Abstract
We previously showed that nuclear export of the large (60S) ribosomal subunit relies on Nmd3 in a Crm1-dependent manner. Recently the general mRNA export factor, the Mtr2/Mex67 heterodimer, was shown to act as an export receptor in parallel with Crm1. These observations raise the possibility that nuclear export of the 60S subunit in Saccharomyces cerevisiae requires multiple export receptors. Here, we show that the previously characterized 60S subunit biogenesis factor, Arx1, also acts as an export receptor for the 60S subunit. We found that deletion of ARX1 was synthetic lethal with nmd3 and mtr2 mutants and was synthetic sick with several nucleoporin mutants. Deletion of ARX1 led to accumulation of pre-60S particles in the nucleus that were enriched for Nmd3, Crm1, Mex67, and Mtr2, suggesting that in the absence of Arx1, 60S export is impaired even though the subunit is loaded with export receptors. Finally, Arx1 interacted with several nucleoporins in yeast two-hybrid as well as in vitro assays. These results show that Arx1 can directly bridge the interaction between the pre-60S particle and the NPC and thus is a third export receptor for the 60S subunit in yeast.
INTRODUCTION
The nuclear pore complex (NPC) is a specialized structure embedded within the nuclear envelope that serves as a conduit for molecules traveling between the nucleus and cytoplasm (for reviews see Pemberton and Paschal, 2005; Tran and Wente, 2006). Molecules less than 40 kDa can freely traverse the NPC by simple diffusion. However, larger molecules or complexes require karyopherins, specialized transport receptors that recognize cargos carrying either nuclear import signal (NLS) or nuclear export signal (NES). The stable interaction of the importin β-like export receptor Crm1 with an NES requires the cooperative binding of RanGTP (reviewed in Kutay and Guttinger, 2005), resulting in a ternary complex that is translocated through the NPC. At the cytoplasmic face of the NPC the export complex is dissociated upon hydrolysis of RanGTP to RanGDP (Bischoff et al., 1994, 1995).
In yeast the large and small ribosomal subunits are assembled in the nucleolus and exported to the cytoplasm separately but in a Crm1-dependent manner (Moy and Silver, 1999; Ho et al., 2000b; Stage-Zimmermann et al., 2000; Gadal et al., 2001). Nmd3 is an essential nucleocytoplasmic shuttling protein that contains a leucine-rich NES and is required for Crm1-dependent export of the 60S subunit (Ho et al., 2000b; Gadal et al., 2001; Thomas and Kutay, 2003; Trotta et al., 2003). However, several questions arise regarding the export of ribosomal subunits. Transport of large cargo molecules is enhanced by the presence of multiple receptors (Ribbeck and Gorlich, 2001). Thus, the ribosomal subunits might also employ multiple receptors. In addition, the channel of the NPC is comprised of disordered FG repeat–containing nucleoporins that form a mesh through weak hydrophobic interactions (Ribbeck and Gorlich, 2001; Denning et al., 2003; Patel et al., 2007). The highly electronegative ribosomal subunits likely require additional proteins to present hydrophobic surfaces that can partition into this milieu. Finally, considering that the size of the 60S subunit approaches the upper limit of what can be accommodated by the NPC (Pante and Kann, 2002), positioning the subunit for entry into the NPC channel may require multiple interactions between the 60S subunit and the NPC. Recently, the general mRNA export Mtr2-Mex67 heterodimer was shown to be required for 60S subunit export in yeast (Yao et al., 2007). An mtr2-33 mutant specifically affects 60S subunit export but not mRNA export (Bassler et al., 2001) and overexpression of MEX67 suppresses nmd3 mutants that are defective in 60S subunit export (Yao et al., 2007; Lo and Johnson, unpublished data). Thus, Mtr2/Mex67 may partially bypass the functional requirement of Nmd3 and Crm1-mediated 60S subunit export.
Here, we demonstrate that export of the 60S subunit also relies on Arx1, a nucleo-cytoplasmic shuttling protein that is associated with pre-60S particles (Belaya et al., 2006; Hung and Johnson, 2006; Lebreton et al., 2006). In the absence of Arx1, 60S particles enriched for Nmd3 and Crm1 as well as Mtr2 and Mex67 accumulate in the nucleus. Arx1 physically interacts with FG-domains of several nucleoporins, suggesting that Arx1 possesses a karyopherin-like feature to facilitate receptor-mediated 60S subunit export.
MATERIALS AND METHODS
Yeast Strains and Plasmids
Strains used in this work are listed in Table 1. Strain AJY1912 was made by homologous recombination. Briefly, arx1Δ::KanMX4 locus was amplified by PCR from strain AJY1901 using primers AJO563 (CTGGGTACCCGGCCGTCATGCCTCTGTGAAGCT) and AJO569 (5′-GCGGAGCTCCCGGGTCGACTGCAAGATTCTGAGCAAATG)¤ and the PCR product was transformed into CH1305. Plasmids used in this work are listed in Table 2. pAJ1029 was made by PCR amplification of ARX1 from wild-type yeast genomic DNA using primers AJO599 (5′-CTGAGCTCCCGGGTCATGCCTCTGTGAAGC) and AJO600 (5′-GCGGAGCTCCCGGGTATGATATACTTATATTATTTATATACTAGCTTTAGAAATGATGAA) and cloned as an SstI fragment into pAJ60. pAJ1481 was made by three part ligation of 1) PCR-amplified ARX1 using primers AJO876 (CTGTCGACGCTCTAGCTATCTCCCACGA) and AJO564 (5′-GCGCCCGGGCTTAATTAACATTTTCATGGTTTCTTCAACTC), 2) SstI- and SalI-digested fragment of pAJ1479 (Dong et al., 2004), and 3) SstI- and PacI-digested vector of pAJ1026. pAJ1484 was made by three part ligation of 1) PacI- and SstI-digested fragment of pAJ1026 (vector), 2) SstI-SalI fragment of pAJ1482 and 3) SalI-PacI fragment of pAJ1481. pAJ1454 was made from PCR amplification of ARX1 from wild-type yeast genomic DNA with primers AJO784 (5′-CTGTCTAGAGGATCCATGGCTCTAGCTATCTCCCA) and AJO785 (GCGAAGCTTGGATCCCTACATTTTCATGGTTTCTTCAACTCCG). The PCR product was digested with BamHI and cloned into the same site of pGAD C-1 (James et al., 1996). pAJ1031 was assembled from the GST-TEV containing PCR product using AJO422 (GGCGTCGACAAACAATGTCCCCTATACTAGGT) and AJO487 (CCGGGATCCGTGATGATGGTGGTGATGGGAACCCTGAAAATACAG) to amplify pGEX-2T and cut with SalI and BamHI; ARX1 amplified with AJO616 (CTGTCGACGGATCCTCCATGGCTCTAGCTATCTC) and AJO617 (GCGTCGACCAGCTGCTAGCTTTAGAAATGATGAAG), cut with BamHI and PvuII; and pAJ251 (LEU2 2μ GAL10-XRN1; Page et al., 1998) and cut with XhoI and PvuII.
Table 1.
Yeast strains
| Strains | Genotype | Reference |
|---|---|---|
| AJY1231 | MATα ade2, his3, leu2, trp1, ura3, mex67::HIS3 (pUN100-mex67-5) | Segref et al. (1997) |
| AJY1608 | MATαhsi3 leu2 ura3 nup120Δ::KanMX4 | This work |
| AJY1901 | MATa his3Δ1 leu2Δ0 ura3Δ0 arx1Δ::KanMX4 | Hung and Johnson (2006) |
| AJY1911 | MATa his3Δ1 leu2Δ0 ura3Δ0 | Hung and Johnson (2006) |
| AJY1912 | MATa ade2 ade3 leu2 lys52-801 ura3-52 arx1Δ::KanMX4 | This work |
| AJY1930 | MATa his3Δ1 leu2Δ0 ura3Δ0 nup84Δ::KanMX4 | Open Biosystemsa |
| AJY1931 | MATa his3Δ1 leu2Δ0 ura3Δ0 nup133Δ::KanMX4 | Open Biosystems |
| AJY2110 | MATa his3 Δ1 leu2Δ0 ura3Δ0 lys2Δ0 nmd3Δ::KanMX4 | This work |
| AJY2470 | MATa his3 Δ1 leu2Δ0 ura3Δ0 nup42Δ::KanMX4 | Open Biosystems |
| AJY2471 | MATa his3Δ1 leu2Δ0 ura3Δ0 nup100Δ::KanMX4 | Open Biosystems |
| AJY2601 | MATα his3Δ1 leu2Δ0 ura3Δ0 arx1Δ::NatMX4 | This work |
| BJ5464 | MATa ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1 | E. Jones |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open Biosystems |
| CH1305 | MATa ade2 ade3 leu2 lys2-801 ura3-52 | Kranz and Holm (1990) |
| PJ69-4A | MATa ade2 ade3 his3 leu2 trp1 ura3gal4Δ gal80ΔLYS2::GAL1-HIS3 GAL2-ADE2 met::GAL7-lacZ | James et al. (1996) |
| PSY412 | MATa ade2 ade3 leu2 lys1 his3 nsp1-10A::URA3 | Nehrbass et al. (1990) |
| PSY1077 | MATα gle2-1 ura3-1 leu2-3,112 his3-11,15 trp1-1 ade2-1 | Stage-Zimmermann et al. (2000) |
| PSY1634 | MATa nup116::HIS3 trp1 leu2 his3 | Stage-Zimmermann et al. (2000) |
| PSY1654 | MATα his3 leu2 lys2 trp1 ura3 nup82Δ::HIS3+pNup82-Δ108 URA3 | Hurwitz and Blobel (1995) |
| PSY1659 | MATa ade2 ura3 leu2 trp1 nic96Δ::HIS3+pUN100-nic96–1 LEU2 | Zabel et al. (1996) |
| Y1029 | MATa ade2 his3 leu2 trp1 ura3 mtr2::HIS3 + pRS316-MTR2 | Santos-Rosa et al. (1998) |
a Open Biosystems, Huntsville, AL.
Table 2.
Plasmids
| Plasmid | Relevant markers | Reference |
|---|---|---|
| pAJ411 | URA3 2μ NMD3 | This work |
| pAJ528 | URA3 2μ MEX67 | This work |
| pAJ538 | LEU2 CEN Nmd3-13myc | Ho et al. (2000b) |
| pAJ582 | LEU2 CEN Nmd3-GFP | Hedges et al. (2005) |
| pAJ739 | URA3 CEN Crm1(T539C)-HA | This work |
| pAJ751 | LEU2 CEN NMD3AA-13MYC | This work |
| pAJ752 | LEU2 CEN NMD3AAA-13MYC | This work |
| pAJ866 | URA3 2μ pGBDU-C1 | James et al. (1996) |
| pAJ908 | URA3 CEN RPL25-eGFP | Kallstrom et al. (2003) |
| pAJ1029 | ADE3 URA3 ARX1 | This work |
| pAJ1031 | LEU2 2μ GAL-GST-TEV-HIS6-ARX1 | This work |
| pAJ1481 | LEU2 CEN GAL-UBI(M)-ARX1-13MYC | This work |
| pAJ1484 | LEU2 CEN GAL-UBI(R)-1RX1-13MYC | This work |
| pAJ1454 | URA3 2μ pGADC-1 ARX1 | This work |
| pAJ1593 | LEU2 CEN NMD3-L505A-13MYC | Hedges et al. (2006) |
| pAJ1594 | LEU2 CEN NMD3-SupraNES-13MYC | West et al. (2007) |
| pAJ1876 | URA3 2μ MTR2 | This work |
| pAJ2060 | URA3 2μ CRM1 | This work |
| pAJ2251 | TRP1 2μ pGBKT7 | Patel et al. (2007) |
| pAJ2252 | TRP1 2μ pGBKT7 NUP116 (165–716) | Patel et al. (2007) |
| pAJ2253 | LEU2 2μ pGADT7 | Patel et al. (2007) |
| pAJ2254 | LEU2 2μ pGADT7 NUP116 (165–458) | Patel et al. (2007) |
| pAJ2255 | LEU2 2μ pGADT7 NUP100 (1–588) | Patel et al. (2007) |
| pAJ2256 | LEU2 2μ pGADT7 NUP57 (1–255) | Patel et al. (2007) |
| pAJ2257 | LEU2 2μ pGADT7 Nsp1 (1–591) | Patel et al. (2007) |
| pAJ2258 | LEU2 2μ pGADT7 NUP159 (441–876) | Patel et al. (2007) |
| pAJ2259 | LEU2 2μ pGADT7 NUP42 (1–364) | Patel et al. (2007) |
Genetic Procedures
Screen for Mutations Synthetic Lethal with arx1Δ.
The synthetic lethal screen was carried out as described using strain AJY1912 containing pAK1029 (Kranz and Holm, 1990). From ∼34,000 colonies screened, four recessive synthetic lethal or synthetic sick mutants were identified. The mutated genes were identified by complementation with a genomic library and complementing clones subcloned to identify the single gene responsible for complementation. We were unable to clone two of the mutants.
Genetic Interactions.
To test for genetic interactions between arx1Δ and nucleoporin mutants or export factors, AJY1901 or AJY2601 was crossed to selected nucleoporin mutants or nmd3(AAA), mtr2-33 or mex67-5 mutant strains (Table 2). Diploids were sporulated, dissected to isolate spore clones¤ and genotyped by appropriate markers.
Yeast Two-Hybrid Analysis.
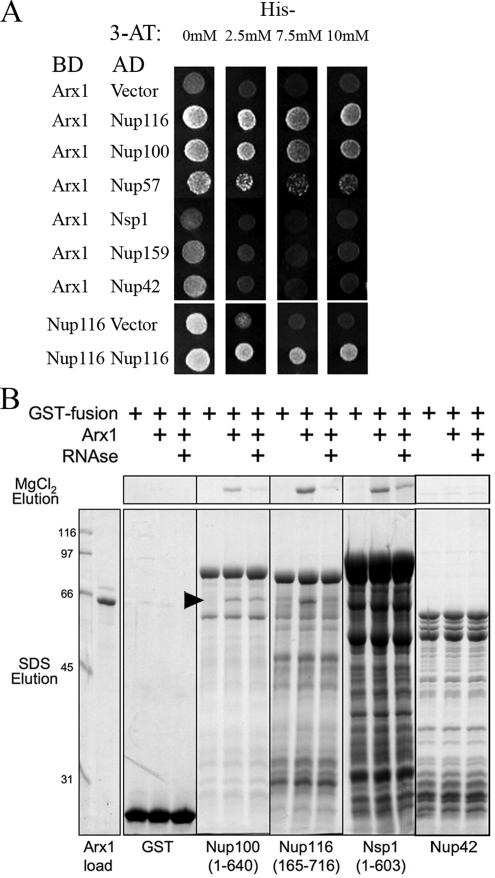
To test for two-hybrid interactions between Arx1 and nucleoporins, the appropriate plasmids (see Figure 7A and Table 2) were introduced into the reporter strain PJ69-4A. Leu+ Trp+ or Leu+ Ura+ transformants were selected and tested in serial dilution assays on ura− trp− his− or leu− ura− his− triple-dropout medium supplemented with 3-AT as indicated. Plates were incubated at 30°C for 4 d.
Figure 7.
Arx1 interacts with nucleoporins. (A) A panel of plasmids that express Arx1 or the Nup116 FG domain, as a control, fused to the Gal4-binding domain (BD) and various Nup FG domains fused to the Gal4 activation domain (AD) were transformed into yeast strain BJ69-4A and spotted onto selective media (Leu− Ura− His− dropout for Arx1-Nup interactions or Leu− Trp− His− dropout for Nup-Nup interactions) to test for potential interactions. Plates were incubated at 30°C for 4 d. Positive interactions should drive expression of the HIS3 reporter. The indicated concentrations of 3-AT were added to increase the stringency of the assay. (B) In vitro interaction of Arx1 with GST-Nup fusions. GST, GST-Nup100(aa 1–640), GST-Nup116(aa 165–716) and GST-Nsp1(aa 1–603)-coated beads (2 μg GST fusion protein in 10 μl of beads) were incubated with purified Arx1 (0.8 μg) in the absence or presence of RNase A (25 μg). After 1 h at 4°C, beads were concentrated by centrifugation, washed twice, and bound proteins eluted first with 0.3 M MgCl2, followed by elution with SDS. Bound proteins were resolved by SDS-PAGE and visualized with Coomassie blue. The bands present below the full-length GST-Nup fusion proteins in the lanes without Arx1 are degradation products of the fusion proteins.
Microscopy
For green fluorescent protein (GFP) microscopy, overnight cultures in selective media containing 2% glucose were fixed with formaldehyde (3.7% final concentration) for 40 min, washed three times in cold 0.1 M potassium phosphate buffer, pH 6.6, and resuspended in 0.1 M potassium phosphate, pH 6.6, 1.2 M sorbitol. For 4′,6′-diamidino-2-phenylindole (DAPI) staining, Triton X-100 was added to fixed cells to a final concentration of 0.1% for 5 min, followed by DAPI at a final concentration of 1 μg/ml for 1 min. Cells were then washed three times with cold phosphate-buffered saline (PBS) and resuspended in PBS with 0.02% NaN3. Fluorescence was visualized on a Nikon Eclipse E800 microscope (Melville, NY) fitted with a 100× objective and a Photometrics CoolSNAP ES digital camera (Woburn, MA) controlled with the NIS-Element AR 2.10 software. Images were prepared using Adobe Photoshop 7.0 (San Jose, CA). Indirect immunofluorescence was performed as described previously (Ho and Johnson, 1999) with anti-Nmd3 antibody.
In Vitro Binding
GST-TEV-HIS6-Arx1 was expressed from pAJ1031 in BJ5464 by growth in the presence of 1% galactose for 7 h at 30°C. All subsequent steps were carried out at 4°C. The cell pellet was washed and resuspended in extraction buffer (50 mM Tris·HCl, pH 8, 300 mM NaCl, 10% glycerol, 1 μM each pepstatin and leupeptin¤ and 1 mM PMSF). Cells were disrupted by vortexing with glass beads. NP40 was added to 0.1% and the extract was clarified by centrifugation for 10 min at 10,000 × g followed by 20 min at 30,000 × g. The extract was passed over a glutathione Sepharose column and washed with extraction buffer¤ followed by extraction buffer with NaCl adjusted to 50 mM¤ and eluted in the same buffer supplemented with 50 mM glutathione. Limiting TEV protease was added to the eluted protein and the sample was dialyzed overnight against 50 mM Tris·HCl¤ pH 8, 50 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol, 1 μM each leupeptin and pepstatin¤ and 1 mM PMSF. The cleaved protein was then passed over a glutathione Sepharose column equilibrated in 50 mM Tris·HCl, 50 mM NaCl, and 10% glycerol to remove glutathione S-transferase (GST) and uncleaved GST-Arx1. Binding of Arx1 to GST-Nups was essentially as described in Allen et al. (2002).
Other Methods
Sucrose gradient sedimentation, Western blotting and immunoprecipitations were as described (Hung and Johnson, 2006). The antibodies used in this work were affinity purified rabbit anti-Nmd3, anti-Rpl8, anti-Mex67 (C. Dargemont, Institut Jacques Monod, Paris, France) and anti-Mtr2 (E. Hurt, Universität Heidelberg, Germany) monoclonal 9e10 anti-myc and 12CA5 anti-HA (Covance Laboratories, Madison, WI).
RESULTS
Genetic Interactions of ARX1
Deletion of ARX1 leads to a cold-sensitive defects in growth and 60S biogenesis (Hung and Johnson, 2006; Lebreton et al., 2006). Although Arx1 binds to the 60S subunit in the nucleus and accompanies 60S subunit export (Nissan et al., 2002), its nuclear function is still poorly understood. To begin to address this, we carried out a small-scale synthetic lethal screen. Subsequent cloning and sequencing of two of the synthetic lethal mutants identified them as NMD3 and NUP120. Nmd3 acts as the Crm1-dependent export adapter for the 60S subunit (Ho et al., 2000b; Gadal et al., 2001), whereas Nup120 is a nucleoporin that has previously been shown to be required for efficient 60S export (Stage-Zimmermann et al., 2000). Sequencing the genomic locus of the nmd3 mutant revealed a T-to-A transversion at nt 1514 (nmd3ΔC14), resulting in a premature stop codon that eliminates the C-terminal 14 aa. Similarly, the genomic mutation in NUP120 was a deletion of nucleotide G3065 (nup120ΔC16), introducing a frame shift that eliminated the C-terminal 16 aa of Nup120. The identification of NMD3 and NUP120 as genetic interactors of ARX1 implies a role for Arx1 in 60S subunit export.
Deletion of the C-terminal fourteen amino acids of Nmd3 resulted in strong nuclear localization of the protein (Figure 1A). We had previously localized the NES of Nmd3 to aa 491–500 based on sequence alignments and functional assays (Ho et al., 2000b). Although the truncation in nmd3ΔC14 did not remove any of the residues previously implicated in export, it was possible to shift the alignment of the NES of Nmd3 five residues toward the C-terminus, to include L505 as the C-terminal hydrophobic residue of the NES (Figure 1B). This alignment gives a consensus leucine-rich NES and suggests that although the NES is highly conserved throughout eukaryotes, it has shifted slightly in position during evolution. Changing leucine 505 to alanine (nmd3L505A) had a strong effect on the localization of Nmd3 and cell growth (Hedges et al., 2006) and was synthetic lethal with arx1Δ (data not shown). Thus, deletion of ARX1 is synthetic lethal with a defect in Nmd3 export function. We also tested the effect of nmd3ΔC14 and nmd3L505A on 60S subunit export. As expected, these mutants showed strongly impaired 60S export, judged by the accumulation of Rpl25-GFP in the nucleus (Figure 1A).
Figure 1.
nmd3ΔC14 is trapped in the nucleus and impairs 60S subunit export. (A) Wild-type (WT) and nmd3ΔC14 strains were cultured in YPD to midlog phase and subjected to indirect immunofluorescence for visualization of endogenous Nmd3 localization. The effect of nmd3ΔC14 on nuclear export of the 60S subunit was monitored by Rpl25-eGFP. The nmd3l505a allele has an effect similar to that of nmd3ΔC14 on blocking nuclear export of the 60S subunit, as monitored by Rpl25-eGFP. (B) Cartoon of the primary structure of Nmd3. CC: Cys-x2-Cys repeats that comprise zinc-binding motifs. Dark gray indicate regions to which mutations that disrupt 60S binding map (Hedges et al., 2006). Amino acid sequence of the C-terminus of Nmd3 with the residues deleted in the nmd3ΔC14 mutant shown in light gray. An alignment of NESs of Nmd3 from various organisms is shown. The canonical leucine-rich NES sequence is shown with two examples from PKI and HIV-1 Rev. (Φ, hydrophobic residues (L,M,I,V); X, any amino acid). Gray bars highlight conserved residues. Sc, S. cerevisiae; Dm, Drosophila melanogaster; and Hs, Homo sapiens. Numbers indicate amino acid positions.
Double Mutants Are Blocked for Nmd3 and 60S Export
As described above, deletion of ARX1 was synthetic lethal with nmd3ΔC14 and strongly synthetic sick with nup120Δ16. To observe the effect of the double mutants on 60S export, we made a conditional allele of ARX1 by fusing ubiquitin (UBI) to the N-terminus of the protein expressed under control of the regulatable GAL promoter (Dong et al., 2004). In this construct, UBI is cleaved in vivo and the stability of the resulting Arx1 depends on the N-terminal residue remaining after cleavage (the N-end rule; Park et al., 1992). Consequently, we made two variants, containing methionine (UBI-M-ARX1) or arginine (UBI-R-ARX1) that are expected to result in stable or unstable proteins, respectively. When expressed on galactose, these two variants complemented the deletion mutant (Figure 2A). Western blot analysis showed that the R variant was present at much lower levels, even in the presence of galactose¤ and diminished further after shift to glucose (Figure 2B).
Figure 2.
Elimination of Arx1 impairs nuclear export of Nmd3 and 60S subunits in the nup120ΔC16 strain. (A) Functional analysis of regulatable Arx1-myc constructs. arx1Δ nup120ΔC16 cells expressing either wild-type Arx1-myc (pAJ1026), Gal-UBI(M)-Arx1-myc (pAJ1481) or Gal-UBI(R)-Arx1-myc (pAJ1484) plasmid as the sole copy of Arx1 were tested for growth on galactose- or glucose-containing dropout medium. (B) Protein expression of regulatable Arx1-myc expression constructs. The arx1Δ nup120ΔC16 cells harboring either Arx1-myc, Gal-UBI(M)-Arx1-myc (pAJ1481) or Gal-UBI(R)-Arx1-myc (pAJ1484) plasmid were cultured in galactose-containing media to midlog phase and transferred to glucose-containing media for the indicated times before they were collected. Extracts were subjected to SDS-PAGE and Western blotting using anti-myc antibody. (C) The arx1Δ nup120ΔC16 stain carrying the unstable Gal-UBI(R)-Arx1-myc construct was transformed with either Rpl25-eGFP (pAJ908) or Nmd3-GFP (pAJ755) plasmid. The strains were then cultured in galactose containing media to midlog phase and transferred to glucose containing media for the indicated times before they were subjected to microscopy. Nuclear DNA was stained with DAPI.
Repression of UBI-R-ARX1 expression in arx1Δ nup120ΔC16 cells severely impaired growth (Figure 2A) and led to nuclear accumulation of Nmd3 (Figure 2C). We also observed nuclear accumulation of Rpl25-GFP after repressing UBI-R-ARX1 (Figure 2C). The nuclear accumulation of Nmd3-GFP appeared faster than that of Rpl25-GFP because Nmd3 shuttles and is rapidly trapped in the nucleus. However, Rpl25-GFP remains associated with ribosomes in the cytoplasm. Consequently, to see a clear change in localization more time is required to deplete the cytoplasmic pool of Rpl25-GFP-containing 60S subunits after export is blocked. In contrast, in the presence of the stable UBI-M-ARX1 construct, Nmd3 began to accumulate in the nucleus only after 4 h of repression and we did not observe any significant nuclear accumulation of Rpl25-GFP (data not shown). We were not able to monitor the effect of ARX1 repression on either Nmd3 or Rpl25 localization in the arx1Δ nmd3ΔC14 mutant as the UBI-R-ARX1 construct was unable to complement this mutant.
ARX1 Shows Genetic Interaction with Other Nucleoporin Mutants
Considering the genetic interaction between ARX1 and NUP120, we asked if ARX1 would show genetic interactions with other nucleoporin mutants as well. From crosses of arx1Δ with a panel of nucleoporin mutants, representing different subcomplexes of the NPC, we found several additional nucleoporin mutants that interacted with arx1Δ (Table 3). In particular, mutations in NUP133 and NUP84, which together with NUP120 form the Nup84 subcomplex, showed strong synthetic interaction. In addition, nup82 and gle2 mutants showed strong genetic interaction, whereas nic96 and nup42 mutants showed weak interaction. Although the Nup84 complex is symmetrically disposed across the nuclear envelope, Nup82 and Gle2 are found on the cytoplasmic face (Suntharalingam and Wente, 2003). Previous work has shown that both the Nup84 and the Nup82 complexes are involved in 60S export (Stage-Zimmermann et al., 2000; Gleizes et al., 2001).
Table 3.
Nucleoporin mutants that were tested for genetic interactions with arx1D
| Nucleoporin mutant | Localization | Genetic interaction with arx1Δ | Affect 60S exporta |
|---|---|---|---|
| nup82-Δ108 | Cytoplasmic | Strong SS | Yes |
| nup84Δ | Symmetric | Strong SS | No |
| nup133Δ | Symmetric | Strong SS | Yes |
| nup42Δ | Cytoplasmic | Weak SS | No |
| nsp1-10A | Symmetric | No | Yes |
| nic96-1 | Symmetric | Weak SS | Yes |
| nup100Δ | Cytoplasmic-biased | No | No |
| Nup116-5 | Cytoplasmic-biased | No | Yes |
| nup120-Δ | Symmetric | Strong SS | Yes |
| gle2-1 | Cytoplasmic | Strong SS | Yes |
SS, synthetic sick.
a From Stage-Zimmermann et al. (2000).
Pre-60S Particles Loaded with Nmd3/Crm1 and Mex67/Mtr2 Accumulate in the Nucleus in arx1Δ Mutants
The physical interaction between Arx1 and the pre-60S subunit as well as the genetic interactions between arx1Δ and nmd3 and nucleoporin mutants strongly suggest that Arx1 is involved in 60S export, possibly interacting with the NPC. If Arx1 were required for a late step in 60S export, we might expect accumulation of the Nmd3-bound 60S subunit in the nucleus. We previously showed that Rpl25-GFP accumulates in the nucleus in arx1Δ cells, indicating a defect in 60S subunit export (Hung and Johnson, 2006). As shown in Figure 3A, Nmd3 also accumulated in the nucleus in arx1Δ cells. Furthermore, the Nmd3 present in the arx1Δ cells was essentially entirely bound to 60S (Figure 3B), demonstrating that export of the Nmd3-60S complex is inhibited in the absence of Arx1. It should be noted that the Nmd3 present in the cytoplasm of wild-type cells is also bound to 60S subunits and must exist only transiently as a free protein during reimport into the nucleus (Ho et al., 2000a). Thus, the change in localization of Nmd3 reflects accumulation of an Nmd3-60S complex in the nucleus in arx1Δ cells but not a change in the ratio of bound to unbound protein.
Figure 3.
Nmd3-bound 60S particles accumulate in the nucleus in an arx1Δ mutant. (A) Cultures of wild-type and arx1Δ cells carrying pAJ582 (Nmd3-GFP) were grown to midlog phase in selective media and the localization of Nmd3 was monitored by fluorescence microscopy. (B) Lysates were prepared from wild-type and arx1Δ cells and fractioned on 7–47% sucrose gradients by ultracentrifugation as described in Materials and Methods. Fractions were collected, and the absorbance at 254 nm was monitored continuously. Proteins were precipitated with trichloroacetic acid, separated by SDS-PAGE, transferred to nitrocellulose membrane, and immunoblotted for Nmd3 or Rpl8p using specific antibodies.
Because Crm1 is the export receptor for Nmd3, the nuclear accumulation of an Nmd3-60S complex in arx1Δ cells could be due to a failure to recruit Crm1. Alternatively, Arx1 could act downstream of Crm1 loading, preventing efficient export of the pre-60S complex. We tested this by immunopurification of Nmd3 from wild-type and arx1Δ cells, probing for Crm1 by Western blotting. Indeed, we observed an increase in Crm1 present in the Nmd3-IP (Figure 4A). Because the interaction of Crm1 with ligands is labile in extracts, due to the loss of RanGTP, we also used a functional mutant Nmd3 (Nmd3-supra NES) in which the NES has been altered to enhance Crm1 binding (Engelsma et al., 2004; West et al., 2007). With this mutant, we also observed an increase in Crm1 levels in the Nmd3-IP from arx1Δ cells (Figure 4A). This result did not necessarily demonstrate that Crm1 was enriched in a complex that contained the 60S subunit. To address this, we asked if Crm1 association with 60S subunits could be detected in sucrose gradients. Indeed, in the absence of Arx1, we observed an increased level of Crm1 cosedimenting with free 60S subunits (Figure 4B, fractions indicated by solid bars). These results indicate that Arx1 is not required for efficient recruitment of Crm1. Rather, Arx1 acts downstream Crm1 and without Arx1, pre-60S particles that are loaded with both Nmd3 and Crm1 accumulate in the nucleus.
Figure 4.
Deletion of Arx1 leads to nuclear accumulation of an Nmd3-Crm1–60S complex. (A) Wild-type and arx1Δ cells transformed with pAJ538 (Nmd3-13myc) or pAJ1594 (Nmd3-supraNES-13myc) and pAJ739 (Crm1T539C-HA) were cultured in selective media and collected at midlog phase. Immunoprecipitations were carried out using anti-myc antibodies and subjected to Western blotting using anti-myc and anti-HA antibodies to monitor Nmd3 and Crm1 levels. N/A, negative control. (B) Lysates from wild-type and arx1Δ cells carrying pAJ856 (Crm1-HA) were prepared in the presence of cycloheximide and fractioned on 7–47% sucrose gradients by ultracentrifugation. Fractions were collected, and the absorbance at 254 nm was monitored continuously. Proteins were precipitated with trichloroacetic acid, separated by SDS-PAGE, transferred to nitrocellulose membrane, and immunoblotted for HA or Rpl8p using specific antibodies.
In addition to Crm1, the Mex67/Mtr2 heterodimer is also required for 60S export (Yao et al., 2007). Consequently, we considered that the pre-60S particle that accumulates in the nucleus in arx1Δ cells might also be loaded with Mex67 and Mtr2. Testing this by coimmunoprecipitation, we found that Mex67 and Mtr2 were strongly increased in the Nmd3-bound pre-60S particles from arx1Δ cells (Figure 5, compare lanes 1 and 3). Mex67 and Mtr2 were also enriched in particles blocked for export by Nmd3(AAA), containing three point mutations within its leucine-rich NES (Figure 5, compare lanes 2 and 4). This mutant does not interact efficiently with Crm1 (West et al., 2007) and accumulates in the nucleus. Thus, the pre-60S particles in arx1Δ cells are enriched for two different export receptors, Crm1 and Mex67/Mtr2, but yet are defective for export. Of the mutations known to trap Nmd3 in the nucleus, all are in factors that act late in the 60S export pathway. These include Crm1 (Ho et al., 2000b), Mex67/Mtr2 (Yao et al., 2007), proteins that regulate Ran and a subset of nucleoporins (Stage-Zimmermann et al., 2000). Thus, Arx1 itself appears to behave as an export receptor or as an effector of NPC function.
Figure 5.
Mex67 and Mtr2 are enriched on pre-60S particles when export is blocked. Cell extracts were prepared from wild-type (BY4741) and arx1Δ (AJY1901) expressing Nmd3-myc (pAJ538) and Nmd3(AAA)-myc (pAJ752). The myc-tagged Nmd3 proteins were immunoprecipitated¤ and proteins that copurifying proteins were detected by Western blotting using antibodies against Mex67, Mtr2, Nmd3-myc, and Rpl8, as a marker for 60S subunits.
Genetic Interactions among Export Factors
If Arx1 acts in parallel with Crm1 and Mex67/Mtr2 as a third export receptor, we would anticipate that it would show genetic interaction with these factors. Because arx1Δ is synthetic lethal with nmd3 mutants that disrupt Crm1 interaction (Table 4A), we tested for pairwise genetic interactions with additional export factors. Indeed, we observed that arx1Δ was synthetic lethal with mtr2-33 (Figure 6A) and was synthetic sick with mex67-5 (Table 4A and data not shown). Conversely, overexpression of factors that are limiting might suppress export defects. For example, overexpression of Mex67 suppresses certain nmd3 mutants (Ho et al., 2000b; Yao et al., 2007). Consequently, we tested for genetic effects of overexpressing Crm1, Arx1, Nmd3 and Mex67/Mtr2 in arx1, nmd3, mex6¤7 and mtr2 mutants. The results of this analysis (Table 4B) are complex. Overexpression of MTR2 or MEX67 suppressed the growth defect of an arx1Δ mutant. However, in many cases overexpression of an export factor was dominant negative in cells in which other 60S subunit export receptors or adapters were mutant. For example, overexpression of CRM1 was dominant negative in nmd3(AAA), arx1Δ, mex67-5¤ and mtr2-33 mutants (Figure 6B). Overexpression of NMD3 was also dominant negative in an arx1 mutant (Table 4B and Figure 6B) and co-overexpression had an additive dominant negative effect (data not shown).
Table 4.
Genetic interactions among export factors
| A. Synthetic effects between export factors |
|
|---|---|
| nmd3(AAA) X arx1Δ | SL |
| nmd3-1 X mex67-5 | SL |
| nmd3(AAA) X mtr2-33 | SL |
| arx1Δ X mex67-5 | SS |
| arx1Δ X mtr2-33 | SL |
| B. High copy expression of export factors | |||||
|---|---|---|---|---|---|
| Mutant |
High copy expression |
||||
|
NMD3 |
ARX1 |
MEX67 |
MTR2 |
CRM1 |
|
| nmd3(AAA) | SUP | SUP | DN | DN | |
| arx1Δ | DN | SUP | SUP | DN | |
| mex67-5 | No effect | No effect | SUP | DN | |
| mtr2-33 | DN | No effect | No effect | DN | |
SL, Synthetic lethal; SS, Synthetic sickness; SUP, suppression; DN, dominant negative.
Figure 6.
ARX1 shows genetic interactions with other 60S export factors. (A) An arx1Δ mutant was mated to mtr2-33 containing MTR2 on a URA3 vector. After sporulation and dissecting, serial dilutions of spore clones of the indicated genotypes were spotted onto 5FOA media to select against the wild-type MTR2 vector. Plates were incubated at 30°C for 4 d. Additional results from high copy expression and synthetic effects of double mutants are summarized in Table 4. (B) NMD3, MEX67, MTR2¤ and CRM1 were expressed from high copy vectors in wild-type (BY4741) and arx1Δ mutant cells (AJY1901). Vector indicates an empty vector control. Serial dilutions were plated onto selective media and incubated for 3 d at 30°C.
Suppression by overexpression is easily explained if the factor being overexpressed is limiting in the mutant condition. Thus, Mex67 appears to be limiting in arx1Δ cells. However, dominant negative interactions are more difficult to interpret. We suggest that a common mechanism of dominant negative effect among these factors is the disruption of bridging interactions by altering the stoichiometry of a bridging factor in a complex. For example, if Arx1 provides a means for the 60S subunit to interact with the NPC, then in the absence of Arx1, recruitment of the pre-60S complex to the NPC will be dependent on Mex67/Mtr2 and Nmd3/Crm1. In the Nmd3/Crm1 pathway, the 60S subunit is recruited to the NPC by bridging interactions between the 60S subunit, Nmd3, Crm1 and the NPC. Overexpression of Nmd3 could drive formation of Nmd3-60S and Nmd3-Crm1 complexes, preventing the formation of a 60S-Nmd3-Crm1 complex. Similarly, overexpression of Crm1 could simultaneously saturate Nmd3 and sites on the NPC, blocking recruitment of the pre-60S to the NPC. Hence, export would become more dependent on Mex67/Mtr2¤ which is required but not sufficient for efficient export.
Arx1 Physically Interacts with Nucleoporins
Arx1 was previously identified among proteins that interacted with Nup42 and Nup100 in vitro (Allen et al., 2001). In these experiments, GST fusions of different nucleoporins were immobilized on beads that were then used as affinity matrices for purification of proteins from yeast whole cell extracts. In this analysis, it was possible that the interactions were indirect, bridged by other proteins or RNAs. As an alternative means to test these interactions, we used yeast two-hybrid analysis. Arx1 was expressed as a fusion to the GAL4 binding domain and was challenged with a panel of nucleoporin fusions to the GAL4 activation domain. We observed interactions between Arx1 and Nup100, Nup116¤ and Nup57 (Figure 7A). As a control we recapitulated the previously reported homotypic interaction of Nup116 FG domains (Figure 7A) as well as interactions of Nup116 with other Nup FG domains (data not shown; Patel et al., 2007). These results support the previous biochemical identification of Arx1 nucleoporin association.
To test the potential Arx1-Nup interaction more directly, we assayed for Arx1 binding to Nups in vitro. Arx1 was purified from yeast as a functional GST fusion protein and the GST moiety was cleaved with TEV protease. Arx1 was then applied to a panel of GST-Nups immobilized on glutathione beads. As shown in Figure 7B, Arx1 bound to Nup100, Nup116 and Nsp1. We did not detect binding to Nup42, as reported previously (Allen et al., 2001) nor to Nup57 for which we detected two-hybrid interaction. The interaction between Arx1 and Nup100 and Nsp1 was resistant to RNase treatment (Figure 7B, arrowhead for Arx1 in the SDS-elution from Nup100, fraction eluted from Nsp1 in MgCl2). The interaction with Nup116 was partially sensitive to RNase, although a small amount of Arx1 was seen in the MgCl2 elution after RNase treatment. We also detected RNase-sensitive interactions with Nup159(aa441–881), Nup49, Nup60 Nup1(aa332–1076) and Nup2 (data not shown). Apparently at least some of the Nup preparations contain RNA that may bridge the interaction with Arx1. However, it is possible that, in vivo, RNA acts as a cofactor for Arx1, helping it fold correctly and/or bind nucleoporins as a means of regulating its binding to the NPC only when associated with the 60S subunit.
DISCUSSION
Here we have shown that the shuttling factor Arx1 acts as a nuclear export receptor for the 60S ribosomal subunit. This conclusion is based on several different lines of evidence. ARX1 shows genetic interaction with multiple genes encoding factors involved in 60S subunit export. These include the 60S export adapter Nmd3 and its receptor Crm1, the mRNA export receptors, Mex67 and Mtr2, which have recently been shown to be required 60S export as well, and various nucleoporins. We found that in the absence of Arx1, export of pre-60S particles was impaired, leading to accumulation of Nmd3-bound pre-60S particles in the nucleus. Surprisingly, the Nmd3-bound subunits also contained elevated levels of Crm1 as well as Mex67 and Mtr2. The nuclear accumulation of pre-60S particles loaded with export receptors suggests that Arx1 functions downstream or in concert with these receptors to facilitate export of the 60S subunit. Finally, we showed that Arx1 interacts with a subset of nucleoporins by two-hybrid assay and by in vitro binding, consistent with a previous report identifying Arx1 among proteins that could be affinity purified from yeast extracts using nucleoporins for bait (Allen et al., 2001). Taken together, these results imply that Arx1 is a third nuclear export receptor for the 60S subunit. Similar work showing that Arx1 acts as an export receptor has recently been reported (Bradatsch et al., 2007).
Sequence comparisons indicate that Arx1 has evolved from the family of type II methionyl aminopeptidases (MetAPs; Hung and Johnson, 2006). However, catalytic residues in the active site are not conserved in Arx1, suggesting that it is not an active peptidase. Indeed, the human ortholog of Arx1, Ebp1, does not have peptidase activity (Monie et al., 2007). These proteins have also diverged from MetAPs by the inclusion of loops and a C-terminal extension that, in Ebp1, provides an RNA-binding domain (Monie et al., 2007). It remains to be determined if human Ebp1, which is involved in pre-60S metabolism in human cells (Squatrito et al., 2004), also plays a role similar to that of Arx1 as an export receptor as the loops in Arx1 are much more extensive than those in Ebp1 and could provide additional interaction surfaces not found in Ebp1. This may be reminiscent of the acquisition of ribosome binding activity by Mex67 and Mtr2, a function specific to protein loops unique to the yeast proteins and not found in the metazoan proteins (Yao et al., 2007). On the other hand, nucleophosmin has been reported to be required for export of 5S rRNA, and by extension, the 60S subunit, in human cells (Yu et al., 2006). Nucleophosmin does not have an obvious ortholog in yeast. Thus, among the export adaptors for the large subunit, only Nmd3 appears to be well conserved in function as an export adapter. Nmd3 may represent the primordial 60S export factor for eukaryotic cells and additional adapters may have evolved independently in different eukaryotic lineages.
The inner channel of the nuclear pore complex is largely composed of FG-repeat–containing nucleoporins that create a hydrophobic meshwork, posing a permeability barrier that selectively controls the translocation of macromolecules (Ribbeck and Gorlich, 2001; Denning et al., 2003). Translocation of hydrophilic cargo through this hydrophobic channel of the NPC requires a mechanism for partitioning the cargo into such an environment. It has been speculated that large cargo molecules require multiple receptors, and this has been demonstrated for protein import in HeLa cells (Ribbeck and Gorlich, 2002). On the other hand, the addition of a second import receptor synergistically stimulated import. The large subunit of the ribosome may be the bulkiest cargo to pass through the NPC. In addition, it is highly electronegative, due to the large amount of RNA on the surface of the subunit. Our results here with Arx1, combined with the previous demonstration that Nmd3 (Ho et al., 2000b; Gadal et al., 2001) and more recently the Mex67/Mtr2 heterodimer (Yao et al., 2007) are required for export suggest that at least three receptors are used for efficient export in yeast. Considering the impact of nmd3 and mtr2 mutants on 60S export, and that Arx1 appears to be a stoichiometric component of the pre-60S complex, it seems likely that these receptors are present simultaneously on the subunit and are required in concert, rather than as alternative export pathways. We might expect these receptors to be distributed over the surface of the large subunit to allow the entire surface of the ribosome to partition into the NPC. Preliminary results suggest that Arx1 binds in the vicinity of the exit tunnel (Hung and Johnson, 2006) whereas Nmd3 appears to bind to the joining surface (Sengupta, Bussiere, Frank and Johnson, unpublished data) on the opposite face of the subunit. The Mex67/Mtr2 heterodimer is suggested to bind 5S rRNA (Yao et al., 2007), again potentially distal to Arx1 and Nmd3. In the case of nuclear import of large cargo molecules in HeLa cells, single import receptors were not sufficient for efficient translocation, but did promote tethering of cargo at the NPC. We have not observed tethering of preribosomal complexes at the NPC under conditions in which export is inhibited.
If multiple receptors are required for export, why are multiple different receptors used rather than multiple copies of a single receptor species? One possibility is that export with multiple different receptors is more efficient than multiple receptors of the same protein species, possibly avoiding competition between receptors for common binding sites. In addition, the different affinities of different receptors for their binding sites on nucleoporins could help to orient the ribosomal subunit with respect to the central channel of the NPC to facilitate its entry.
The presence of multiple receptors on the large subunit also raises the question of how subunit export is regulated. We have proposed previously that Nmd3 may act as a structural proofreading factor whose binding would depend on the proper assembly of a complex binding site that is presented only upon proper maturation of the subunit (Johnson et al., 2002). Thus, the loading of Nmd3 would dictate the time of export. However, with multiple receptors, this seems a more complicated proposition. Would their loading be coordinated, or is there a hierarchy in their function, i.e., would one factor, such as Nmd3, be the primary determinant for release from the nucleolus or docking at the NPC whereas the other factors load later or perhaps function only at the NPC? Our preliminary results suggest that there is not a rigid hierarchy to the loading of these proteins. However, the accumulation of particles in the nucleus when one export receptor is disrupted could result in accumulation of Crm1 and Mex67/Mtr2 on these particles, possibly during abortive cycles of docking and release from the NPC.
ACKNOWLEDGMENTS
We thank E. Hurt and C. Dargemont for anti- Mtr2 and Mex67 antibodies, respectively, the Rexach lab at the University of California, Santa Cruz for plasmids and J. Hedges and X. Luo for assistance in preparation of plasmids. This work was supported by National Institutes of Health Grant RO1 GM53655 to A.W.J.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-09-0968) on December 12, 2007.
REFERENCES
- Allen N. P., Huang L., Burlingame A., Rexach M. Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 2001;276:29268–29274. doi: 10.1074/jbc.M102629200. [DOI] [PubMed] [Google Scholar]
- Allen N. P., Patel S. S., Huang L., Chalkley R. J., Burlingame A., Lutzmann M., Hurt E. C., Rexach M. Deciphering networks of protein interactions at the nuclear pore complex. Mol. Cell Proteom. 2002;1:930–946. doi: 10.1074/mcp.t200012-mcp200. [DOI] [PubMed] [Google Scholar]
- Bassler J., Grandi P., Gadal O., Lessmann T., Petfalski E., Tollervey D., Lechner J., Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- Belaya K., Tollervey D., Kos M. FLIPing heterokaryons to analyze nucleo-cytoplasmic shuttling of yeast proteins. RNA. 2006;12:921–930. doi: 10.1261/rna.2301806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F. R., Klebe C., Kretschmer J., Wittinghofer A., Ponstingl H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F. R., Krebber H., Smirnova E., Dong W., Ponstingl H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 1995;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradatsch B., et al. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol. Cell. 2007;27:767–779. doi: 10.1016/j.molcel.2007.06.034. [DOI] [PubMed] [Google Scholar]
- Denning D. P., Patel S. S., Uversky V., Fink A. L., Rexach M. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. USA. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Lai R., Nielsen K., Fekete C. A., Qiu H., Hinnebusch A. G. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J. Biol. Chem. 2004;279:42157–42168. doi: 10.1074/jbc.M404502200. [DOI] [PubMed] [Google Scholar]
- Engelsma D., Bernad R., Calafat J., Fornerod M. Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J. 2004;23:3643–3652. doi: 10.1038/sj.emboj.7600370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O., Strauss D., Kessl J., Trumpower B., Tollervey D., Hurt E. Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 2001;21:3405–3415. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleizes P. E., Noaillac-Depeyre J., Léger-Silvestre I., Teulières F., Dauxois J. Y., Pommet D., Azum-Gelade M. C., Gas N. Ultrastructural localization of rRNA shows defective nuclear export of preribosomes in mutants of the Nup82p complex. J. Cell Biol. 2001;155:923–936. doi: 10.1083/jcb.200108142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges J., Chen Y. I., West M., Bussiere C., Johnson A. W. Mapping the functional domains of yeast NMD3, the nuclear export adapter for the 60 S ribosomal subunit. J. Biol. Chem. 2006;281:36579–36587. doi: 10.1074/jbc.M606798200. [DOI] [PubMed] [Google Scholar]
- Hedges J., West M., Johnson A. W. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 2005;24:567–579. doi: 10.1038/sj.emboj.7600547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J., Johnson A. W. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:2389–2399. doi: 10.1128/mcb.19.3.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J. H., Kallstrom G., Johnson A. W. Nascent 60S ribosomal subunits enter the free pool bound by Nmd3p. RNA. 2000a;6:1625–1634. doi: 10.1017/s1355838200001291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J. H., Kallstrom G., Johnson A. W. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 2000b;151:1057–1066. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung N. J., Johnson A. W. Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:3718–3727. doi: 10.1128/MCB.26.10.3718-3727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz M. E., Blobel G. NUP82 is an essential yeast nucleoporin required for poly(A)+ RNA export. J. Cell Biol. 1995;130:1275–1281. doi: 10.1083/jcb.130.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. W., Lund E., Dahlberg J. Nuclear export of ribosomal subunits. Trends Biochem. Sci. 2002;27:580–585. doi: 10.1016/s0968-0004(02)02208-9. [DOI] [PubMed] [Google Scholar]
- Kallstrom G., Hedges J., Johnson A. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 2003;23:4344–4355. doi: 10.1128/MCB.23.12.4344-4355.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz J. E., Holm C. Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc. Natl. Acad. Sci. USA. 1990;87:6629–6633. doi: 10.1073/pnas.87.17.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Guttinger S. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 2005;15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Lebreton A., Saveanu C., Decourty L., Rain J. C., Jacquier A., Fromont-Racine M. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J. Cell Biol. 2006;173:349–360. doi: 10.1083/jcb.200510080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monie T. P., Perrin A. J., Birtley J. R., Sweeney T. R., Karakasiliotis I., Chaudhry Y., Roberts L. O., Matthews S., Goodfellow I. G., Curry S. Structural insights into the transcriptional and translational roles of Ebp1. EMBO J. 2007;26:3936–3944. doi: 10.1038/sj.emboj.7601817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy T. I., Silver P. A. Nuclear export of the small ribosomal subunit requires the Ran-GTPase cycle and certain nucleoporins. Genes Dev. 1999;13:2118–2133. doi: 10.1101/gad.13.16.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U., Kern H., Mutvei A., Horstmann H., Marshallsay B., Hurt E. C. NSP1: a yeast nuclear envelope protein localized at the nuclear pores exerts its essential function by its carboxy-terminal domain. Cell. 1990;61:979–989. doi: 10.1016/0092-8674(90)90063-k. [DOI] [PubMed] [Google Scholar]
- Nissan T. A., Bassler J., Petfalski E., Tollervey D., Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A. M., Davis K., Molineux C., Kolodner R. D., Johnson A. W. Mutational analysis of exoribonuclease I from Saccharomyces cerevisiae. Nucleic Acids Res. 1998;26:3707–3716. doi: 10.1093/nar/26.16.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pante N., Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol. Biol. Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E. C., Finley D., Szostak J. W. A strategy for the generation of conditional mutations by protein destabilization. Proc. Natl. Acad. Sci. USA. 1992;89:1249–1252. doi: 10.1073/pnas.89.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. S., Belmont B. J., Sante J. M., Rexach M. F. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- Pemberton L. F., Paschal B. M. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- Ribbeck K., Gorlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K., Gorlich D. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 2002;21:2664–2671. doi: 10.1093/emboj/21.11.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Moreno H., Simos G., Segref A., Fahrenkrog B., Pante N., Hurt E. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol. 1998;18:6826–6838. doi: 10.1128/mcb.18.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A., Sharma K., Doye V., Hellwig A., Huber J., Luhrmann R., Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squatrito M., Mancino M., Donzelli M., Areces L. B., Draetta G. F. EBP1 is a nucleolar growth-regulating protein that is part of pre-ribosomal ribonucleoprotein complexes. Oncogene. 2004;23:4454–4465. doi: 10.1038/sj.onc.1207579. [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann T., Schmidt U., Silver P. A. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell. 2000;11:3777–3789. doi: 10.1091/mbc.11.11.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam M., Wente S. R. Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell. 2003;4:775–789. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- Thomas F., Kutay U. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 2003;116:2409–2419. doi: 10.1242/jcs.00464. [DOI] [PubMed] [Google Scholar]
- Tran E. J., Wente S. R. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Trotta C. R., Lund E., Kahan L., Johnson A. W., Dahlberg J. E. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 2003;22:2841–2851. doi: 10.1093/emboj/cdg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M., Hedges J. B., Lo K. Y., Johnson A. W. Novel interaction of the 60S ribosomal subunit export adapter Nmd3 at the nuclear pore complex. J. Biol. Chem. 2007;282:14028–14037. doi: 10.1074/jbc.M700256200. [DOI] [PubMed] [Google Scholar]
- Yao W., Roser D., Kohler A., Bradatsch B., Bassler J., Hurt E. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol. Cell. 2007;26:51–62. doi: 10.1016/j.molcel.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Yu Y., Maggi L. B., Jr, Brady S. N., Apicelli A. J., Dai M. S., Lu H., Weber J. D. Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol. Cell. Biol. 2006;26:3798–3809. doi: 10.1128/MCB.26.10.3798-3809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel U., Doye V., Tekotte H., Wepf R., Grandi P., Hurt E. C. Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p. J. Cell Biol. 1996;133:1141–1152. doi: 10.1083/jcb.133.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]