transgenic mice were verified by PCR from tail DNA using primers 5′-GCTCTAGAACTAGTGGATCC-3′ (forward) and 5′-CTAGAGTCTCTTGAACTCTAGG-3′ (reverse). The sh-20 and sh-25 lines contain three and seven copies of the transgene, respectively (data not shown). Transgenic mice were bred with wild-type C57BL/6 mice. IGHD II mice were also bred with wild-type C57BL/6 mice, and transgenic animals were identified by PCR, as described (5). Progeny resulting from crosses of short hairpin RNA (shRNA)-17.5 mice and IGHD II mice were identified by PCR, using the same primers as above. Mice were weighed once a week from wk 4–16 after weaning. All mouse work was performed according to VU IACUC guidelines and in accordance with VU IACUC protocol number M/05/075.
Pituitary dissections
Pituitaries were dissected from 8-wk-old mice and fixed in 4% paraformaldehyde in PBS. Pituitaries were visualized with a Leica MZ16F scope and QImaging Retiga EXi camera.
Immunohistochemistry and imaging
The 20-μm cryostat sections from pituitaries of 10-wk-old mice were subjected to fluorescent immunohistochemical staining using goat antimouse GH antibodies and Cy-3-conjugated antigoat antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). Pituitary sections were mounted in 50% glycerol and imaged with a Zeiss LSM510 Meta Laser Scanning microscope. Stacks were acquired with LSM510 software, and Z-projections and contrast adjustments were made with NIH ImageJ. See supplemental information for additional information concerning image acquisition (published as supplemental data on The Endocrine Society’s Journals Online web site at http://endo.endojournals.org).
Electron microscopy
Pituitaries from 10-wk-old mice were fixed overnight in 2% glutaraldehyde. After washes in 1× PBS, pituitaries were transferred to 1% OsO4 for 1 h before washing with dH2O. Preparations were then stained en bloc in 1% aqueous uranyl acetate and dehydrated in an ethanol series, passed through propylene oxide, transferred to a 1:1 Spurr:propylene oxide mixture, and removed and embedded in Spurr’s embedding reagent. Ultrathin serial sections (50–60 nm) were obtained on a Leica UCT Ultracut microtome, transferred to formvar-coated grids, and examined on a CM10 transmission electron microscope (FEI, Hillsboro, OR), operating at 80 kV and images captured with an AMT 2 mega pixel camera (Advanced Microscopy Techniques, Danvers, MA).
IGF-I serum analysis
Serum was obtained by tail vein bleeding at wk 8 and 16 after weaning. Mouse IGF-I levels were determined using a mouse IGF-I ELISA (Immunodiagnostic Systems Inc., Fountain Hills, AZ).
Statistical analysis
Statistical significance between mice was determined for data points from wk 16 for all growth curves and for IGF-I serum data using a one-way ANOVA followed by a Tukey-Kramer honestly significant difference analysis (JMP, version 5.01). Normal distributions for all data were determined by a Shapiro-Wilk’s test. A summary of values used to determine statistical significance is listed in supplemental Tables 2–4 (published as supplemental data on The Endocrine Society’s Journals Online web site at http://endo.endojournals.org).
Results
Allele-specific targeting of mutant GH-1 transcripts by RNAi
Skipping of exon 3 produces GH-1 transcripts containing a unique sequence at the junction of exon 2 and exon 4. We previously showed that shRNAs complementary to this unique sequence specifically target transcripts encoding the 17.5-kDa isoform (Δ3 transcripts) but do not alter wild-type transcript levels (2). However, we did not examine the effects of the shRNAs on protein levels. Therefore, we transfected At-T20 cells, a murine neuroendocrine cell line that does not produce endogenous GH, with the vector encoding shRNA-17.5 (Fig. 1B) along with vectors expressing either wild-type hGH or the IVS3 + 1 mutant that expresses only the 17.5-kDa isoform. Cells were harvested after 48 h and analyzed for hGH protein and mRNA levels. As shown, the shRNAs effectively decreased the levels of Δ3 transcripts without affecting full-length transcripts (Fig. 1C). Interestingly, despite the fact that the experiment shown in Fig. 1 did not lead to complete loss of Δ3 transcripts, there was a total absence of the 17.5-kDa isoform (Fig. 1D). In other experiments, we do not always observe complete loss of the 17.5-kDa isoform. Despite slight experimental variability, we conclude that expression of shRNA-17.5 results in significant allele-specific silencing of Δ3 transcripts and abrogation of the 17.5-kDa isoform.
Generation of shRNA-17.5 transgenic mice
To test whether shRNA-17.5 targets the mutant GH-1 allele in vivo, we generated mice expressing the shRNA by pronuclear microinjection of a 347-bp DNA restriction fragment of pSUPER-sh17.5 (Fig. 1B). Ten independent lines of shRNA-positive mice were generated (supplemental Table 1). These mice show no differences in weight or lifespan compared with wild-type littermates and display no obvious phenotypes, and nine of the 10 lines are fertile (supplemental Fig. 1). Although overexpression of shRNAs can lead to toxicity and lethality due to oversaturation of the endogenous micro-RNA pathway or due to possible off-target effects (12,13), the presence of the shRNA-17.5 transgene did not induce any discernable effects in at least nine of the 10 lines. Also, we did not observe any induction of interferon responses, at least as measured by levels of 2′,5′-oligoadenylate synthetase (OAS1), a key interferon-responsive gene (14) (supplemental Fig. 2).
Rescue of IGHD II in vivo by RNAi
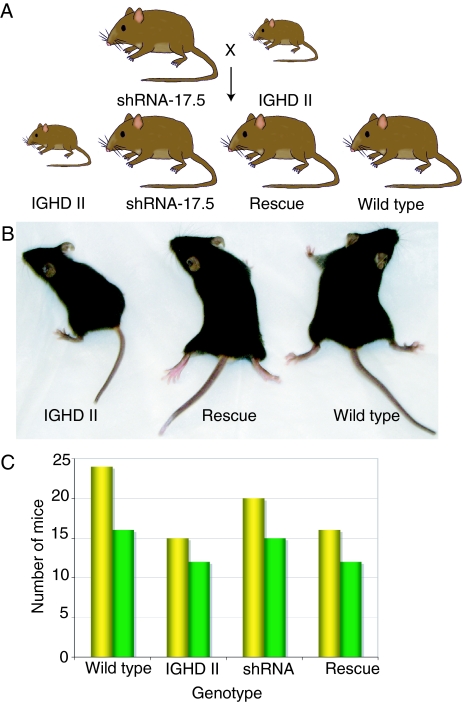
Mice expressing shRNA-17.5 were bred with the IGHD II mice to determine whether the IGHD II phenotype could be rescued in vivo (Fig. 2A). Mice expressing both hGH-17.5 kDa and shRNA-17.5 showed rescue of the growth deficit compared with IGHD II littermates (Figs. 2B and 3A). Of the 10 founder shRNA-17.5 transgenic lines, five have thus far been crossed, and all rescue growth (supplemental Table 1). Below, we more completely characterize two rescue lines. The shRNA line 20 (sh-20) completely rescued the growth deficit with no difference in weight compared with wild-type littermates. The shRNA line 25 (sh-25) significantly improved weight gain compared with IGHD II mice but did not fully attain normal levels (Fig. 3A). Both lines had lower weights at weaning, but with time these increased to be significantly greater than the IGHD II mice. For both lines, weight rescue was observed in both male and female progeny and was obvious at weaning (particularly in the sh-20 rescue line), as would be expected because this is when growth deficits become manifest in rodents. The genotypes assorted close to the predicted Mendelian ratios among the progeny for both lines (Fig. 2, A and C).
Figure 2.
Mating strategy. A, Expected offspring from matings between IGHD II and shRNA-17.5 mice; B, 5-wk-old IGHD II, rescue (20R), and wild-type littermates; C, genetic assortment of progeny from matings of sh-20 (yellow) or sh-25 (green) with IGHD II mice.
Figure 3.
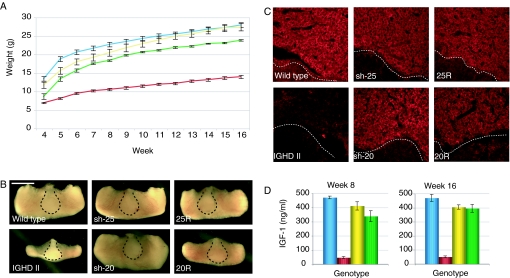
Rescue of IGHD II mice A, Growth curves of wild-type (blue, n = 10), IGHD II (red, n = 8), 20R (yellow, n = 6), and 25R (green, n = 4) mice; B, pituitary dissections from 8-wk-old mice. The posterior pituitary is indicated within the dashed line (scale bar, 1 mm); C, fluorescent immunohistochemistry staining with anti-mGH on 20-μm pituitary sections taken from 10-wk-old mice. The posterior pituitary is indicated below the dashed line; D, serum IGF-I levels were measured by ELISA from 8- and 16-wk-old mice (n = 3 for each genotype). Wild-type, blue; IGHD II, red; 20R, yellow; 25R, green. Error bars are ±sem. From ANOVA for A and D, P < 0.0001. For complete statistical analyses, see supplemental Tables 3 and 4.
We next analyzed pituitary morphology and function for the sh-20 and sh-25 rescue (20R and 25R) lines. Pituitary dissections revealed that the severe anterior pituitary hypoplasia observed in the IGHD II mice was not detected in either the 20R or 25R lines (Fig. 3B). Pituitaries shown in Fig. 3 are from 8-wk-old mice, but the increase in pituitary size was observable at weaning (3 wk; data not shown). Interestingly, considering the degree of weight rescue by the different lines, the anterior pituitaries from the 20R mice were smaller than the 25R pituitaries and remained so past 6 months in age. No significant differences in anterior pituitary size were observed between wild-type mice and the sh-20 and sh-25 transgenic lines. As expected, the posterior pituitary was similar in size for all mice. Because the IGHD II model mice exhibit pronounced somatotroph loss, we next sectioned pituitaries and performed fluorescent immunohistochemistry using an antibody that specifically recognizes mGH. As shown in Fig. 3C, the IGHD II mice had a severe decrease in mGH production, consistent with dominant-negative effects exerted by the IVS3 + 1 mutant. In contrast, the sh-20, sh-25, 20R, and 25R lines all expressed mGH at levels indistinguishable from wild-type. Quantitative measurement of serum GH in single samples is uninformative due to the pulsatile nature of GH secretion. However, long-term restoration of GH output should correct the low IGF-I levels observed in IGHD II. IGF-I is secreted in the liver in response to GH stimulation, and low serum IGF-I levels indicate GH deficiency (15). Accordingly, serum IGF-I levels were measured in groups of mice at 8 and 16 wk of age. As shown in Fig. 3D, both rescue lines showed IGF-I levels that were similar to wild-type levels and significantly higher than the IGHD II mice. At wk 8, the IGF-I serum levels in the 25R mice were slightly lower than the 20R and wild-type mice, but by wk 16, there were no significant differences in IGF-I levels between the three genotypes. These results are concordant with the weight trends where the 25R line rescues the IGHD II phenotype slightly less than the 20R line. Interestingly, although the 20R mice have slightly greater weights and initially higher serum IGF-I levels than the 25R mice, the 20R pituitaries are consistently smaller though than the 25R pituitaries.
Morphological rescue of IGHD II
GH is packaged into granules, forming dense-core secretory vesicles (DCSVs). The specific mechanism responsible for the dominant-negative nature of the 17.5-kDa isoform is not certain but may be due to formation of 17.5:22-kDa dimers and oligomers that disrupt granule packaging, thus preventing the secretion of either isoform from somatotrophs, ultimately triggering an unfolded protein response (5). These complexes apparently overwhelm the degradative capacity of the proteasome, leading to the accumulation of toxic aggregates in the cytosol, endoplasmic reticulum, and Golgi apparatus. Consistent with this model, electron micrographs of anterior pituitary sections from IGHD II mice show Golgi and endoplasmic reticulum defects, loss of DCSVs, and dramatically increased levels of intracellular lipid and vacuolation (5) (Fig. 4, A and B). Wild-type cells show a large accumulation of mature DCSVs that are stored and released upon appropriate hypothalamic stimulation. As is evident from the electron micrographs, secretory cells from the rescue mice have abundant DCSVs, comparable to wild-type, with normal morphology (Fig. 4, A and B). Thus, our data from gross morphology to electron micrographs indicate that genetic delivery of shRNAs against transcripts encoding the dominant-negative 17.5-kDa isoform is able to rescue growth and somatotroph function in IGHD II mice.
Figure 4.
Morphological rescue of IGHD II. Electron micrographs are of the anterior pituitaries from 10-wk-old mice of the indicated genotypes. Scale bar, 2 μm; magnification, ×10,500 (A) and ×3,400 (B).
Silencing of the 17.5-kDa isoform
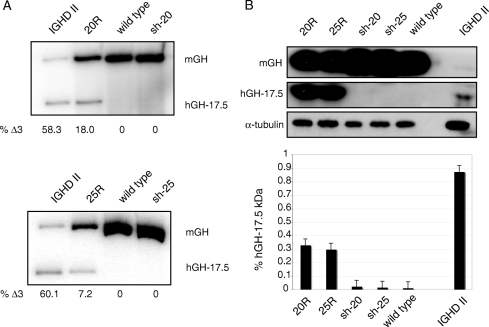
Interestingly, despite the fact that we observed rescue of IGHD II with several transgenic shRNA-17.5 lines (supplemental Table 1), we have not been able to directly detect expression of the shRNAs. We assume this is due to expression at levels below our current limit of detection and/or instability of the shRNAs. Indeed, such low levels may preclude lethality and/or toxicity as reported by Grimm et al. (12). Nevertheless, we were able to show that the shRNAs are directly altering the levels of the 17.5-kDa isoform by examining pituitary GH RNA and protein levels in the rescue mice. First, using primers that are complementary to both human and mGH transcripts, we performed RT-PCR analysis to determine the relative levels of wild-type, full-length mGH mRNA compared with the levels of the Δ3 transcripts encoding the hGH 17.5-kDa isoform. As shown in Fig. 5A, there was a dramatic change in the ratio of the two transcripts between the IGHD II mice and the rescue mice, consistent with the notion that rescue involves reducing levels of the Δ3 transcripts. Bearing in mind that IGHD II mice have severely hypoplastic anterior pituitaries, Western blots using antibodies that recognize both mGH and hGH showed that what little GH was produced was mainly the 17.5-kDa isoform (Fig. 5B). For the rescue mice, even though significant levels of the 17.5-kDa isoform were detected, the dramatic increase in full-length, wild-type mGH was apparently able to overcome any dominant-negative effects exerted by the mutant, thereby rescuing growth and pituitary function (Fig. 5B and supplemental Fig. 3). Thus, our biochemical and genetic results are completely consistent with functional reduction of the 17.5-kDa hGH isoform to rescue the dominant-negative phenotype.
Figure 5.
Silencing of the 17.5-kDa isoform. A, Total RNA was isolated from pituitaries of the indicated genotypes, and RT-PCR was performed with identical primers that distinguish between mGH and hGH. Bands corresponding to the full-length spliced product from the mouse alleles (mGH) and the 17.5-kDa human transgene (hGH-17.5) are as indicated. Quantitation of the two transcripts is based on the ratio of products within a single lane and was calculated by phosphorimager densitometry. B, Western blots of pituitary lysates from the same genotypes as in A were performed with an antibody that recognizes both hGH and mGH. The two upper panels were deliberately overexposed to allow visualization of the GH isoforms in IGHD II mice. A shorter exposure of the same gel is shown in supplemental Fig. 4. α-Tubulin was used as a loading control. The relative amounts of the 17.5-kDa isoform present in the different genotypes are shown in the graph and represent three independent experiments. Error bars are sem.
Discussion
RNAi has many potential advantages over traditional therapies, including increased specificity and versatility (16,17). Many recent advances in RNAi therapeutics have been used to successfully knock down viral genes or disease alleles, although to our knowledge, none have been used to knock out a dominant-negative disease allele in an animal model. These approaches have used either small interfering RNAs (siRNAs) or shRNAs to target relatively accessible tissues such as subretinal injection into the eye (18), intranasal delivery to the lung (19,20), transfection via Lipofectamine complexes to the vaginal epithelium (21), tail vein injection to the liver (22,23), viral delivery to the liver (24), and viral delivery via intraspinal or intracranial injections (25,26,27). Zimmermann et al. (28) used systemic delivery of siRNAs in primates targeting apolipoprotein B in the liver by saphenous vein injection. By comparison, although it is not protected by the blood-brain barrier, the pituitary is relatively inaccessible to direct targeting due to its location. We therefore decided to deliver shRNA-17.5 via genetic means by making transgenic mice expressing the shRNAs. Using this approach, we were able to specifically reduce expression of the dominant-negative 17.5-kDa hGH isoform, allowing recovery of wild-type GH levels to rescue an autosomal dominant mouse model of human IGHD II.
A potential caveat to our results concerns the inability to directly detect expression of shRNA-17.5 precursors or mature siRNAs. This could be used to argue that the effects we observe are indirect. However, several lines of evidence argue against such a conclusion. First, we have shown genetically that when IGHD II mice are crossed with shRNA mice, only the progeny that contain both the shRNA transgene and the Δ3 transgene exhibit the rescue phenotype. From mating and genotyping over 400 mice, we observed rescue only in the double transgenics. Also, the extent of the rescue varied between the different shRNA lines; the 25R lines exhibit slightly lower growth rates and initial IGF-I serum levels compared with the 20R lines (Figs. 3, A and D, and supplemental Table 1). Second, the transgenic shRNA lines show no overt phenotype that could be responsible for or contribute to the rescue phenotype observed in the IGHD II background. Third, the genotypes assort in the expected Mendelian ratios (Fig. 2C). Thus, the genetics argue strongly against any indirect effect. Biochemically, we also showed that in the IGHD II mice, the predominant isoform is the dominant-negative hGH 17.5-kDa protein, whereas in the rescue mice, there is a switch, and the major protein detected is the wild-type mGH (Fig. 5B). Together, these data support our conclusion that the rescue of IGHD II is a direct effect of shRNA-17.5.
Future work will be directed toward a viable therapeutic strategy using exogenous delivery of siRNAs, but the results from this report show that employing RNAi provides a promising approach to treat IGHD II in humans. Although GH treatment can counteract GH deficits in children and adults with GH deficiency, the underlying somatotroph destruction continues with additional bystander effects that can evolve to damage other pituitary hormone axes in some individuals (29). A particularly encouraging feature of our results is the restoration of macroscopically and microscopically normal somatotroph populations in the rescue mice without any overt phenotype in other cells. Because normalization of somatotroph function offers a selective advantage in survival and replacement during somatotroph turnover, this promotes complete functional recovery without necessarily achieving complete suppression of the aberrant allele in every cell. More generally, many human diseases result from aberrant splicing and/or dominant-negative isoforms and our results show that RNAi offers a promising way to specifically degrade mutant alleles while sparing wild-type, functional alleles.
Supplementary Material
Acknowledgments
We thank Priscilla Dannies for wild-type and Δ3 hGH cDNA constructs. We also thank Alex Flynt for helpful discussions, Melissa Prince and Cindy Holladay for assistance with mouse maintenance, Elvin A Woodruff III for electron microscopy, Bryan Harris for help with mouse screening, and Scott P. Egan for help with statistical analyses. Fluorescent confocal microscopy was possible through use of Vanderbilt Cell-Imaging Shared Resource equipment, and transgenic mice were created in the Vanderbilt Transgenic core.
Footnotes
This work was supported by National Institutes of Health Grant DK 035592 and by funds from the Vanderbilt Mouse Metabolic Phenotyping Center (DK 59637).
Disclosure Statement: The authors have nothing to declare.
First Published Online November 15, 2007
Abbreviations: DCSV, Dense-core secretory vesicle; hGH, human GH; IGHD II, isolated GH deficiency type II; mGH, mouse GH; 20R and 25R, sh-20 and sh-25 rescue; RNAi, RNA interference; shRNA, short hairpin RNA; siRNA, small interfering RNA; VU IAUIC, Vanderbilt University Institutional Animal Use and Care Committee.
References
- Mullis PE 2007 Genetics of growth hormone deficiency. Endocrinol Metab Clin North Am 36:17–36 [DOI] [PubMed] [Google Scholar]
- Ryther RCC, Flynt AS, Harris BD, Phillips III JA, Patton JG 2004 GH1 splicing is regulated by multiple enhancers whose mutation produces a dominant-negative GH isoform that can be degraded by allele-specific small interfering RNA (siRNA). Endocrinology 145:2988–2996 [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Yamamoto M, Ohmori S, Kamijo T, Ogawa M, Seo H 1999 Inhibition of growth hormone (GH) secretion by a mutant GH-I gene product in neuroendocrine cells containing secretory granules: an implication for isolated GH deficiency inherited in an autosomal dominant manner. J Clin Endocrinol Metab 84:2134–2139 [DOI] [PubMed] [Google Scholar]
- Lee M, Wajnrajch M, Kim S, Plotnick L, Wang J, Gertner J, Leibel R, Dannies P 2000 Autosomal dominant growth hormone (GH) deficiency type II: the Del32-71-GH deletion mutant suppresses secretion of wild-type GH. Endocrinology 141:883–890 [DOI] [PubMed] [Google Scholar]
- McGuinness L, Magoulas C, Mathers K, Carmignac D, Manneville JB, Christian H, Phillips III JA, Robinson ICAF 2003 Autosomal dominant growth hormone deficiency disrupts secretory vesicles: in vitro and in vivo studies in transgenic mice. Endocrinology 144:720–731 [DOI] [PubMed] [Google Scholar]
- Ryther RCC, McGuinness L, Phillips III JA, Moseley CT, Magoulas CB, Robinson ICAF, Patton JG 2003 Disruption of exon definition produces a dominant-negative growth hormone isoform that causes somatotroph death and IGHD II. Hum Genet 113:140–148 [DOI] [PubMed] [Google Scholar]
- Procter A, Phillips III J, Cooper D 1998 The molecular genetics of growth hormone deficiency. Hum Genet 103:255–272 [DOI] [PubMed] [Google Scholar]
- Millar D, Lewis M, Horan M, Newsway V, Easter T, Gregory J, Fryklund L, Norin M, Crowne E, Davies S, Edwards P, Kirk J, Waldron K, Smith P, Phillips III J, Scanlon M, Krawczak M, Cooper D, Procter A 2003 Novel mutations of the growth hormone 1 (GH1) gene disclosed by modulation of the clinical selection criteria for individuals with short stature. Hum Mutat 21:424–440 [DOI] [PubMed] [Google Scholar]
- Monson J 2003 Long-term experience with GH replacement therapy: efficacy and safety. Eur J Endocrinol 148:S9–S14 [DOI] [PubMed] [Google Scholar]
- Mullis PE, Robinson ICAF, Salemi S, Eble A, Besson A, Vuissoz J-M, Deladoey J, Simon D, Czernichow P, Binder G 2005 Isolated autosomal dominant growth hormone deficiency: an evolving pituitary deficit? A multicenter follow-up study. J Clin Endocrinol Metab 90:2089–2096 [DOI] [PubMed] [Google Scholar]
- Deladoey J, Stocker P, Mullis P 2001 Autosomal dominant GH deficiency due to an Arg183His GH-1 gene mutation: clinical and molecular evidence of impaired regulated GH secretion. J Clin Endocrinol Metab 86:3941–3947 [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA 2006 Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441:537–541 [DOI] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS 2006 Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12:1179–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams B 2003 Activation of the interferon system by short-interfering RNAs. Nat Cell Biol 5:834–839 [DOI] [PubMed] [Google Scholar]
- Clemmons DR 2007 IGF-I assays: current assay methodologies and their limitations. Pituitary 10:121–128 [DOI] [PubMed] [Google Scholar]
- Dykxhoorn DM, Palliser D, Lieberman J 2006 The silent treatment: siRNAs as small molecule drugs. Gene Ther 13:541–552 [DOI] [PubMed] [Google Scholar]
- Kim DH, Rossi JJ 2007 Strategies for silencing human disease using RNA interference. Nat Rev Genet 8:173–184 [DOI] [PubMed] [Google Scholar]
- Reich SJ FJ, Kuroki A, Tang W, Yang X, Maguire AM, Bennett J, Tolentino MJ 2003 Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol Vis 9:210–216 [PubMed] [Google Scholar]
- Bitko V, Musiyenko A, Shulyayeva O, Barik S 2005 Inhibition of respiratory viruses by nasally administered siRNA. Nat Med 11:50–55 [DOI] [PubMed] [Google Scholar]
- Zhang W, Yang H, Kong X, Mohapatra S, San Juan-Vergara HS, Hellermann G, Behera S, Singam R, Lockey RF, Mohapatra SS 2005 Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nat Med 11:56–62 [DOI] [PubMed] [Google Scholar]
- Palliser D, Chowdhury D, Wang Q-Y, Lee SJ, Bronson RT, Knipe DM, Lieberman J 2006 An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature 439:89–94 [DOI] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Rohl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher HP 2004 Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432:173–178 [DOI] [PubMed] [Google Scholar]
- Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, Vaish N, Zinnen S, Vargeese C, Bowman K, Shaffer CS, Jeffs LB, Judge A, MacLachlan I, Polisky B 2005 Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol 23:1002–1007 [DOI] [PubMed] [Google Scholar]
- Grimm D, Kay MA 2006 Therapeutic short hairpin RNA expression in the liver: viral targets and vectors. Gene Ther 13:563–575 [DOI] [PubMed] [Google Scholar]
- Ralph GS, Radcliffe PA, Day DM, Carthy JM, Leroux MA, Lee DC, Wong LF, Bilsland LG, Greensmith L, Kingsman SM, Mitrophanous KA, Mazarakis ND, Azzouz M 2005 Silencing mutant SOD1 using RNAi protects against neurodegeneration and extends survival in an ALS model. Nat Med 11:429–433 [DOI] [PubMed] [Google Scholar]
- Raoul C, Abbas-Terki T, Bensadoun JC, Guillot S, Haase G, Szulc J, Henderson CE, Aebischer P 2005 Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat Med 11:423–428 [DOI] [PubMed] [Google Scholar]
- Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, Orr HT, Paulson HL, Yang L, Kotin RM, Davidson BL 2004 RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med 10:816–820 [DOI] [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Rohl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher HP, MacLachlan I 2006 RNAi-mediated gene silencing in non-human primates. Nature 441:111–114 [DOI] [PubMed] [Google Scholar]
- Salemi S, Yousefi S, Baltensperger K, Robinson ICAF, Eble A, Simon D, Czernichow P, Binder G, Sonnet E, Mullis PE 2005 Variability of isolated autosomal dominant GH deficiency (IGHD II): impact of the P89L GH mutation on clinical follow-up and GH secretion. Eur J Endocrinol 153:791–802 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.