Abstract
The two large cytoplasmic domains (C1 and C2) of adenylyl cyclases (AC), when expressed separately and mixed together, reconstitute enzyme activity that can be regulated by various modulators. Therefore, we have used the C1 or its C1a subdomain and C2 regions from type I AC (ACI) and type V AC (ACV) to identify the region on ACI that interacts with βγ subunits of heterotrimeric G proteins. In addition, we also used a chimeric C1 domain (VC1aIC1b) in which the C1a region was derived from ACV and the C1b region was from ACI. By mixing the C1 or C1a or VC1aIC1b domains with C2 regions of ACI or ACV, we have shown that the C1a region (amino acids 236–471) of ACI is sufficient to observe βγ-mediated inhibition of enzyme activity, which is stimulated by either constitutively active Gsα (Gsα*) or Ca2+/calmodulin (CaM). Although the C1b region and C2 domain of ACI were by themselves not sufficient for inhibition of activity by βγ subunits, the presence of both of these regions formed another βγ interaction site that was sufficient to observe Gsα*- or Ca2+/CaM-stimulated activity. Inhibition of AC activity attributable to interaction of βγ subunits at either of the two sites was blocked by a peptide (QEHA) that has previously been shown to inhibit the effects of βγ on various effectors. Moreover, the C1 region of ACI was sufficient to observe Giα1-elicited inhibition of Ca2+/CaM-stimulated activity. Although the C1a region of ACV was sufficient for inhibition of activity by Giα1, the presence of C1b region from either ACI or ACV increased sensitivity to inhibition by the inhibitory G protein. Thus, the inhibitory influences of Giα1 are mediated on the C1 regions of both ACI and ACV. The effects of βγ on ACI can be mediated by interactions with the C1a region and a βγ interacting site formed by the C1b and C2 domains of this enzyme.
Mammalian adenylyl cyclases (ACs) that catalyze the conversion of ATP to cAMP can be divided into membrane-bound and cytosolic forms (1–3). The recently characterized mammalian cytosolic AC is not regulated by heterotrimeric G proteins or forskolin (3). On the other hand, the membrane bound mammalian ACs are regulated not only by G protein α-subunits and forskolin but also by βγ subunits of heterotrimeric G proteins and other modulators such as Ca2+ and Ca2+/calmodulin (see refs. 1 and 2 for reviews). To date, nine distinct forms (types I–IX; ACI–ACIX) of membrane-bound adenylyl cyclases have been cloned and characterized (see refs. 1 and 2 for reviews). In addition, two splice variants of type VIII isoform have been reported (4).
Although the nine membrane-bound forms of AC share considerable sequence homology, the various modulators regulate their activity differently. Thus, type I, III, and VIII isoforms are stimulated by Ca2+/calmodulin (4–8b). The closely related type V and VI forms of AC are inhibited by low concentrations of calcium without the involvement of calmodulin (9–12). Similarly, although Giα (the inhibitory GTP binding protein of AC) inhibits type V and VI ACs, it does not alter the activity of type II enzyme (see ref. 1 and 2 for reviews). On the other hand, the βγ subunits of heterotrimeric G proteins conditionally stimulate type II and IV ACs, provided that active Gsα (the stimulatory GTP binding protein of AC) is present (13–15). However, βγ subunits inhibit the activity of type I adenylyl cyclase (5, 13, 15).
Because the full-length ACs cannot be expressed in large amounts and because large scale purification of these enzymes is problematic, several groups have expressed the two large cytosolic domains (C1 and C2) of AC in bacteria and have used these to study regulation of the enzyme (16–20). Mixtures of C1 and C2 domains of AC have been shown to reconstitute enzyme activity that can be regulated by G protein α subunits, forskolin, and Ca2+ in a manner similar to the full-length enzyme (16–20). Therefore, using the C1 and C2 domains from type I and V ACs (ACI and ACV)‡, as well as a chimeric C1 region derived from these enzyme, we have identified the regions on ACI and ACV that are involved in the inhibition of enzyme activity by βγ and Giα subunits, respectively. Our data demonstrate that the C1a region (amino acids 236–471) of ACI is sufficient to observe βγ-mediated inhibition of enzyme activity. Additionally, we demonstrate that, although neither the C1b (amino acids 472–607) nor the C2 (amino acids 809-1133) regions of ACI by themselves are sufficient to observe inhibition of enzyme activity by βγ subunits, when present together, these regions are also sufficient to form a βγ interacting site. Concerning inhibition of AC activity by the type 1 isoform of Giα (Giα1), our data show that the C1a region of ACV (amino acids 322–571) and C1 region of ACI (amino acids 236–607) are sufficient to observe inhibition of enzyme activity by the G protein. Moreover, when C1b regions of ACI or ACV are connected to the C1a region (amino acids 322–571) of ACV, sensitivity to inhibition by Giα1 is augmented.
MATERIALS AND METHODS
Plasmid Construction.
The vector pTrcHisB (Invitrogen) was used for expression of the soluble forms of canine ACV and bovine ACI subunits in Escherichia coli. Plasmid construction of the recombinant forms of ACV subunits was performed as described (17, 18). cDNAs encoding the C1a (IC1a; amino acids 236–471), C1 (IC1; amino acids 236–607), and C2 (IC2; amino acids 809-1133) regions of ACI were obtained by PCR using ACI cDNA as template and the following primers: IC1a, primer A: 5′ ATAATATGGATCCGGCTGAGCGCGCCCAG 3′, primer B: 5′ ATATATAGCGCTATGAGTTTTCAGAAAACTGTTCCTCTC 3′; IC1, primer C: 5′ ATATATGGATCCGGCTGAGCGCGCCCAG 3′, primer D: 5′ ATATATAAGCTTCTAGTCCTGAAGCTGGTGGTACTTTCGCTCTCG 3′; IC2, primer E: 5′ ATATATAGATCTGTCAAGCTGCGGCTG 3′, primer F: 5′ ATATATAAGCTTCTAAGCCTCCTTCCCAGAGGC 3′. The PCR products were cloned into the BamHI/HindIII sites (IC1 and IC1a regions) and the BglII/HindIII sites (IC2 domain) of the plasmid pTrcHisB.
To generate the chimeric C1 domain consisting of C1a region (amino acids 322–571) from ACV and C1b region (amino acids 472–607) of ACI (VC1aIC1b), unique Eco47III and BglII restriction sites were introduced at the 3′ end of VC1a (silent T > C mutation at nucleotide 1,674) using PCR methodology; the 5′ primer contained a BamHI site. This PCR fragment was then cloned into BamHI/BglII-treated plasmid pTrcHisB, which contained the cDNA encoding C1 and C2 regions of ACV joined by an artificial linker (17). This construct was digested with Eco47III, and the PCR-generated IC1b cDNA (encoding amino acids 472–607 of ACI), engineered to contain EcoRV sites at the 5′ and 3′ ends was then inserted into this new restriction site. By using PCR methodology, this latter construct was then used as a template to generate the chimeric C1 region in which the C1a region was derived from ACV and the C1b domain was from ACI; the 5′ and 3′ primers were designed to include unique BamHI and HindIII sites, respectively. The cDNA encoding the chimeric C1 region was cloned into the BamHI/HindIII sites in plasmid pTrcHisB.
To clone the rat Giα1 subunit in the expression vector pQE60, the complete Giα1 cDNA was first generated by PCR using cDNA of Giα1 in pGEM-2 (from R. Reed, Johns Hopkins Univ. School of Medicine, Baltimore) as template and the primers (5′ TATATACCATGGGCTGCACACTG 3′) and (5′ TATATAGAATTCTTAGAAGAGGAGACCACAG 3′). This PCR-generated cDNA was subjected to NcoI digestion to obtain a 261-bp fragment (encoding amino acids 1–87), which was then cloned into the NcoI site of pQE60. The resulting construct was sequenced to ensure the correct orientation of the NcoI/NcoI fragment, and following treatment with Eco47III/HindIII was ligated with the Eco47III/HindIII fragment of Giα1 cDNA (amino acids 7–354) to generate the complete Giα1 (amino acids 1–354) in plasmid pQE60. All of the constructs were verified by sequencing.
Expression of Recombinant Proteins in E. coli.
The AC subunits IC1, IC2, VC1, VC2, and VC1aIC1b were expressed in TP2000 strain of E. coli, which does not contain endogenous AC. Expression of protein and cell lysis were performed as described (17) except that the induction with isopropyl β-d-thiogalactoside was performed at 23°C for 15 h. All of the G protein α subunits were expressed in E. coli (JM109 DE3) as described by Lee et al. (21). Giα1 was coexpressed with yeast N-myristoyltransferase to ensure synthesis of myristoylated G-protein as described by Linder et al. (22). Purification of the myristoylated, recombinant Giα1 protein was achieved by the method of Mumby and Linder (23). Bovine brain βγ subunits of heterotrimeric G proteins were purified to homogeneity as described by Mumby et al. (24) with the modifications of Neer et al. (25).
Adenylyl-Cyclase Assays.
AC activity assays were performed for 15 min at 30°C in a volume of 150 μl in the presence of 5 mM MgCl2 as described (17). The constitutively active Q213L mutant of Gsα (Gsα*; ref. 26) was used to stimulate enzyme activity. To ensure maximal activation of Gsα*, the G protein was activated by incubation with 100 nM guanosine 5′-[γ-thio]triphosphate at room temperature for 30 min. Likewise, Giα1 was activated by a similar treatment for 1 h. Guanosine 5′-(O-(2-thiodiphosphate) (GDPβS)-bound form of Giα1 was obtained by incubation with 10 μM GDPβS for 1 h at room temperature. Purified bovine brain calmodulin was mixed with Ca2+ and was added to AC assays to yield final concentrations of 500 nM and 30 μM, respectively. To monitor inhibition of AC activity by Giα1 or βγ-subunits, the G-protein subunits and AC were preincubated for 2 min at room temperature before the start of the assay. When the effect of G-protein βγ subunits were studied, the final concentration of the detergent, lubrol, in the assay was maintained at 0.001% (vol/vol) and was added to all controls. Wherever peptides QEHA and SKEE were used, they were preincubated with βγ subunits for 1 h on ice, and the mixture then was added into AC assays.
RESULTS AND DISCUSSION
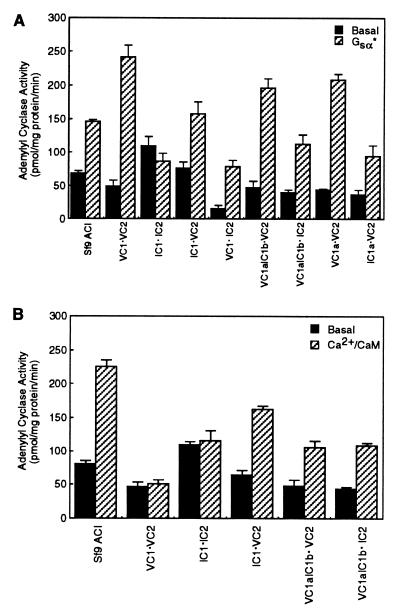
Among the AC isoforms, ACI is unique in that its enzymatic activity is inhibited by βγ subunits of heterotrimeric G proteins (5, 13, 15). Therefore, the initial aim of our study was to identify the regions on this enzyme that are necessary and/or sufficient for inhibition of enzyme activity by βγ subunits. Because the major cytoplasmic regions (C1 and C2) of ACs are sufficient to reconstitute enzyme activity that can be regulated by modulators such as Gsα, forskolin, and Giα (16–20), our approach entailed the use of C1 and C2 regions of ACI and ACV. Therefore, as a starting point, we determined whether mixing the recombinant C1 or its shorter C1a region from ACV or ACI with the C2 domains of these enzymes reconstituted activity that could be stimulated by Gsα. The ability of the chimeric C1 protein (VC1aIC1b) to reconstitute enzyme activity with C2 domains of ACI or ACV also was tested. The abbreviations used to designate the domains of AC that were derived from either ACI or ACV to reconstitute enzyme activity are presented in Fig. 1. As demonstrated in Fig. 2A, with the exception of IC1⋅IC2, the constitutively active mutant (Q213L) of Gsα, Gsα* stimulated activity of all combinations of C1 or C1a or the chimeric C1 (VC1aIC1b) in the presence of C2 region of either ACI or ACV. The inability of Gsα* to stimulate activity of IC1⋅IC2 cannot be attributed to the lack of interaction between the domains of ACI to form an active enzyme because basal activity of this mixture was measurable and higher than other combinations of C1 and C2 domains (Fig. 2A). More likely, the complex formed by IC1 and IC2 does not allow access to the Gsα* binding site. It should be noted that the optimal concentrations of Gsα* required to observe maximal stimulation of enzyme activity with the different combinations shown in Fig. 2A were empirically determined and were found to be different. Thus, VC1⋅IC2 required 120 nM Gsα* for maximal stimulation; all other combinations required 80 nM Gsα*.
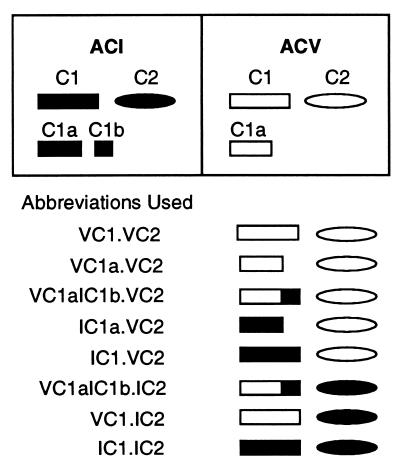
Figure 1.
Schematic representation of the abbreviations used to describe the various forms of AC derived by mixing the cytosolic C1 or C1a or chimeric C1 regions with C2 domains of either ACV or ACI. The various domains of ACI (black) and ACV (white) are represented in the boxes. The amino acid residues encompassing the domains in bovine ACI are as follows: C1, 236–607; C1a, 236–471; C1b, 472–607; C2, 809–1,133. In canine ACV, the domains shown comprise the following amino acid residues: C1, 322–683; C1a, 322–571; C2, 933–1,184. In the list of abbreviations, the roman numeral preceding the C1 or C2 domains and their subregions denote the AC isoform from which that particular region is derived.
Figure 2.
Stimulation of adenylyl cyclase activity reconstituted by mixing the cytosolic C1 and C2 regions of ACI and/or ACV. (A) Adenylyl cyclase activity was measured under basal conditions or in the presence of Gsα*. Either membranes of Sf9 cells expressing full-length ACI or bacterial lysates expressing the C1 or C1a or chimeric C1 region (VC1aIC1b) were mixed with lysates expressing C2 domains of ACI or ACV to reconstitute enzyme activity. For all forms of AC shown, the amount of Gsα* used was that required to maximally stimulate activity (empirically determined). Except for mixtures of VC1⋅IC2 and full-length ACI in which the maximally effective concentration of Gsα* was 120 nM, all other AC forms shown required 80 nM G protein. Shown is the mean ± SEM (n = 3 experiments). (B) Membranes of Sf9 cells expressing the full-length ACI and various mixtures of C1 or chimeric C1 and C2 domains of ACI or ACV were assayed for adenylyl cyclase activity in the presence and absence of CaM (500 nM). Ca2+ was added to these assays at a final concentration of 30 μM and was present under basal conditions in the absence of CaM. The mean ± SEM (n = 3 experiments) is shown.
Because the C1b region of ACI has previously been demonstrated to be the site of calmodulin (CaM) binding and necessary for activation of the enzyme by Ca2+/CaM (27), we determined whether Ca2+/CaM could stimulate adenylyl cyclase activity that had been reconstituted by mixing C1 domain of ACI or C1 domain containing IC1b with C2 regions of ACI or ACV. These experiments demonstrated that, like the full-length ACI, the activity of the soluble forms of AC could be stimulated when either the C1 region or the C1b domain of ACI were present (Fig. 2B). Again, the exception was the soluble AC comprising both C1 and C2 regions of ACI (IC1⋅IC2; Fig. 2B). Because the activity of IC1⋅IC2 could not be elevated by either Gsα* or Ca2+/CaM, this form of the soluble AC was not further studied; similar results, i.e., lack of modulation, also have been observed with a linked form of IC1a-IC2a AC (16). As expected, when the C1 region of ACV was used, Ca2+/CaM did not stimulate enzyme activity. These data demonstrate that, as determined in the context of the full-length ACI and ACII chimeras (27), the activity of soluble forms of AC also can be modulated by Ca2+/CaM provided that the C1b region of ACI is present.
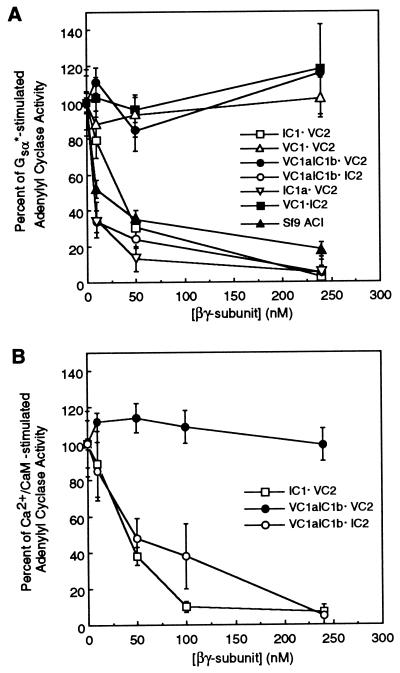
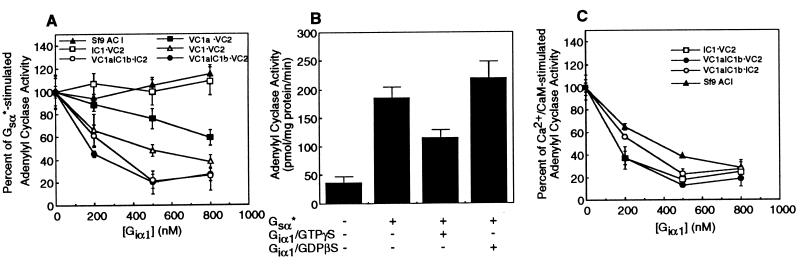
Next, we investigated whether the different forms of soluble ACs could be regulated by βγ subunits. As demonstrated previously by others (5, 13, 15), βγ subunits, in a concentration-dependent manner, inhibited the activity of full-length ACI (Fig. 3A). Likewise, consistent with previous findings that ACV is not inhibited by βγ subunits (reviewed in refs. 1 and 2), the soluble form of ACV (VC1⋅VC2) consisting of both C1 and C2 domains from ACV was not inhibited by βγ subunits (Fig. 3A). To determine which portion(s) of ACI is necessary to observe the inhibition of activity mediated by βγ subunits, experiments were performed with the C1 or C1a and C2 regions of ACI mixed with the complementary domains of ACV. When AC activity was reconstituted with C1 domain or its N-terminal C1a region from ACI and C2 region of ACV (IC1⋅VC2 and IC1a⋅VC2, respectively), the Gsα*-stimulated activity was inhibited by βγ subunits in a manner similar to that observed with the full-length ACI (Fig. 3A). On the other hand, when the C2 region of ACI was reconstituted with C1 domain of ACV (VC1⋅IC2), Gsα*-stimulated activity was not altered by βγ subunits (Fig. 3A). These data demonstrate that the C1a region of ACI is sufficient to observe inhibition of enzyme activity by βγ subunits of G proteins. To determine whether the C1b region of ACI also contributed to inhibition of activity by βγ subunits of G proteins, the chimeric C1 region comprising C1a of ACV and C1b of ACI (VC1aIC1b) was reconstituted with C2 region of ACV (VC1aIC1b⋅VC2). The Gsα*-stimulated AC activity of this enzyme was not altered by βγ subunits, indicating that the IC1b region alone is not sufficient to observe βγ-mediated inhibition. Interestingly, however, when the C2 region of type I AC is used to reconstitute AC activity with VC1aIC1b, βγ subunits inhibited the Gsα*-stimulated enzyme as effectively as that observed with the full-length ACI and IC1a⋅VC2 (Fig. 3A). These findings, coupled with the observations that neither C1b nor C2 regions of ACI by themselves are sufficient to observe βγ-mediated inhibition of enzyme activity, suggest that the C1b and C2 domains of ACI interact with each other to form a βγ interacting site. This contention also is supported by the data in Fig. 3B. Hence, when the activity of VC1aIC1b⋅IC2 was elevated by Ca2+/CaM, βγ subunits of heterotrimeric G proteins inhibited activity. However, Ca2+/CaM-stimulated activity of the VC1aIC1b⋅VC2 enzyme was not altered by βγ subunits (Fig. 3B). As expected, Ca2+/CaM stimulated the activity of IC1⋅VC2 AC, and this activity also was inhibited by βγ subunits in a concentration-dependent manner (Fig. 3B). Taken together, the data in Fig. 3 demonstrate that the C1a region of ACI is sufficient to observe βγ-mediated inhibition of enzyme activity and that the C1b and C2 domains of ACI cooperate to form a βγ interacting site that also permits these latter G protein subunits to inhibit enzyme activity. The requirement for both C1b and C2 regions of ACI to observe βγ effects is reminiscent of our previous findings that the C1b region of ACV interacts with 10 amino acid regions on its C2 domain and that this intramolecular interaction modulates the ability of Gsα to stimulate enzyme activity (18).
Figure 3.
Inhibition of activity of different AC forms by βγ subunits of heterotrimeric G proteins. (A) AC activity in either membranes of Sf9 cells expressing the full-length ACI or in mixtures of subdomains from ACI and ACV was stimulated with Gsα*. The Gsα* concentrations used were identical to those in Fig. 2A. The effect of various concentrations of βγ subunits to modulate AC activity was monitored. Data are presented as percent inhibition of Gsα*-stimulated activity and are the mean ± SEM (n = 3 experiments). (B) Same as A except that the AC activity was stimulated by addition of CaM (500 nM). Ca2+ was present at a final concentration of 30 μM. Percent inhibition of Ca2+/CaM-stimulated activity is shown as the mean ± SEM (n = 3 experiments).
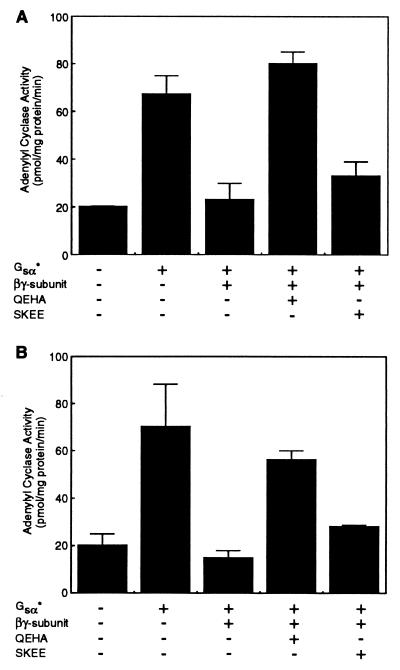
Previously, studies from Iyengar’s laboratory have shown that the conditional stimulation of type II AC (ACII) by βγ subunits can be inhibited by a peptide (QEHA) corresponding to amino acids 956–982 in the C2 domain of ACII (28). This peptide also inhibited the ability of βγ subunits to activate β-adrenergic receptor kinase, muscarinic K+ channels, and phospholipase Cβ (28). In addition, the peptide QEHA attenuated the ability of βγ subunits to inhibit calmodulin-stimulated AC in rat brain membranes (28); whether the peptide QEHA modulated βγ subunit-mediated inhibition of ACI was not determined in that study. Therefore, we investigated whether the peptide QEHA could block the actions of βγ subunits on the soluble AC forms containing regions of ACI. The peptide QEHA attenuated the ability of βγ to inhibit activity of the IC1a⋅VC2 enzyme (Fig. 4A). Chen et al. (28) demonstrated that the effects of βγ on β-adrenergic receptor kinase, phospholipase Cβ, ACII, and muscarinic K+ channels were not modulated by a control peptide, SKEE, corresponding to the region (residues 1,000–1,026) on ACIII that is cognate to that encompassed by QEHA in ACII. Consistent with these observations, the SKEE peptide did not alter the ability of βγ subunits to inhibit the IC1a⋅VC2 (Fig. 4A). The peptide QEHA also obliterated the ability of βγ subunits to inhibit the activity of VC1aIC1b⋅IC2 (Fig. 4B); the control peptide, SKEE, did not modulate the effects of βγ (Fig. 4B). These data (Fig. 4) demonstrate that the peptide QEHA, which attenuates the effects of βγ on several effectors (28), can also obliterate the effects of βγ on the C1a region of ACI and the βγ interacting site formed by C1b and C2 regions of ACI. These observations are consistent with recent reports that have shown that the effector-interacting domains on the β subunit have overlapping regions (29, 30).
Figure 4.
The peptide QEHA corresponding to amino acid residues 956–982 in ACII attenuates the ability of βγ subunits to inhibit AC activity. (A) IC1a⋅VC2 form of AC was stimulated by Gsα* (80 nM). The ability of βγ subunits (200 nM) to inhibit Gsα*-stimulated activity was monitored in the presence and absence of 200 nM each of the peptides QEHA and SKEE; peptide SKEE corresponds to residues 1,000–1,026 in ACIII, the cognate region of peptide QEHA in ACII. The mean ± SEM (n = 3 experiments) are shown. (B) Same as A except that the VC1aIC1b·IC2 form of AC was used. The mean ± SEM (n = 3 experiments) is presented.
Studies from Gilman’s laboratory have demonstrated that Giα can inhibit Ca2+/CaM-stimulated activity of ACI (31); inhibition of Gsα-stimulated ACI by Giα was very modest (≤10%; ref. 31). These findings, coupled with the recent report of Dessauer et al. (32) that the C1a region on ACV is the site of Giα interaction, prompted us to investigate whether the Giα site on ACI was also located on the same region. As demonstrated by data in Fig. 5A, Gsα*-stimulated activity of the full-length ACI expressed in Sf9 cells was not inhibited by Giα1. Although a very modest inhibition by Giα of Gsα-stimulated ACI has been reported (31), we could not observe this. This difference probably relates to the fact that the Giα1 inhibition of Gsα-stimulated ACI is very modest. Like the full-length ACI, Gsα*-stimulated activity of IC1⋅VC2 also was not inhibited by Giα1 (Fig. 5A). On the other hand, Gsα*-stimulated activity of VC1aIC1b⋅VC2 and VC1aIC1b⋅IC2 was inhibited by Giα1 in a concentration-dependent manner (Fig. 5A). These findings demonstrate that the Gsα*-stimulated activity of ACI is not altered by Giα1 and that Giα1 can inhibit Gsα*-stimulated activity when the C1a region of ACV is present. Therefore, consistent with the findings of Dessauer et al. (32), our data also demonstrate that the VC1a region is sufficient to observe Giα1-mediated inhibition of Gsα*-stimulated activity. This contention is further supported by the data that VC1⋅VC2 and VC1a⋅VC2 forms of the soluble AC also were inhibited by Giα1 when the activity of these enzymes was stimulated by Gsα* (Fig. 5A). Notably, the VC1a⋅VC2 form of AC was less sensitive to inhibition by Giα1 than the VC1⋅VC2 form (Fig. 5A). Similar results (data not shown) were also obtained with soluble ACs in which the C1a or C1 domain of ACV is linked by an artificial linker to the VC2 domain (17). These findings, like those of Dessauer et al. (32), demonstrate that the C1b region of ACV increases the sensitivity to inhibition by Giα. However, our observations that VC1aIC1b⋅VC2 and VC1aIC1b⋅IC2 forms of AC are more sensitive to inhibition by Giα1 than VC1a⋅VC2 show that the C1b region of ACV can be swapped with a very dissimilar but equivalent region from ACI to restore sensitivity to Giα1-mediated inhibition. Thus, it would appear that the C1b region of either ACI or ACV may alter the conformation of the VC1a domain and permit better interactions with Giα1.
Figure 5.
Inhibition of activity of various forms of AC by Giα1. (A) The activity of full-length ACI and mixtures of C1 or C1a or chimeric C1 (VC1aIC1b) and C2 regions of ACV or ACI was stimulated by Gsα* as described in Fig. 2A. The effects of various concentrations of recombinant, myristoylated Giα1 on AC activity were monitored. Data are presented as percent inhibition of Gsα*-stimulated activity and are the mean ± SEM (n = 3 experiments). (B) VC1⋅VC2 form of AC was stimulated by Gsα* (150 nM), and the effect of 500 nM concentration of Giα1 bound to either guanosine 5′-[γ-thio]triphosphate or GDPβS was monitored. Adenylyl cyclase activities presented are the mean ± SEM (n = 3 experiments). (C) Same as A except that the activity of different AC forms was stimulated by Ca2+ (30 μM) plus CaM (500 nM). Percent inhibition of Ca2+/CaM-stimulated activity is shown as mean ± SEM (n = 3 experiments).
To demonstrate that the recombinant, myristoylated Giα1 was inhibiting AC activity in a specific manner, the experiment depicted in Fig. 5B was performed. Essentially, Giα1 was incubated with either guanosine 5′-[γ-thio]triphosphate or GDPβS, and its ability to inhibit the VC1⋅VC2 form of soluble AC that had been stimulated by Giα was monitored. As shown in Fig. 5B, only the guanosine 5′-[γ-thio]triphosphate-bound Giα1, and not GDPβS-bound Giα1, inhibited AC activity.
Next, we investigated whether Giα1 could inhibit ACI activity that was stimulated by Ca2+/CaM. As shown in Fig. 5C, Giα1 in a concentration-dependent manner inhibited either the full-length ACI or IC1⋅VC2 activity that had been stimulated by Ca2+/CaM. IC1a⋅VC2 form of AC is not stimulated by Ca2+/CaM (data not shown), and, therefore, experiments concerning inhibition by Giα1 of this form of AC in the presence of Ca2+/CaM were not possible. Interestingly, when the C1a region of ACV was linked to the C1b region of ACI and mixed with C2 domain of either enzyme (i.e., VC1aIC1b⋅VC2 or VC1aIC1b⋅IC2), Giα1 inhibited Ca2+/CaM-stimulated activity (Fig. 5C). Overall, therefore, the data in Fig. 5C demonstrate that the C1 region of ACI is necessary for inhibition by Giα1 of Ca2+/CaM-stimulated activity. Moreover, the data in Fig. 5C with VC1aIC1b plus either VC2 or IC2 also support the notion that the C1a region of ACV is sufficient to observe inhibition of activity by Giα1.
In summary, the data presented here demonstrate that the C1a region of ACI is sufficient to observe inhibition of its activity by βγ subunits of heterotrimeric G proteins. Moreover, the C1b region of ACI interacts with its C2 domain, and the presence of these two regions is also sufficient to observe βγ subunit-mediated inhibition of enzyme activity. Notably, neither the C1b nor the C2 domains of ACI by themselves are sufficient to interact with βγ subunits and inhibit enzyme activity. Concerning the inhibition of ACI activity by Giα1, our data demonstrate that the C1 region of the enzyme is involved with interactions with the G protein. As demonstrated by Dessauer et al. (32), we also found that the C1a region of ACV is sufficient to observe inhibition by Giα1; sensitivity to inhibition by Giα1 could be augmented in the presence of C1b region of either ACV or ACI. Unfortunately, we could not determine whether the C1a region alone of ACI was sufficient to inhibit enzyme activity by Giα1 because Gsα*-stimulated activity of ACI was not inhibited by Giα1 and the C1b region was required to stimulate enzyme activity by Ca2+/CaM so that the inhibitory actions of Giα1 could be studied. Nevertheless, our data demonstrate that, in the two different isoforms of AC that are inhibited by Giα1, the G protein interacts with the C1 domain.
Acknowledgments
We thank Dr. Ravi Iyengar for reading this manuscript as well as for the baculovirus to express ACI in Sf9 cells and the generous gift of peptides QEHA and SKEE. We are also deeply indebted to the following individuals who provided us with several of the reagents: Dr. A. G. Gilman for the Gsα and ACI cDNAs; Dr. Randall Reed for the Giα1 cDNA; Dr. W.-J. Tang for the TP2000 strain of E. coli; Dr. Jeffrey I. Gordon for providing us with the plasmid pBB131 encoding Saccharomyces cerevisiae N-myristoyltransferase; and Dr. Harry Jarrett for his gift of purified calmodulin. This research was supported by Grant HL 59679 from the National Institutes of Health.
ABBREVIATIONS
- AC
adenylyl cyclase
- ACI
type I AC
- ACV
type V AC
- GDPβS
guanosine 5′-O-(2-thiodiphosphate)
- CaM
calmodulin
Footnotes
For explanation of abbreviations of ACI and ACV domains, see Fig. 1.
References
- 1.Iyengar R. FASEB J. 1993;7:768–775. doi: 10.1096/fasebj.7.9.8330684. [DOI] [PubMed] [Google Scholar]
- 2.Sunahara R K, Dessauer C W, Gilman A G. Annu Rev Pharmacol. 1997;36:461–480. doi: 10.1146/annurev.pa.36.040196.002333. [DOI] [PubMed] [Google Scholar]
- 3.Buck J, Sinclair M L, Schapal L, Cann M J, Levin L R. Proc Natl Acad Sci USA. 1999;96:79–84. doi: 10.1073/pnas.96.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cali J J, Parekh R S, Krupinski J. J Biol Chem. 1996;271:1089–1095. doi: 10.1074/jbc.271.2.1089. [DOI] [PubMed] [Google Scholar]
- 5.Tang W-J, Gilman A G. J Biol Chem. 1991;266:8595–8603. [PubMed] [Google Scholar]
- 6.Cali J J, Zwaagstra J C, Mons N, Cooper D M F, Krupinski J. J Biol Chem. 1994;269:12190–12195. [PubMed] [Google Scholar]
- 7.Choi E-J, Xia Z, Storm D R. Biochemistry. 1992;31:6492–6498. doi: 10.1021/bi00143a019. [DOI] [PubMed] [Google Scholar]
- 8a.Bakalyar H A, Reed R R. Science. 1990;250:1403–1406. doi: 10.1126/science.2255909. [DOI] [PubMed] [Google Scholar]
- 8b.Yoshimura M, Cooper D M F. Proc Natl Acad Sci USA. 1992;89:6716–6720. doi: 10.1073/pnas.89.15.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishikawa Y, Shuichi K, Chen L, Halnon N J, Kawabe J-I, Homcy C. J Biol Chem. 1992;267:13553–13557. [PubMed] [Google Scholar]
- 10.Yoshimura M, Cooper D M F. Proc Natl Acad Sci USA. 1992;89:6716–6720. doi: 10.1073/pnas.89.15.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Premont R T, Chen J, Ma H-W, Ponnapalli M, Iyengar R. Proc Natl Acad Sci USA. 1992;89:9809–9813. doi: 10.1073/pnas.89.20.9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katsushika S, Chen L, Kawabe J-I, Nilakantan R, Halnon N J, Homcy C J, Ishikawa Y. Proc Natl Acad Sci USA. 1992;89:8774–8778. doi: 10.1073/pnas.89.18.8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang W-J, Gilman A G. Science. 1991;254:1500–1503. doi: 10.1126/science.1962211. [DOI] [PubMed] [Google Scholar]
- 14.Federman A D, Conklin B R, Schrader K A, Reed R R, Bourne H R. Nature (London) 1992;356:159–161. doi: 10.1038/356159a0. [DOI] [PubMed] [Google Scholar]
- 15.Taussig R, Quarmby L M, Gilman A G. J Biol Chem. 1993;268:9–12. [PubMed] [Google Scholar]
- 16.Tang W-J, Gilman A G. Science. 1995;268:1769–1772. doi: 10.1126/science.7792604. [DOI] [PubMed] [Google Scholar]
- 17.Scholich K, Barbier A J, Mullenix J B, Patel T B. Proc Natl Acad Sci USA. 1997;94:2916–2920. doi: 10.1073/pnas.94.7.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scholich K, Wittpoth C, Barbier A J, Mullenix J B, Patel T B. Proc Natl Acad Sci USA. 1997;94:9602–9607. doi: 10.1073/pnas.94.18.9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sunahara R K, Dessauer C W, Whisnant R E, Kleuss C, Gilman A G. J Biol Chem. 1997;272:22265–22271. doi: 10.1074/jbc.272.35.22265. [DOI] [PubMed] [Google Scholar]
- 20.Dessauer C W, Gilman A G. J Biol Chem. 1996;271:16967–169674. doi: 10.1074/jbc.271.28.16967. [DOI] [PubMed] [Google Scholar]
- 21.Lee E, Linder M E, Gilman A G. Methods Enzymol. 1994;237:146–164. doi: 10.1016/s0076-6879(94)37059-1. [DOI] [PubMed] [Google Scholar]
- 22.Linder M E, Pang I-H, Duronio, Gordon J I, Sternweis P J, Gilman A G. J Biol Chem. 1991;266:4654–4659. [PubMed] [Google Scholar]
- 23.Mumby S M, Linder M E. Methods Enzymol. 1994;237:254–268. doi: 10.1016/s0076-6879(94)37067-2. [DOI] [PubMed] [Google Scholar]
- 24.Mumby S, Pang I-H, Gilman A G, Sternweis P C. J Biol Chem. 1988;263:2020–2026. [PubMed] [Google Scholar]
- 25.Neer E J, Lok J M, Wolf L G. J Biol Chem. 1984;259:14222–14229. [PubMed] [Google Scholar]
- 26.Masters S B, Tyler Miller R, Chi M-H, Chang F-H, Beiderman B, Lopez N G, Bourne H. J Biol Chem. 1989;264:15467–15474. [PubMed] [Google Scholar]
- 27.Levin L, Reed R R. J Biol Chem. 1995;270:7573–7579. doi: 10.1074/jbc.270.13.7573. [DOI] [PubMed] [Google Scholar]
- 28.Chen J, DeVivo M, Dingus J, Harry A, Li J, Sui J, Carty D J, Blank J L, Exton J H, Stoffel R H, et al. Science. 1995;268:1166–1169. doi: 10.1126/science.7761832. [DOI] [PubMed] [Google Scholar]
- 29.Ford C E, Skiba N P, Bae H, Daaka Y, Reuveny E, Shekter L R, Rosal R, Weng G, Yang C-S, Iyengar R, et al. Science. 1998;280:1271–1274. doi: 10.1126/science.280.5367.1271. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Sternweis P M, Charnecki S, Smith T F, Gilman A G, Neer E J, Kozasa T. J Biol Chem. 1998;273:16265–16272. doi: 10.1074/jbc.273.26.16265. [DOI] [PubMed] [Google Scholar]
- 31.Taussig R, Tang W-J, Helper J R, Gilman A G. J Biol Chem. 1994;269:6093–6100. [PubMed] [Google Scholar]
- 32.Dessauer C W, Tesmer J J G, Sprang S R, Gilman A G. J Biol Chem. 1998;273:25831–25839. doi: 10.1074/jbc.273.40.25831. [DOI] [PubMed] [Google Scholar]