Abstract
The basic leucine (Leu) zipper (bZIP) proteins compose a family of transcriptional regulators present exclusively in eukaryotes. The bZIP proteins characteristically harbor a bZIP domain composed of two structural features: a DNA-binding basic region and the Leu zipper dimerization region. They have been shown to regulate diverse plant-specific phenomena, including seed maturation and germination, floral induction and development, and photomorphogenesis, and are also involved in stress and hormone signaling. We have identified 89 bZIP transcription factor-encoding genes in the rice (Oryza sativa) genome. Their chromosomal distribution and sequence analyses suggest that the bZIP transcription factor family has evolved via gene duplication. The phylogenetic relationship among rice bZIP domains as well as with bZIP domains from other plant bZIP factors suggests that homologous bZIP domains exist in plants. Similar intron/exon structural patterns were observed in the basic and hinge regions of their bZIP domains. Detailed sequence analysis has been done to identify additional conserved motifs outside the bZIP domain and to predict their DNA-binding site specificity as well as dimerization properties, which has helped classify them into different groups and subfamilies, respectively. Expression of bZIP transcription factor-encoding genes has been analyzed by full-length cDNA and expressed sequence tag-based expression profiling. This expression profiling was complemented by microarray analysis. The results indicate specific or coexpression patterns of rice bZIP transcription factors starting from floral transition to various stages of panicle and seed development. bZIP transcription factor-encoding genes in rice also displayed differential expression patterns in rice seedlings in response to abiotic stress and light irradiation. An effort has been made to link the structure and expression pattern of bZIP transcription factor-encoding genes in rice to their function, based on the information obtained from our analyses and earlier known results. This information will be important for functional characterization of bZIP transcription factors in rice.
Several families of transcription factors have been identified both in animals as well as plants. The basic Leu zipper (bZIP) transcription factor family is among the largest and most diverse dimerizing transcription factor families. bZIP transcription factors owe their name to their highly conserved bZIP domain composed of a basic region and a Leu zipper (Hurst, 1994). The bZIP domain is 60 to 80 amino acids in length. The basic region consists of about 16 amino acid residues characterized by the presence of an invariant N-x7-R/K motif, whereas the Leu zipper is composed of heptad repeats of Leu or other bulky hydrophobic amino acids (Ile, Val, Phe, or Met) positioned exactly nine amino acids toward the C terminus. The basic and Leu zipper regions are functionally distinct. The basic region is responsible for sequence-specific DNA binding and is highly conserved. The Leu zipper is an amphipathic sequence in the form of coiled-coil, which confers dimerization specificity and is less conserved. This Leu zipper sequence mediates homo- and/or heterodimerization in bZIP proteins. bZIP monomers are in the form of long α-helices, and, at the time of DNA binding, the N-terminal half binds in the major groove to double-stranded DNA, whereas the C-terminal half mediates dimerization to form a superimposed coiled-coil structure called the Leu zipper (Landschulz et al., 1988; Ellenberger et al., 1992). bZIP transcription factor-encoding genes have been identified exclusively in eukaryotes. Their number is 17 in Saccharomyces cerevisiae, 31 in Caenorhabditis elegans, 27 in Drosophila, 56 in humans, and 67 in Arabidopsis (Arabidopsis thaliana) genomes (Riechmann et al., 2000; Fassler et al., 2002; Vinson et al., 2002; Deppmann et al., 2004).
In plants, like other transcription factor genes, members of the bZIP transcription factor family are also either expressed constitutively or in an organ-specific (Schindler et al., 1992a; Rodriguez-Uribe and O'Connell, 2006), stimulus-responsive (de Vetten and Ferl, 1995), development-dependent (Chern et al., 1996), and cell-cycle-specific (Minami et al., 1993) manner. So far, over 120 bZIP transcription factors have been identified in different plants (Supplemental Table S1). In many plants, bZIP transcription factors have been reported to function in the regulation of seed maturation and germination (Izawa et al., 1994; Chern et al., 1996). Available functional information suggests their role not only in the integration of spatial and temporal information during floral induction, but also in flower development (Chuang et al., 1999; Walsh and Freeling, 1999; Strathmann et al., 2001; Abe et al., 2005; Thurow et al., 2005; Wigge et al., 2005; Muszynski et al., 2006). Genetic and molecular studies have revealed their regulatory role in photomorphogenesis and light signaling (Holm et al., 2002; Jakoby et al., 2002; Mallappa et al., 2006). They also participate in defense against pathogens (Zhang et al., 2003; Thurow et al., 2005). Many bZIP transcription factors function in abscisic acid (ABA) and/or stress signaling (de Vetten and Ferl, 1995; Lopez-Molina et al., 2002; Fujita et al., 2005), and a few of them have been shown to be hormone responsive (Fukazawa et al., 2000; Niggeweg et al., 2000).
Fourteen bZIP transcription factors have been identified or functionally characterized from rice (Oryza sativa). Among these, OSBZ8 and TRAB1 are possibly involved in the regulation of transcription by ABA-mediated signaling (Nakagawa et al., 1996; Hobo et al., 1999a). OsZIP-1a has a putative role in induction of the Em gene promoter by ABA (Nantel and Quatrano, 1996), and LIP19 and OsOBF1 are involved in cold signaling (Aguan et al., 1993; Shimizu et al., 2005). RISBZ1 is required for endosperm-specific expression of storage protein genes and is thought to be involved in the regulation of some metabolic enzymes expressed in developing seeds (Onodera et al., 2001; Yamamoto et al., 2006), RITA-1 is assumed to be involved in the regulation of rice genes expressed during seed development (Izawa et al., 1994), and REB is a major DNA-binding protein for an α-globulin gene promoter in rice endosperm, later shown to be a transcriptional activator in rice grains (Nakase et al., 1997; Yang et al., 2001). OsZIP-2a and OsZIP-2b are expected to have putative roles in selective inactivation of some G-box binding factors (Nantel and Quatrano, 1996). RF2a and RF2b function in vascular development and also play a role in symptom development of rice tungro disease (Yin et al., 1997b; Dai et al., 2003, 2004). Two other rice bZIP proteins, RISBZ4 and RISBZ5, have unknown functions (Onodera et al., 2001).
In this study, we report the identification and characterization of 89 genes encoding bZIP transcription factors in the rice genome. These bZIP transcription factors in rice have been classified on the basis of their putative DNA-binding-site specificity and dimerization properties. Their domain organization and gene structure have been analyzed. Duplication events likely to contribute to the expansion of the bZIP family in rice were also identified. Phylogenetic analyses have been done to reveal the relationship pattern among the bZIP transcription factor genes in rice, as well as with other known plant bZIP proteins. An elaborate analysis of the spatial and temporal expression pattern of bZIP transcription factors, during various developmental stages, including vegetative growth, floral transition, and panicle and seed development, has been performed. Moreover, it has been demonstrated that expression of bZIP transcription factor-encoding genes is regulated by various environmental cues, such as light and abiotic stress.
RESULTS AND DISCUSSION
Identification and Nomenclature of bZIP Transcription Factors in Rice
To identify bZIP transcription factor genes in rice, BLAST searches of the rice (Oryza sativa subsp. japonica ‘Nipponbare’) genome were performed using the hidden Markov model (HMM) profile of the bZIP domain as query. Further analysis based on the presence of the bZIP domain with confidence (E value <1.0) as given by SMART revealed 89 potential nonredundant bZIP transcription factor genes, which were designated as OsbZIP genes. The OsbZIP genes were given a number designation from 1 to 89 to provide a unique identifier for each bZIP transcription factor gene as proposed for bZIP transcription factors in Arabidopsis (Jakoby et al., 2002). The nomenclature was based on their position on rice chromosomes 1 to 12 and from top to bottom. The OsbZIP gene number, The Institute of Genomic Research (TIGR) locus, open reading frame (ORF) length, protein length, and chromosomal location of all 89 OsbZIP genes are listed in Supplemental Table S2. Domain analysis revealed that all but five of the potential bZIP transcription factors had a typical bZIP domain having an invariant N-x7-R/K motif in the basic region and a heptad repeat of Leu or other bulky hydrophobic amino acids positioned exactly nine amino acids upstream of R/K toward the C terminus. Of the remaining five, in the case of OsbZIP34, the heptad repeat of Leu was positioned 30 amino acids instead of the usual nine amino acids toward the C terminus, whereas, in the case of OsbZIP80, the conserved Arg/Lys in the basic region was replaced by an Ile residue. This is similar to the earlier reported rice bZIP transcription factor OsZIP-1a (Nantel and Quatrano, 1996). On the other hand, among the remaining three unusual OsbZIP transcription factors, both OsbZIP21 and OsbZIP82 have a Lys, and OsbZIP85 has a Phe in place of the conserved Asn in the basic region. Such an unusual basic domain has also been reported in the case of the yeast Met-4 bZIP transcription factor (Thomas et al., 1992).
Structure of OsbZIP Genes
To obtain some insight into the gene structure of the 89 OsbZIP genes, their exon/intron organization was analyzed. It was found that 17 (19.1%) of the total OsbZIP genes were intronless (Supplemental Table S2). This is in agreement with the total percentage (19.9%) of rice genes predicted to be intronless (Jain et al., 2008). Among those having introns, the number of introns in the ORF varied from one to 12. Further, the pattern of intron positions within the basic and hinge regions of the bZIP domain (because these regions are the most conserved in the bZIP domain) as well as intron phases with respect to codons were analyzed. Of the 72 intron-containing OsbZIP genes, 62 had introns in the bZIP domain region. On the basis of intron presence, position, and splicing phase, OsbZIP genes were divided into seven patterns, a to g (Fig. 1; Supplemental Fig. S1; Supplemental Table S2). Patterns a and b were the most prevalent. Both had one intron each, but the position and phase were different. Pattern a had an intron in phase 2 (P2) in the basic region, whereas pattern b had an intron in phase 0 (P0) in the hinge region. Pattern c, having two introns each in phase 0, was seen in most of the members (13/16) of group VII only as described later. Patterns d to f were uncommon and each was found in one OsbZIP gene only. Pattern g was devoid of any intron in the basic or hinge region. Pattern g was found in 27 OsbZIP genes, of which only 17 were intronless, whereas the remaining 10 had introns in regions other than their basic and hinge regions. It should also be noted that, whenever an intron was present in the hinge region, both its phase and position were always conserved. However, introns present in the basic region were at variable positions either having splicing phase 0 (P0) or 2 (P2). Further, it was observed that there were more phase 0 introns and symmetric exons (with the same phase on both ends). Such a condition might aid in exon shuffling and recombinational fusion events in the case of the OsbZIP gene family members (Patthy, 1987). The overall pattern of intron position acts as an index to the phylogenetic relationships in a gene family. The splicing phase has remained well conserved during the course of evolution of OsbZIP genes.
Figure 1.
Intron distribution within the basic and hinge regions of the bZIP domains of OsbZIP proteins. Intron distribution patterns (a–g) are depicted. An example of a sequence in the basic and hinge regions is shown at the top with arrows indicating the position of two introns (as observed in the majority of OsbZIP proteins), interrupted by black vertical lines in the sequence and the bars (representing the sequence in different intron patterns). The number of introns and the number of OsbZIP proteins having a particular pattern are also indicated. P0 and P2 indicate the splicing phases of the basic and hinge regions of the bZIP domains, where P0 and P2 refer to phase 0 and phase 2, respectively.
Chromosomal Distribution of OsbZIP Genes
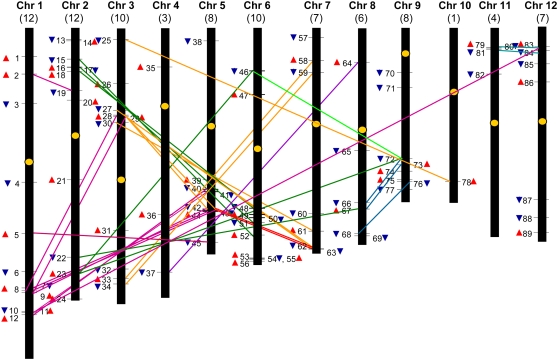
To determine the chromosomal location of all 89 OsbZIP genes, the position (in base pair) of each OsbZIP gene was determined on rice chromosome pseudomolecules available at TIGR (release 5). Figure 2 is a diagrammatic representation of the chromosomal distribution of OsbZIP genes on the 12 rice chromosomes (the exact position in base pair of each OsbZIP gene on rice chromosome pseudomolecules is given in Supplemental Table S2). Although the OsbZIP genes are distributed on each of the 12 rice chromosomes, their distribution is not uniform. Their chromosomal distribution pattern reveals that certain chromosomes and chromosomal regions have a relatively high density of OsbZIP genes. For instance, 12 OsbZIP genes are located each on chromosomes 1 and 2, whereas there is a single OsbZIP gene present on chromosome 10.
Figure 2.
Genomic distribution of OsbZIP genes on rice chromosomes. OsbZIP genes are numbered 1 to 89. Yellow ovals on the chromosomes (vertical bars) indicate the position of centromeres. Chromosome numbers are indicated at the top of each bar and the number in parentheses corresponds to the number of OsbZIP genes present on that chromosome. The OsbZIP genes present on duplicated chromosomal segments are connected by colored lines (one color per chromosome). The red and blue triangles indicate the upward and downward direction of transcription, respectively. The position of OsbZIP genes on TIGR rice chromosome pseudomolecules (release 5) is given in the Supplemental Table S2.
In the case of plants, during the course of its evolution, a gene family has generally undergone either tandem duplication or large-scale segmental duplication to maintain the high number of family members (Cannon et al., 2004). To comprehend the mechanisms underlying OsbZIP gene family evolution, both tandem and segmental duplication events were examined. Surprisingly, duplication in the case of OsbZIP genes was confined to only chromosomal block duplications in rice because none of the OsbZIP genes were found to be arranged in tandem. Forty-nine OsbZIP genes were found to be located on the duplicated segmental regions of rice chromosomes mapped by TIGR (Fig. 2; Supplemental Table S3) at a maximal length distance permitted between collinear gene pairs of 500 kb. All OsbZIP genes located on the duplicated segments of chromosomes belong to the same group, subdivided on the basis of the DNA-binding properties as described later. Most of the members of groups VI (10 of 14), VII (9 of 16), and IX (8 of 11) were found to be present on the duplicated segments of rice chromosomes. These results suggest that segmental genome duplication events have contributed largely to the expansion of the bZIP transcription factor gene family in rice.
Identification of Additional Structural Features in OsbZIP Transcription Factors
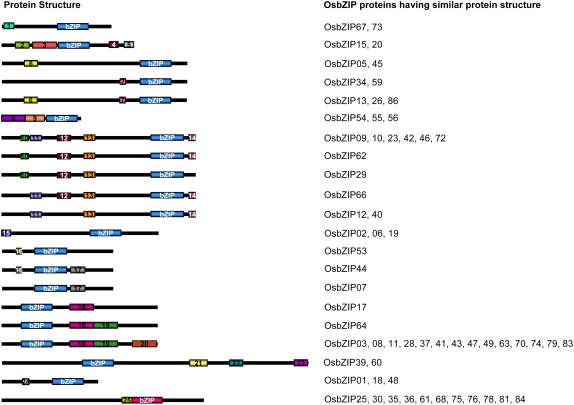
Few bZIP transcription factors, like lotus (Lotus japonicus) LjBzf1 and soybean (Glycine max) STF1 (Cheong et al., 1998; Nishimura et al., 2002), contain domains other than the characteristic bZIP domain. In the case of the AtbZIP transcription factor family, some of the members have been shown to have additional conserved motifs in addition to the bZIP domain and have been classified into 10 groups on the basis of the sequence similarities of their basic region and the presence of additional conserved motifs (Jakoby et al., 2002). The search for the presence of additional domains in the OsbZIP transcription factors using the SMART database did not predict any other domain of known function. Members of the OsbZIP transcription factor family were also searched for the presence of any conserved motifs using the Multiple EM (Expectation Maximization) for Motif Elicitation (MEME) analysis tool. A total of 25 additional conserved motifs, outside the bZIP domain, could be identified in 63 OsbZIP transcription factors (Fig. 3). The multilevel consensus sequence and the amino acid length of these conserved motifs are given in Supplemental Table S4. However, the biological significance of only a few of these motifs is known. Five OsbZIP proteins have motif 6, characterized by the presence of small stretches of Pro and aromatic amino acids Tyr, Phe, and Trp, and is a part of the Pro-rich domain. Similar motifs have also been identified in the N-terminal region of group G bZIP transcription factors in Arabidopsis, which have been shown to have transcriptional activation potential (Schindler et al., 1992b; Jakoby et al., 2002). Motif 7 identified in five OsbZIP transcription factors corresponds to GCB (GBF-conserved box; consensus NLNIGMDxW), which is a part of the trans-activation domain, conserved among plant HBP-1a/GBF-type bZIP factors (Meier and Gruissem, 1994; Okanami et al., 1996). Interestingly, all seven OsbZIP factors possessing motif 6 and/or motif 7 are also predicted to be G-box binding factors as described later.
Figure 3.
Protein structure of representative OsbZIP proteins based on the position of the bZIP domain and the presence of additional conserved motifs outside the bZIP domain as identified by MEME. The bZIP domains are shown in blue, except the unusual bZIP domain, which is shown in pink. Different motifs are highlighted in different color boxes with numbers 1 to 25, where the same number refers to the same motif present in the different OsbZIP proteins. OsbZIP protein numbers having similar protein structure are given in front of each structure on the right side. The details of predicted conserved motifs are given in Supplemental Table S4.
A part of motifs 10, 12, and 13 represents potential casein kinase II (CKII) phosphorylation sites (S/TxxD/E, where x represents any amino acid), indicated by the presence of conserved TVDE and TLED/E in motifs 12 and 13, respectively. Motif 14 also contains a phosphorylation site for Ca2+-dependent protein kinase (R/KxxS/T), which is found in two residues C-terminal to a conserved Leu within the motif. Such motifs have been identified in members of group A bZIP transcription factors in Arabidopsis, which include AREBs/ABFs (Bensmihen et al., 2002; Jakoby et al., 2002). Eight, nine, 11, and 10 OsbZIP proteins containing motifs 10, 12, 13, and 14, respectively, have such consensus phosphorylation sites for various protein kinases. Among these, OsbZIP66 corresponds to already-identified rice ABA-inducible bZIP transcription factor TRAB1 (Hobo et al., 1999a).
Motif 24, identified in three OsbZIP proteins, corresponds to the COP1 interaction motif through which the Arabidopsis bZIP proteins like HY5 and HYH interact with the WD40 domain of COP1 (Holm et al., 2001, 2002). Certain other motifs of unknown function are common to AtbZIP and OsbZIP transcription factors (Jakoby et al., 2002). For example, OsbZIP07 and OsbZIP44 share motif 17 with group F; OsbZIP39 and OsbZIP60 have a part of motif 21 in common with group B; and 14 OsbZIP proteins share a part of motif 20 with group D AtbZIP proteins. Although the function of these motifs is not yet known, the presence of these conserved motifs certainly reflects functional similarities among the OsbZIP proteins sharing these common motifs with the AtbZIP proteins.
Predicted DNA-Binding-Site Specificity of OsbZIP Transcription Factors
bZIP transcription factors owe their DNA-binding ability to the basic region of their bZIP domain. Their DNA-binding specificity, in turn, is determined by the presence of certain key amino acid residues present in the basic and hinge regions of the bZIP domain that contact DNA bases at cis-acting elements (Suckow et al., 1993a; Niu et al., 1999). A majority of the plant bZIP proteins isolated to date recognize elements with an ACGT core (Foster et al., 1994). On the basis of their binding preferences, plant bZIP proteins have been classified into three groups (Izawa et al., 1993). To predict the DNA-binding-site specificity of the OsbZIP proteins, sequence alignment of the amino acids present in the basic and hinge regions of the 89 OsbZIP proteins was done. Based on this alignment and the type of amino acid residues present at four different subregions, as defined by Niu et al. (1999; nomenclature based on Suckow et al., 1993b)—A (−31 to −27), B (−25 and −24), and C (−21 to −11) within the basic region, and D (−6 and −1) in the hinge region—the binding pattern of these OsbZIP proteins could be classified into 11 groups (I–XI; Supplemental Fig. S2). The characteristic features of the OsbZIP proteins classified into different groups are described in Table I.
Table I.
Characteristic features of the OsbZIP transcription factors classified into 11 groups (I–XI) according to their DNA-binding specificity and amino acid sequences in basic and hinge regions
| Group (No. of Members) | Characteristic Features | Putative Binding Site | Known Binding Sites in Rice | Known Binding Sites in Other Species |
|---|---|---|---|---|
| I (7) | Conserved residues in positions −18 (N), −15 (S), −14 (A), −11 (S), and −10 (R). Have a QA(/Q)EC(/A,T) E(/D)E(/D,Q) hinge sequence specific to GBFs (Foster et al., 1994) and the RKQS conserved sequence in the basic region | G-box and/or G-box-like sequences | G-box and/or G-box-like sequences for OSBZ8 (OsbZIP05) and OsZIP-1a (OsbZIP86; Nakagawa et al., 1996; Nantel and Quatrano, 1996) | Wheat EmBP-1 (Niu et al., 1999) |
| II (3) | NRVSAQQAR sequence in their basic region | TGACGT-containing sequences (e.g. CREG/A, Hex, Hexm2), some G-box-like sequences (e.g. G-box and Hexm1) | Not known | Soybean STF1 (Cheong et al., 1998) |
| III (6) | Key residues in the basic region (RNRE/DSAxxSR) and a distinct hinge region [i.e. YVE(/K)E(/D)] | G- and C-boxes with equal affinity | Not known | Tobacco TGA1b (Katagiri et al., 1989; Niu et al., 1999) |
| IV (22) | Specific hinge region sequence Q(/A)A(/Q,H)H (/R,Y,A)L(/M,V) E(/T,A,N,K,)E(/D) | Hybrid ACGT elements like G/C, G/A, C/G boxes | G-, A-, and C-box for RITA-1 (OsbZIP20; Izawa et al., 1994; Niu et al., 1999), GCCACGT(C/A)AG (G/C and A/G hybrid box) for REB (OsbZIP33; Nakase et al., 1997), RISBZ1 (OsbZIP58), RISBZ4 (OsbZIP15), and RISBZ5 (OsbZIP52) recognize GCN4 motif (TGA(G/C)TCA; Onodera et al., 2001), C/G hybrid for LIP19 (OsbZIP38)/OsOBF1 (OsbZIP87) heterodimer, G/C hybrid (hex) for OsOBF1 (OsbZIP87) homodimer (Shimizu et al., 2005) | Maize OPAQUE2 (Lohmer et al., 1991; Schmidt et al., 1992) and Antirrhinum AmbZIP910 (Martinez-Garcia et al., 1998) |
| V (3) | Basic region has A residue at −19 position and hinge region has a conserved QYISE sequence | Relaxed specificity or may bind to other unknown sequences | Not known | AtbZIP34 and AtbZIP61 |
| VI (14) | Highly conserved motifs like M/A-I/M-K and Q/R-A-Y/R in the basic and hinge regions, respectively | ABREs with a consensus sequence (T/G/C/)ACGT(G/T)GC or others containing GCGT or AAGT instead of ACGT (Ono et al., 1996; Hobo et al., 1999b) | ABREs for TRAB1 (OsbZIP66; Hobo et al., 1999a) | Not known |
| VII (16) | Conserved residues in positions −21 (L), −20 (A), −19 (Q), −18 (N), −15 (A), −14 (A), −12 (K), −11 (S), and −10 (R). Possess a KAY(/H)V(/I)QQ(/N) hinge sequence specific to CBFs (Foster et al., 1994) | C-box sequence | Not known | Tobacco TGA1a (Niu et al., 1999) |
| VIII (3) | Conserved residue in position −15 (A) specific to CBFs | C-box elements preferentially | Not known | Not known |
| IX (11) | Conserved Lys substitution at −10 position of the basic region instead of Arg | Sequences other than those containing a palindromic ACGT core | CCA(N)nTGG for RF2a (OsbZIP75; Yin et al., 1997a), Box II for RF2a (OsbZIP75) and RF2b (OsbZIP30; Yin et al., 1997b; Dai et al., 2004) | Arabidopsis PosF21 (Aeschbacher et al., 1991), tomato VSF-1 (Ringli and Keller, 1998), tobacco RSG (Fukazawa et al., 2000) |
| X (3) | Lack the conserved Asn in position −18 | Might not be able to bind to DNA as homodimers | Not known | Not known |
| XI (1) | Hydrophobic Ile residue at position −10 instead of a conserved Arg/Lys | Might not be able to bind DNA or else possess a uniquely different DNA-binding specificity | Corresponds to OsZIP-2a reported earlier (Nantel and Quatrano, 1996) | Not known |
It may be reasoned that promoters of different genes may have variations in their target sites and that sometimes binding affinity could be low in certain cases in order to have a desirably low expression level of a particular gene; hence, these predictions are made to facilitate further studies on DNA-binding patterns of OsbZIP transcription factors. Such a prediction of DNA-binding ability, followed by experimental verification, would definitely be of great utility in identifying which genes are selectively activated by different OsbZIP transcription factors.
Predicted Dimerization Properties of OsbZIP Transcription Factors
bZIP transcription factors are known to bind to DNA predominantly as homo- or heterodimers mediated by the Leu zipper region of the bZIP domain (Landschulz et al., 1988). The amino acid sequence of the Leu zipper determines the homo- and/or heterodimerization of bZIP proteins (O'Shea et al., 1992). The Leu zipper region responsible for dimerization is composed of heptad repeats of amino acids, identified as a, b, c, d, e, f, and g, depending upon their position in the heptad. Leu zipper dimerization stability and specificity are determined by the amino acids present at the a, d, e, and g positions because they are near the Leu zipper interface. Dimerization specificity of Arabidopsis, human, and Drosophila bZIP transcription factors has been predicted on the basis of the presence of attractive or repulsive interhelical g↔e′ electrostatic interactions and the presence of polar or charged amino acids in the a and d positions of the hydrophobic interface of the Leu zipper region (Fassler et al., 2002; Vinson et al., 2002; Deppmann et al., 2004).
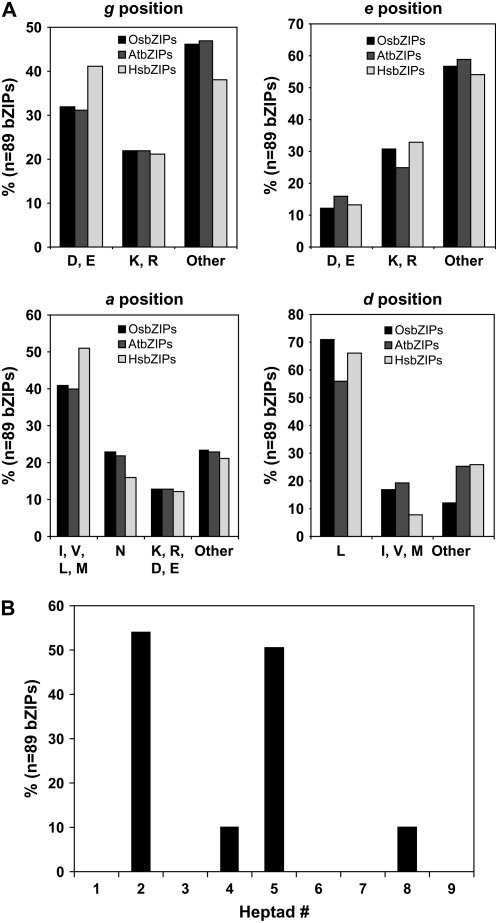
To predict the dimerization specificity of 89 OsbZIP proteins, the amino acid sequence of their bZIP domains was manually arranged in the form of heptads starting from four amino acids (corresponding to the g position in the first heptad) before the occurrence of the first Leu (N-x7-R/K-x9-L) in the bZIP domain (Supplemental Fig. S3). N-terminal and C-terminal boundaries of the OsbZIP Leu zippers have been demarcated following the criteria used for Arabidopsis bZIP proteins (Deppmann et al., 2004). It was observed that Leu zipper length was variable, ranging from two to nine heptads in the case of OsbZIP proteins. Detailed analysis of the type of amino acids present at the a, d, e, and g positions in OsbZIP proteins was carried out and a cross-comparison was made with that of Arabidopsis and human (HsbZIPs). A comparison of the charged amino acids present in the g and e positions of different heptads showed that OsbZIPs, like AtbZIPs, have a lower frequency of charged amino acids in these positions compared to HsbZIPs (Fig. 4A). However, the frequency of charged residues was more in OsbZIPs than AtbZIPs. This indicates that interactions between g↔e′ pairs may be more prominent in regulating dimerization specificity in OsbZIPs as compared to AtbZIPs. About 23% of amino acids present at the a position were Asns (Fig. 4A), which is almost equal to the frequency observed in AtbZIPs (Deppmann et al., 2004). This indicates that, in the case of OsbZIPs, there will be a greater number of homodimerizing Leu zippers because N-N interhelical interaction is preferred over interaction with other amino acids at the a position similar to AtbZIPs (Acharya et al., 2002). Interestingly, the frequency of Leu, responsible for dimer stability, in the d position in OsbZIPs was found to be 71%, which is significantly greater than in AtbZIPs (56%; Fig. 4A). However, the abundance of other aliphatic amino acids at the d position was comparable in OsbZIPs (17%) and AtbZIPs (19%). The presence of Leu might be responsible for dimer stability of longer zippers as indicated by its higher frequency in heptads 7 to 9 of the Leu zipper. Frequency of Asn residues in the a position is the highest in heptad 2 followed by heptad 5 (Fig. 4B), as observed earlier for AtbZIP proteins (Deppmann et al., 2004). This further supports the assumption that a larger number of OsbZIPs may prefer to homodimerize. Polar amino acids, like Ser and Thr, and charged amino acids were also found in the a position of some heptads in a few OsbZIP proteins (Supplemental Fig. S3).
Figure 4.
Amino acid analysis at the g, e, a, and d positions of the Leu zipper. A, Histogram of frequency of amino acids in the g, e, a, and d positions of the Leu zipper for the OsbZIP, AtbZIP, and HsbZIP proteins. B, Histogram of the frequency of Asn residue in the a position of the Leu zippers for all OsbZIP proteins.
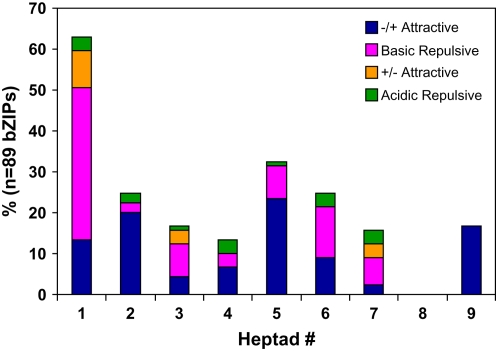
To analyze the contribution of charged residues in governing dimerization properties of OsbZIP proteins, the frequency of attractive and repulsive g↔e′ pairs in each heptad of OsbZIP Leu zippers was calculated and the corresponding histogram is represented in Figure 5. This analysis was based on the frequency of attractive basic-acidic pairs (R↔E and K↔E), attractive acidic-basic pairs (E↔R, E↔K, D↔R, and D↔K), repulsive acidic pairs (E↔E, E↔D, E↔Q, and Q↔E), and repulsive basic pairs (K↔K, R↔K, Q↔K, R↔Q, and K↔Q) in the heptads. It was observed that the frequency of interactive g↔e′ pairs was the maximum in the first heptads, with a sharp decrease in the next three heptads. The frequency of attractive g↔e′ pairs increases in the fifth heptad. Moreover, only attractive g↔e′ pairs were found in the ninth heptad. However, repulsive g↔e′ pairs are predominant in the sixth and seventh heptads, thereby suggesting the chances of heterodimerization. It should be noted that few OsbZIP proteins had multiple repulsive g↔e′ pairs, which were found to be completely absent in Arabidopsis Leu zippers.
Figure 5.
Histogram of the frequency of attractive or repulsive g↔e′ pairs per heptad for all OsbZIP proteins.
Based on the above analyses, OsbZIP proteins were classified into 29 subfamilies (BZ1–BZ29) on the basis of similar predicted dimerization specificities. Various properties of these subfamilies have been described in detail in Supplemental Table S5. Known interaction patterns between different bZIP transcription factors in rice obey the predictions made in this study (Supplemental Table S5). It is evident from this analysis that OsbZIP proteins are expected to display complex and varied dimerization patterns, with the potential to homodimerize with themselves or with the same subfamily members as well as heterodimerize with the members of other subfamilies. Because many of the OsbZIP transcription factors show the potential to heterodimerize, it should be pointed out that most of the OsbZIP proteins, which do not show differential expression profiles in our microarray analysis (described later) and are rather ubiquitously expressed, might assume specific functions when they heterodimerize in planta.
Phylogenetic Analysis of OsbZIP Genes
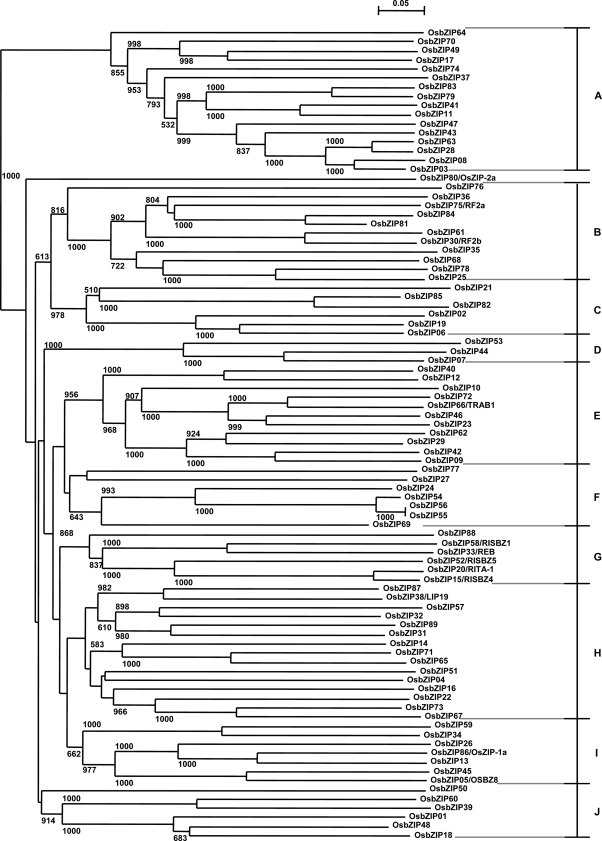
To analyze the evolutionary history of the OsbZIP genes, an unrooted phylogenetic tree was generated using the sequence alignments of the OsbZIP proteins (Fig. 6). Excluding one (i.e. OsbZIP80), all the other OsbZIP transcription factors could be subdivided into 10 clades, designated A to J, with well-supported bootstrap values. It was observed that a majority of the members, predicted to have similar DNA-binding properties, clustered together. However, certain members of groups III, IV, and VI were exceptions because they clustered apart into different clades. To find the relatedness at the amino acid level, three more phylograms, based on (1) bZIP domain, (2) basic and hinge regions, and (3) Leu zipper region of OsbZIP proteins, were generated (Supplemental Fig. S4). These three phylograms are similar yet have some interesting differences (marked in Supplemental Fig. S4), highlighting the fact that more than one OsbZIP protein might recognize the same DNA sequence for binding but may have different dimerization properties, thereby capable of controlling a wide range of transcriptional responses. Most of the members belonging to one clade also shared one or more conserved motifs outside the bZIP domain. Further, 62 OsbZIP proteins formed 31 sister pairs, of which 17 pairs were located on the duplicated segmental regions of rice chromosomes mapped by TIGR. Because the remaining 14 sister pairs have a high degree of similarity in their protein sequences, some of these might be present on the unidentified duplicated chromosomal segments.
Figure 6.
Phylogenetic relationship among the OsbZIP proteins. The phylogenetic tree is based on the sequence alignments of the OsbZIP proteins. The unrooted tree was generated using ClustalX by the neighbor-joining method. Bootstrap values from 1,000 replicates are indicated at each node. OsbZIP proteins are grouped into 10 distinct clades (A–J).
To examine the phylogenetic relationships of rice and other known bZIP transcription factors, another unrooted phylogenetic tree was constructed from the sequence alignments of their proteins (Supplemental Fig. S5). All the bZIP transcription factors could be divided into distinct clades. Interspecies clustering was observed, indicating that homologous bZIP transcription factors exist in rice and other plants. For example, it was found that OsbZIP proteins predicted to be G-box (group I) and C-box (group VII) binding in our analysis also clustered together with other known G-box (e.g. EmBP-1, ROM2, and SGBF-1) and C-box (e.g. TGA1a, MBF2, and HBP-1b) binding bZIP proteins from other plants, respectively (Supplemental Fig. S5). Most of the clades had both OsbZIP and AtbZIP proteins. This indicates that bZIP transcription factor genes have appeared before divergence between monocots and dicots. It is also worth mentioning that most of the OsbZIP proteins, which clustered together with AtbZIP proteins in the same clade, shared one or more additional conserved motifs outside the bZIP domain. Therefore, a majority of the OsbZIP proteins are expected to be orthologs of the AtbZIP proteins. Furthermore, a majority of the clades contain members from different plants, suggesting that the structure and function of most of the bZIP genes has probably remained conserved during angiosperm evolution.
Full-Length cDNA, EST, and Microarray-Based Expression Profiling of OsbZIP Genes in Various Tissues/Organs
To extract expression information for the rice bZIP transcription factors, a survey for the availability of any full-length cDNA (FL-cDNA) and/or EST in databases was done. This analysis was based on the information given on the gene expression evidence search page available at TIGR. It was found that one or more FL-cDNA and/or ESTs were available for 75 of 89 OsbZIP transcription factor genes (Supplemental Table S6). This shows that a very large percentage (84.27%) of these transcription factor genes is expressed. The total number of mapped EST sequences for OsbZIP transcription factors was highly variable, ranging from a minimum of one for a few OsbZIP genes (e.g. OsbZIP07) to a maximum of 277 (for OsbZIP38; Supplemental Table S6). This expression profiling was based on EST sequences obtained from various rice tissue/organ libraries, which in turn suggests that OsbZIP genes show organ/tissue-specific differential expression. Most of the 14 OsbZIP proteins for which we did not find any FL-cDNA and/or EST evidence in TIGR showed significant similarity with bZIP proteins from other plant species, as revealed by BLASTP and TBLASTN searches in various databases (Supplemental Table S6).
Another approach was used for expression profiling, which centered on the analysis of organ-specific expression of OsbZIP genes. This analysis was based on the use of microarray data from an earlier study (Ma et al., 2005), which described the expression pattern in several representative rice organs/tissues, including cultured cell, seedling, shoot, root, heading-stage panicle, and filling-stage panicle at the whole-genome level. Expression information for a total of 54 OsbZIP genes could be extracted. Further analysis demonstrated that 50 OsbZIP genes were expressed in at least one of the above-mentioned rice organs/tissues (Supplemental Table S7). Only 22 of these were expressed in all six organ/cell types and very few of them exhibited tissue/organ-specific expression (Supplemental Fig. S6). Overall expression data analysis signifies that bZIP transcription factors may play regulatory roles at multiple developmental stages in rice.
Microarray Analysis of Expression Patterns of OsbZIP Genes during Floral Transition, Panicle, and Seed Development
Microarray analysis was performed using Affymetrix rice whole-genome arrays to look at the gene expression profiles of OsbZIP transcription factor genes. Different rice tissues/organs and developmental stages (panicle and seed) were selected for microarray analysis, including seedling, seedling root, mature leaf, Y leaf (leaf subtending the shoot apical meristem [SAM]), SAM, and various stages of panicle (P1–P6) and seed (S1–S5) development, as described earlier (Jain et al., 2007). However, in addition to the six (P1–P6) different stages of panicle development, three more stages of early panicle development (P1-I–P1-III) were included in this analysis. These stages were characterized as follows: 0.5 to 2 mm (P1-I), branch meristem, spikelet meristem, formation of secondary and tertiary branches, and differentiation of rudimentary glumes; 2 to 5 mm (P1-II), floral meristem and floral organ initiation; and 5 to 10 mm (P1-III), early floral organ differentiation. These stage specifications are also approximations based on information from Itoh et al. (2005).
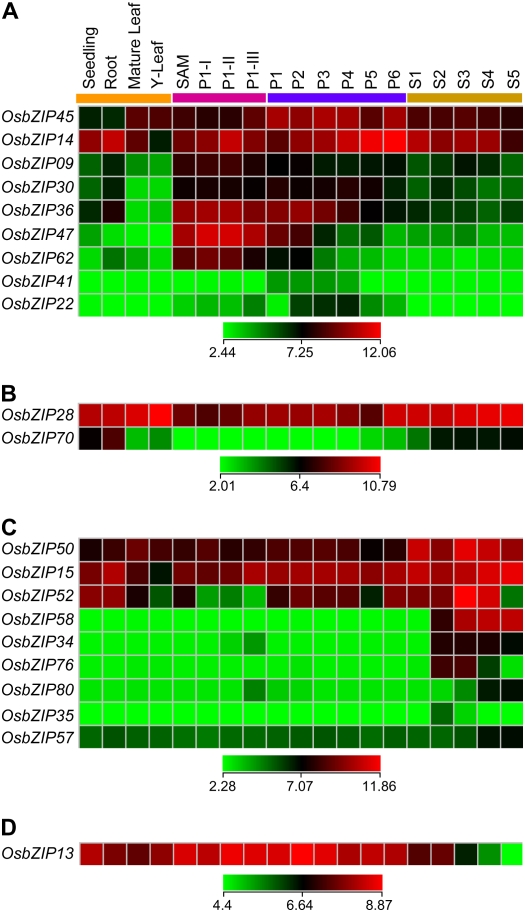
Whole-chip data were processed as described in “Materials and Methods.” To analyze the expression profiles of the OsbZIP genes, the log signal values of 85 OsbZIP genes represented on the array were extracted. Average log signal values for all 85 OsbZIP genes from three biological replicates of each sample are given in Supplemental Table S8 and the corresponding hierarchical cluster display of these genes is depicted in Supplemental Figure S7. On the basis of the signal values, it was evident that the majority of the OsbZIP genes are expressed in at least one of the rice vegetative organs and/or stages of development analyzed. This also includes at least nine of the 14 OsbZIP genes for which no EST or FL-cDNA could be detected from the data available at TIGR as described earlier. Of the remaining five OsbZIP genes, the Affymetrix IDs were not available for three genes and the other two had very low expression (Supplemental Fig. S7). With the aim of revealing OsbZIP gene expression profiles, both during panicle and seed development stages, differential expression analysis was performed. A gene was defined as differentially expressed at a given stage only if the expression level of the gene at that stage was significantly higher (>2-fold) with P value <0.05 in comparison to the levels at all the other stages. To facilitate the analysis, two separate data subsets were created having genes differentially expressed during panicle and seed development in comparison to seedling, root, mature leaf, and Y leaf as controls. We found that a total of 28 and 25 OsbZIP genes were showing differential expression patterns in at least one of the stages of panicle and seed development, respectively, as compared to vegetative organs. Out of these, there was an overlap of 13 OsbZIP genes between panicle and seed development. After excluding these 13 OsbZIP genes, those remaining were analyzed for preferential differential expression in any stage of panicle development as compared to seed development stages and vice versa. This resulted in the identification of 11 and 10 OsbZIP genes, which displayed a preferential differential expression profile in at least one of the stages of early/late panicle and seed development, respectively (Fig. 7). Further confirmation of some of the representative genes in panicle and seed development stages was done by real-time PCR analysis (Supplemental Fig. S8, A and B). This analysis highlights the potential role of different OsbZIP transcription factor genes during different stages of early and/or late panicle and seed development. These data have been discussed in some detail below in light of the earlier observations on the role of bZIP proteins in regulating reproductive development in plants.
Figure 7.
Expression profiles of OsbZIP genes differentially expressed during panicle and seed development. Expression profiles of nine up-regulated (A) and two down-regulated (B) OsbZIP genes in SAM or any stage of panicle development (P1-I–P1-III and P1–P6) as compared to vegetative reference tissues/organs (seedling, root, mature leaf, and Y leaf) and seed developmental stages (S1–S5) are presented. Expression profiles of nine up-regulated (C) and one down-regulated (D) OsbZIP genes in any stage of seed development (S1–S5) compared to vegetative reference tissues/organs (seedling, root, mature leaf, and Y leaf), SAM, and panicle developmental stages (P1-I–P6) are presented. The average log signal values of OsbZIP genes in various tissues/organs and developmental stages (mentioned at the top of each lane) are presented by cluster display. The color scale (representing log signal values) is shown at the bottom.
Molecular studies in both monocots and dicots have reported the involvement of bZIP transcription factors in transition from the vegetative to the reproductive phase, namely, FD in Arabidopsis (Abe et al., 2005; Wigge et al., 2005) and LIGULELESS2 (LG2) and DELAYED FLOWERING1 (DLF1) in maize (Zea mays; Walsh and Freeling, 1999; Muszynski et al., 2006). Our analysis also highlights a few OsbZIP genes with specific expression in SAM and early panicle (P1-I, P1-II, P1-III, and P1) development stages (Fig. 7, A and B). Such expression profiles suggest the possible involvement of OsbZIP transcription factors in regulating the expression pattern of genes involved in floral transition, determination of floral meristem, floral organ identity, and early panicle development in rice. A few bZIP transcription factors have also been linked with other aspects of flower development, namely, the PERIANTHIA (PAN) gene of Arabidopsis linked with floral organ number as well as their relative positions (Chuang et al., 1999); BZI proteins of tobacco (Nicotiana tabacum) involved in regulation of flower organ size (Strathmann et al., 2001); and TGA2.1 implicated in defining organ identity in tobacco flowers (Thurow et al., 2005). A few OsbZIP genes were also found to be differentially expressed in one or more stages of panicle development (P2–P6) as well (Fig. 7, A and B). These OsbZIP transcription factors, in turn, might be involved in the regulation of certain genes whose expression governs similar processes in rice.
In plants, quite a large number of the bZIP proteins characterized to date have been shown to play a role in the regulation of seed-specific genes, thereby linking them with different seed developmental processes (Izawa et al., 1994; Chern et al., 1996; Onodera et al., 2001). Interestingly, in cereals, bZIP proteins function as regulatory factors for genes encoding storage proteins in the endosperm (Schmidt et al., 1992; Vicente-Carbajosa et al., 1998; Onate et al., 1999). Our analysis also resulted in the identification of 10 OsbZIP genes that depicted differential expression in at least one or more of the developmental stages of seed in rice (Fig. 7, C and D), as described earlier. Out of these, nine OsbZIP genes displayed >2-fold increase in the transcript level during different stages of seed development (Fig. 7C), whereas a single OsbZIP gene (i.e. OsbZIP13) showed a decrease in the transcript level in the late stages (S4 and S5) of seed development (Fig. 7D). The seed-specific high-abundance expression profile of these nine OsbZIP genes could be correlated to their specific role in regulating the expression of genes involved in different stages of seed development, from embryo and endosperm development to seed maturation. OsbZIP58, up-regulated in stages from S2 to S5 in our study, corresponds to an earlier characterized seed-specific rice bZIP transcription factor, RISBZ1 (Onodera et al., 2001). On similar lines, genes like OsbZIP34 and OsbZIP76 can be proposed to have an important role in seed development. Two other rice bZIP transcription factors, one of them being RITA-1, have been shown to be highly expressed during seed development (Izawa et al., 1994); the other, REB, isolated from maturing seeds, has been shown to bind to the promoter of rice seed storage protein gene α-globulin (Nakase et al., 1997). Surprisingly, the OsbZIP transcription factor genes corresponding to RITA-1 and REB in our study (i.e. OsbZIP20 and OsbZIP33, respectively) do not show seed-specific expression. Their expression profiles (Supplemental Fig. S7) reveal that both are highly expressed throughout rice plant development. This probably indicates that these might either play multifunctional roles in other developmental processes in rice apart from being involved in seed development or may gain seed specificity as a result of heterodimerization and hence function to regulate seed-specific genes. This assumption is further supported by the fact that REB has a high degree of sequence similarity with maize bZIP factor OHP1, whose expression in not confined to seed only and is known to heterodimerize with another maize bZIP factor, OPAQUE2 (O2), to regulate storage protein synthesis in the endosperm (Schmidt et al., 1992; Nakase et al., 1997). Recently, it has been demonstrated that synergistic interaction exists between a rice Dof transcriptional activator, RPBF, and a bZIP activator, RISBZ1, which in turn regulates the expression of endosperm-specific genes in a quantitative manner (Yamamoto et al., 2006). Such interactions between other bZIP transcription factors and transcriptional regulators from other groups might exist in nature as well.
Furthermore, the relationship between the expression pattern of OsbZIP genes in one or more stages of panicle and/or seed development and sequence features as well as phylogeny was analyzed (Supplemental Table S9). It was found that a number of OsbZIP genes having the same conserved motifs and/or lying close in the phylogenetic tree had similar expression patterns. Few duplicated OsbZIP genes and those having same intron patterns were also found to be coexpressed in one or more stages of panicle and/or seed development. Some OsbZIP genes predicted to have similar binding affinities were also found to exhibit similar expression profiles in various panicle and/or seed developmental stages.
Light Regulation
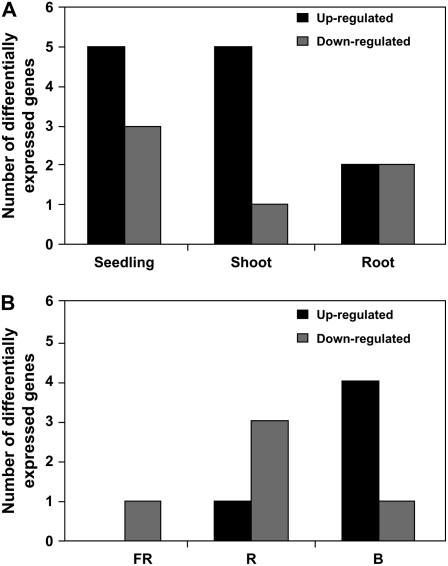
bZIP transcription factors have also been shown to regulate promoters of light-responsive genes. Among these are the Arabidopsis GBFs and parsley CPRF proteins, which have been shown to be light regulated either at the expression level or in their intracellular localization (Jakoby et al., 2002). To test whether such a light-responsive relationship exists in the case of OsbZIP genes and to identify the OsbZIP transcription factor genes regulated by light, analysis of the microarray data generated in an earlier study by Jiao et al. (2005) was carried out. This genomic study of rice gene expression in response to light revealed that different light qualities regulate a large number of rice genes. Expression values of only 47 OsbZIP genes could be retrieved under different light conditions in three different rice tissues/organs (i.e. seedling, shoot, and root). Applying a 1.5-fold threshold with a P value <0.05, examination of expression ratios between light- and dark-grown rice tissues/organs resulted in the identification of 14 differentially expressed OsbZIP genes, with nine up- and five down-regulated OsbZIP transcription factor genes in white light in comparison to dark (Fig. 8A; Supplemental Table S10). In contrast to white light, fewer (eight) OsbZIP transcription factor genes were regulated under specific monochromatic lights in rice seedlings with the maximal number of differentially expressed genes in the case of blue light, followed by red and far-red light (Fig. 8B; Supplemental Table S10). Among the different activated and/or repressed OsbZIP genes, certain genes are differentially regulated similarly under different light treatments (Supplemental Fig. S9, A and B). Two bZIP transcriptional activators, HY5 and HYH, have been shown to mediate transcription of light-inducible genes in Arabidopsis. HY5 has been well documented as a positive regulator of photomorphogenesis (Jakoby et al., 2002). Likewise, genetic analysis has linked HYH to blue-light-regulated expression of genes (Holm et al., 2002). The OsbZIP proteins OsbZIP01, OsbZIP18, and OsbZIP48 might represent closely related orthologs of Arabidopsis HY5 and/or HYH, as evident from the phylogenetic analysis as well as the presence of the additional conserved COP1 interaction motif specific to HY5 and HYH, and might perform similar functions in rice. Incidentally, expression data for OsbZIP01 and OsbZIP48 could not be retrieved because these genes were not represented on the microarray chip. However, OsbZIP18 was found to be down-regulated under red light. Expression data were also correlated with pattern of introns, gene duplications, motif analysis, predicted DNA-binding specificity, and phylogeny. A link could be seen in the case of OsbZIP23 and OsbZIP40 as well as OsbZIP28 and OsbZIP41 (Supplemental Table S9).
Figure 8.
Differential expression of OsbZIP genes under different light conditions based on microarray analysis from an earlier study (Jiao et al. 2005). A, Number of genes differentially expressed in various organs (mentioned below each bar) under white light. B, Number of differentially expressed genes in seedlings under different light conditions (mentioned below each bar; F, far-red; R, red; B, blue).
Because a reasonable number of OsbZIP genes were displaying light-dependent differential expression, it can be assumed that other OsbZIP transcription factors, which might be involved in light signal transduction, may respond differently to light as depicted by some other bZIP transcription factors. The above analysis is, however, a first step toward understanding the light-responsive nature of OsbZIP genes in terms of their expression level. Extensive studies on the light-dependent localization as well as binding patterns of OsbZIP transcription factors would be important to understand their role in regulating expression of light-responsive genes.
Expression of OsbZIP Genes under Abiotic Stress Conditions
The role of the phytohormone ABA in the adaptation of plants to various abiotic environmental stress conditions, like drought, high salinity, and cold stress, has been well characterized. The action of ABA is dependent on the modification of ABA-regulated genes, which possess an ABA-responsive element (ABRE) or certain ABRE-like sequences in their promoter region (Busk and Pages, 1998). Interestingly, all the transcription factors reported to date, which bind to ABRE or ABRE-like sequences, come under the bZIP class of transcription factors (Busk and Pages, 1998). It is worth mentioning that a recent study reports that three bZIP transcription factor genes, AREB1/ABF2, AREB2/ABF4, and ABF3/DPBF5, depict ABA-, drought-, and high-salinity-inducible expression in vegetative tissues in Arabidopsis (Fujita et al., 2005).
To explore the possible involvement of OsbZIP transcription factors in the regulation of abiotic stress-related genes, the expression pattern of OsbZIP genes was analyzed under abiotic stress conditions, based on microarray analysis performed with total RNA isolated from rice seedlings subjected to dehydration, salinity, and cold treatment. Examination of microarray data indicated that at least 37 OsbZIP genes displayed differential expression. We found that 26 genes were up-regulated and 11 genes were down-regulated by at least 2-fold, with a P value <0.05, under one or more of the above-mentioned stress treatments (Fig. 9, A and B). Two OsbZIP genes were up-regulated under all three stress conditions. One of these, OsbZIP45, represents the closely related ortholog of maize GBF1, which has been shown to be induced by hypoxia (de Vetten and Ferl, 1995). Interestingly, of the 18 OsbZIP genes differentially regulated under any two stress conditions, 16 OsbZIP genes were differentially expressed under dehydration and salt stresses but not under cold stress (Fig. 9; Supplemental Table S11). As far as alteration in expression to independent stress treatments was concerned, 13 OsbZIP genes showed differential expression (10 up- and three down-regulated) under dehydration stress and four OsbZIP genes showed differential expression (two up- and two down-regulated) under cold, whereas no OsbZIP gene depicted differential expression under salt stress alone (Fig. 9, C and D). It should be noted that OsbZIP38 (up-regulated under cold treatment) corresponds to the earlier identified rice low-temperature-induced LIP19 gene (Shimizu et al., 2005). This confirms the reliability of our expression data. Additionally, the differential expression of some representative genes under abiotic stress conditions (as deduced from microarray data) was confirmed by real-time PCR results (Supplemental Fig. S8C).
Figure 9.
Expression profiles of OsbZIP genes differentially expressed under abiotic stress conditions. Expression profiles of 26 significantly up-regulated (A) and 11 significantly down-regulated (B) OsbZIP genes as compared to the control seedlings. The average log signal values of OsbZIP genes under control and various stress conditions (mentioned at the top of each lane) are presented by cluster display (values are given in Supplemental Table S11). Only those genes that exhibited 2-fold or more differential expression with a P value ≤0.05 under any of the given abiotic stress conditions are shown. The color scale (representing log signal values) is shown at the bottom of each. C and D, Venn diagram of the number of differentially expressed OsbZIP genes under abiotic stress conditions. Number of up-regulated (C) and down-regulated (D) OsbZIP genes in any of the abiotic stress conditions as compared to control seedlings are given.
The transcript levels of ABA-inducible bZIP transcription factor genes OSBZ8 (OsbZIP05) and TRAB1 (OsbZIP66; Nakagawa et al., 1996; Hobo et al., 1999a) were up-regulated under dehydration and salt stress conditions in our study, indicating the involvement of these genes in the ABA-dependent stress signal transduction pathway. This observation is consistent with earlier reports in the literature on drought stress-inducible genes also being activated by ABA (Ingram and Bartels, 1996). Further, a characteristic link was observed between sequence features, phylogeny, and expression pattern of the OsbZIP genes whose expression was altered under stress conditions (Supplemental Table S9). It was found that a majority of OsbZIP genes possessing additional conserved motifs representing potential CKII and/or Ca2+-dependent protein kinase phosphorylation sites (motifs 10, 11, 12, 13, 14), confined to group VI of predicted DNA-binding specificity and proposed to be involved in stress and/or ABA signaling, are actually up-regulated under stress conditions. Thus, it would be interesting to analyze these OsbZIP transcription factors in the context of stress signaling.
Recently, it has been demonstrated that an extensive gene set is shared among pollination/fertilization and/or seed development and stress responses like dehydration and wounding in rice (Cooper et al., 2003). To find out whether OsbZIP transcription factors are also a part of such interrelated genetic programs, a cross-comparison between the differentially expressed OsbZIP genes under different stress treatments and different stages of panicle and/or seed development was made. Such a cross-comparison resulted in the identification of 11 OsbZIP genes up-regulated under stress conditions (OsbZIP05, 10, 25, 30, 34, 45, 46, 57, 68, 78, and 80), which had an overlapping expression pattern at one or more stages of panicle and/or seed development. Two other genes, OsbZIP23 (up-regulated under all the three stress conditions) and OsbZIP65 (down-regulated under dehydration and cold stresses), were found to be down-regulated in one or more stages of both panicle and seed development. Thus, it is evident that OsbZIP genes might form a subset within the genes shared among stress and developmental responses and these overlapping OsbZIP transcription factors might be common to more than one signal transduction pathway.
To find out whether OsbZIP genes coexpressed during abiotic stress and various stages of panicle and/or seed development encode for potential heterodimerizing partners, their expression pattern was integrated with predicted dimerization properties. It was observed that genes depicting coexpression profiles, whether belonging to the same or different subfamily, represent good candidates for heterodimerization in planta (Supplemental Table S12). However, the possibility of heterodimerization of products of OsbZIP genes, which do not show coexpression, cannot be ruled out completely because the features in the Leu zipper region governing heterodimerization are equally important. Only experimental evidence will provide credibility to these presumptions.
CONCLUSION
bZIP transcription factors have been characterized in different plant species and linked to various developmental and physiological processes. An extensive study of 89 rice bZIP genes has been performed in terms of structure, phylogeny, sequence, and expression analyses. These bZIP transcription factors could be divided into 11 groups on the basis of their predicted DNA-binding specificity, many of them supported by the presence of additional conserved motifs outside the bZIP domain. We also defined 29 different subfamilies based on their complex predicted dimerization patterns. Their predicted DNA-binding specificity and dimerization patterns would facilitate both in vitro and in vivo studies to reveal their binding and dimerization patterns. Structural differences between different OsbZIP members have been studied and an attempt made to link them to their functional roles in rice in a meaningful manner. Phylogenetic analysis of bZIP transcription factor-encoding genes in rice and other plant species also provides useful information on their conserved functional roles. Expression data further support the role of bZIP proteins in performing diverse developmental and physiological functions during floral transition, panicle, and seed development, as well as abiotic stresses and light signaling in rice. Although the function of only a few rice bZIP genes is known to date, our analyses provide a platform for a more detailed functional analysis of bZIP genes in rice.
MATERIALS AND METHODS
Database Search for Rice bZIP Transcription Factors
For identifying the possible bZIP transcription factor genes in rice (Oryza sativa), a BLAST search of all the annotated proteins in the rice genome at TIGR (release 5) was carried out using the HMM profile (build 2.3.2) of the bZIP domain as query. The HMM profile of the 65-amino acid-long bZIP domain was generated by alignments of 73 known Arabidopsis (Arabidopsis thaliana) bZIP proteins and 71 rice protein sequences that were showing the presence of a bZIP domain. This search resulted in the identification of 130 proteins (including proteins corresponding to different gene models present at the same locus) in rice, with a positive score cutoff of 0.0. Of these 130 proteins, those corresponding to different gene models present at the same locus were removed and those remaining were checked by SMART for the presence of the bZIP domain. Ninety proteins were predicted to have a bZIP domain with confidence (E value <1.0). One protein (i.e. LOC_Os12g06010), annotated as a retrotransposon, was also removed from the analysis.
Sequence Analysis
Additional conserved motifs outside the bZIP domain were identified by the MEME analysis tool (http://meme.sdsc.edu/meme/meme.html), version 3.5.4, which is widely used for analyzing a group of protein sequences to identify motifs in them. At first, all 89 OsbZIP protein sequences were given together as input and a limit of 50 motifs was specified with all other parameters set to default. These motifs were analyzed manually based on E-value cutoff <e-001 and those motifs were considered significant that were shared by the majority of OsbZIP proteins classified into the same group, according to their DNA-binding-site specificity. The ClustalX (version 1.83) program was used to perform multiple sequence alignments. The unrooted phylogenetic trees were generated by the neighbor-joining method and displayed using the NJPLOT program. To obtain information on the intron/exon structure, cDNA sequences of OsbZIP proteins were aligned with their corresponding genomic sequences using Spidey (http://www.ncbi.nlm.nih.gov/spidey). Information on intron distribution pattern and intron splicing phase within the basic and hinge regions of the bZIP domains was also derived from the aligned cDNA sequences.
Localization of OsbZIP Genes on Rice Chromosomes and Segmental Duplication
Each of the OsbZIP genes was positioned on rice chromosome pseudomolecules available at TIGR (http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml) by the BLASTN search. Segmental genome duplication of rice available at TIGR (http://www.tigr.org./tdb/e2k1/osa1/semental_dup/index.shtml) was used to determine the presence of OsbZIP genes on duplicated chromosomal segments, with the maximal length distance permitted between collinear gene pairs of 500 kb.
Plant Material and Growth Conditions
Rice (sp. indica var. IR64) seeds obtained from the Indian Agricultural Research Institute were used for this study. Plant material for different developmental stages and their growth conditions for microarray analysis are the same as described previously (Jain et al., 2007). Three additional early panicle development stages (i.e. 0.5–2.0 mm, P1-I; 2.1–5.0 mm, P1-II; and 5.1–10.0 mm, P1-III) were taken. For collection of different stages of early panicle development, leaves and stem were carefully removed and growth stage of each sample was measured in millimeters as well as carefully examined under the microscope before sampling. For stress series, 7-d-old light-grown rice seedlings were dried for 3 h (dehydration stress), treated with 200 mm NaCl for 3 h (salt stress), and kept at 4°C ± 1°C for 3 h (cold stress) as described previously (Jain et al., 2007).
Microarray Hybridization and Data Analysis
Isolation of total RNA and quality controls was done as described previously (Jain et al., 2006). Microarray analysis was performed using one-cycle target-labeling and control reagents (Affymetrix) using 5 μg of high-quality total RNA as starting material for each sample as described earlier (Jain et al., 2007). Affymetrix GeneChip Rice Genome Arrays (Gene Expression Omnibus platform accession no. GPL2025) were used for microarray analysis.
For microarray data analysis, the image (cel) files were imported into ArrayAssist (version 5.0) software. Three biological replicates of each sample with an overall correlation coefficient value >0.94 were selected for final analysis. Data from 66 hybridizations were included in the final analysis. Two separate projects were created, one for the developmental series that included data from 57 chips (three biological replicates for each of seedling, root, mature leaf, Y leaf, SAM, P1-I, P1-II, P1-III, P1, P2, P3, P4, P5, P6, S1, S2, S3, S4, and S5) and the other for stress series that included data from 12 chips (three biological replicates for each of control seedling and seedlings subjected to salt, dehydration, and cold stress). Data for three hybridizations corresponding to control seedlings were common to both experiments (developmental and stress).
The normalization and probe summarization for all the rice genes present on the chip in all the tissue samples analyzed was performed by Gene Chip Robust Multiarray Analysis algorithm followed by log transformation of data. Average log signal intensity values of three biological replicates for each sample were then computed and these were used for further analysis. To identify differentially expressed genes, Student's t test was performed with microarray data obtained for mRNA derived from seedling, mature leaf, Y leaf, and root as reference tissues/organs. After this, the log signal intensity values for rice probe set IDs corresponding to OsbZIP genes were extracted into a separate dataset. IDs of probe sets present on the Affymetrix rice genome array representing OsbZIP genes were extracted using the genome browser tool available at TIGR (http://www.tigr.org/tigr-scripts/osa1-web/gbrowse/rice). Data for only one probe set for each OsbZIP gene were used for expression analysis. The probes for 85 (of 89) OsbZIP genes could be identified that were represented on the Affymetrix rice genome array (probe set IDs corresponding to each OsbZIP gene have been listed in Supplemental Table S8). Two genes, namely, OsbZIP55 and OsbZIP56, showed 100% sequence identity. Hence, both have the same probe set on the GeneChip. All subsequent analyses were performed on this subset of data only. Genes that were up- or down-regulated ≥2-fold with a P value ≤0.05 (after applying Benjamini and Hochberg correction) in any stage of panicle and/or seed development as compared to reference tissues/organs were considered to be differentially expressed significantly. A gene was defined as specifically enriched in a given organ only if the expression level of the gene in the organ was significantly higher (>2-fold) than the levels in all the other organs. A similar approach was followed in the case of stress series.
Real-Time PCR Analysis
Verification of microarray data was done by checking the differential expression profiles of a few of the representative OsbZIP genes in various rice tissues/developmental stages and stress treatments by real-time PCR analysis using gene-specific primers as described earlier (Jain et al., 2006). Real-time PCR analysis was based on at least two biological replicates of each sample and three technical replicates of each biological replicate. Supplemental Table S13 includes the primer sequences used for real-time PCR analysis.
FL-cDNA and EST-Based Expression Profiling
For FL-cDNA and EST-based expression profiling, the gene expression evidence search page (http://www.tigr.org/tdb/e2k1/osa1/locus_expression_evidence.shtml) available at TIGR rice genome annotation was used. TIGR locus IDs corresponding to the OsbZIP genes were searched to find the availability of corresponding FL-cDNA and ESTs. FL-cDNA and EST sequences showed minimal alignment over 90% length of the transcript with 95% identity.
Microarray data from this article can be found in the Gene Expression Omnibus database at the NCBI under accession numbers GSE6893 and GSE6901.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Position and pattern of introns within the basic and hinge regions of the bZIP domains of the OsbZIP transcription factors.
Supplemental Figure S2. Alignment of basic and hinge regions of OsbZIP proteins.
Supplemental Figure S3. Amino acid sequence alignment of the Leu zipper region of 89 OsbZIP proteins.
Supplemental Figure S4. Phylogenetic relationship among the OsbZIP proteins based on (a) bZIP domain; (b) basic and hinge regions; and (c) Leu zipper defined from the first Leu.
Supplemental Figure S5. Phylogenetic relationship among the OsbZIP proteins and other known plant bZIP proteins.
Supplemental Figure S6. Organ-specific expression of OsbZIP genes based on the microarray analysis from an earlier study (Ma et al., 2005).
Supplemental Figure S7. Hierarchical clustering display of 85 OsbZIP genes represented on an Affymetrix rice genome array in various rice organs and developmental stages (mentioned at the top of each lane).
Supplemental Figure S8. Real-time PCR analysis of representative OsbZIP genes differentially expressed in various tissues/developmental stages and stress treatments.
Supplemental Figure S9. Venn diagrams representing a number of differentially regulated OsbZIP genes in various organs under different light conditions based on microarray analysis from an earlier study (Jiao et al., 2005).
Supplemental Table S1. bZIP transcription factors identified in plants.
Supplemental Table S2. OsbZIP transcription factor genes.
Supplemental Table S3. OsbZIP genes present on duplicated chromosomal segments of rice.
Supplemental Table S4. Additional conserved motifs in OsbZIP proteins as predicted by MEME.
Supplemental Table S5. Classification of OsbZIP proteins into subfamilies with similar predicted dimerization specificity.
Supplemental Table S6. Availability of FL-cDNA and/or ESTs corresponding to OsbZIP genes.
Supplemental Table S7. OsbZIP genes expressed in at least one of the six organs or cell types.
Supplemental Table S8. Average log signal values of 85 OsbZIP genes from three biological replicates of each sample.
Supplemental Table S9. Relationship between expression pattern of OsbZIP genes and intron pattern, conserved motifs, gene duplication, predicted DNA-binding specificity, and phylogenetic analysis.
Supplemental Table S10. OsbZIP genes differentially expressed under different light conditions.
Supplemental Table S11. Average log signal values of 37 OsbZIP genes differentially expressed under various abiotic stress conditions.
Supplemental Table S12. Relationship between expression pattern of OsbZIP genes and heterodimerization of OsbZIP transcription factors.
Supplemental Table S13. Primer sequences used for real-time PCR analysis.
Supplementary Material
This work was supported by the Department of Biotechnology, Government of India, the University Grants Commission, and the Council of Scientific and Industrial Research, New Delhi (research fellowship to A.N.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jitendra P. Khurana (khuranaj@genomeindia.org).
The online version of this article contains Web-only data.
Open Access article can be viewed online without a subscription.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052–1056 [DOI] [PubMed] [Google Scholar]
- Acharya A, Ruvinov SB, Gal J, Moll JR, Vinson C (2002) A heterodimerizing leucine zipper coiled coil system for examining the specificity of a position interactions: amino acids I, V, L, N, A, and K. Biochemistry 41 14122–14131 [DOI] [PubMed] [Google Scholar]
- Aeschbacher RA, Schrott M, Potrykus I, Saul MW (1991) Isolation and molecular characterization of PosF21, an Arabidopsis thaliana gene which shows characteristics of a b-Zip class transcription factor. Plant J 1 303–316 [PubMed] [Google Scholar]
- Aguan K, Sugawara K, Suzuki N, Kusano T (1993) Low-temperature-dependent expression of a rice gene encoding a protein with a leucine-zipper motif. Mol Gen Genet 240 1–8 [DOI] [PubMed] [Google Scholar]
- Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F, Giraudat J, Parcy F (2002) The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk PK, Pages M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37 425–435 [DOI] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Yoo CM, Park JM, Ryu GR, Goekjian VH, Nagao RT, Key JL, Cho MJ, Hong JC (1998) STF1 is a novel TGACG-binding factor with a zinc-finger motif and a bZIP domain which heterodimerizes with GBF proteins. Plant J 15 199–209 [DOI] [PubMed] [Google Scholar]
- Chern MS, Eiben HG, Bustos MM (1996) The developmentally regulated bZIP factor ROM1 modulates transcription from lectin and storage protein genes in bean embryos. Plant J 10 135–148 [DOI] [PubMed] [Google Scholar]
- Chuang CF, Running MP, Williams RW, Meyerowitz EM (1999) The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev 13 334–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper B, Clarke JD, Budworth P, Kreps J, Hutchison D, Park S, Guimil S, Dunn M, Luginbuhl P, Ellero C, et al (2003) A network of rice genes associated with stress response and seed development. Proc Natl Acad Sci USA 100 4945–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S, Petruccelli S, Ordiz MI, Zhang Z, Chen S, Beachy RN (2003) Functional analysis of RF2a, a rice transcription factor. J Biol Chem 278 36396–36402 [DOI] [PubMed] [Google Scholar]
- Dai S, Zhang Z, Chen S, Beachy RN (2004) RF2b, a rice bZIP transcription activator, interacts with RF2a and is involved in symptom development of rice tungro disease. Proc Natl Acad Sci USA 101 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vetten NC, Ferl RJ (1995) Characterization of a maize G-box binding factor that is induced by hypoxia. Plant J 7 589–601 [DOI] [PubMed] [Google Scholar]
- Deppmann CD, Acharya A, Rishi V, Wobbes B, Smeekens S, Taparowsky EJ, Vinson C (2004) Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: a comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res 32 3435–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger TE, Brandl CJ, Struhl K, Harrison SC (1992) The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell 71 1223–1237 [DOI] [PubMed] [Google Scholar]
- Fassler J, Landsman D, Acharya A, Moll JR, Bonovich M, Vinson C (2002) B-ZIP proteins encoded by the Drosophila genome: evaluation of potential dimerization partners. Genome Res 12 1190–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster R, Izawa T, Chua NH (1994) Plant bZIP proteins gather at ACGT elements. FASEB J 8 192–200 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J, Sakai T, Ishida S, Yamaguchi I, Kamiya Y, Takahashi Y (2000) Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobo T, Asada M, Kowyama Y, Hattori T (1999. b) ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J 19 679–689 [DOI] [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hattori T (1999. a) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci USA 96 15348–15353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Hardtke CS, Gaudet R, Deng XW (2001) Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J 20 118–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst HC (1994) Transcription factors. 1: bZIP proteins. Protein Profile 1 123–168 [PubMed] [Google Scholar]
- Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47 377–403 [DOI] [PubMed] [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46 23–47 [DOI] [PubMed] [Google Scholar]
- Izawa T, Foster R, Chua NH (1993) Plant bZIP protein DNA binding specificity. J Mol Biol 230 1131–1144 [DOI] [PubMed] [Google Scholar]
- Izawa T, Foster R, Nakajima M, Shimamoto K, Chua NH (1994) The rice bZIP transcriptional activator RITA-1 is highly expressed during seed development. Plant Cell 6 1277–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Khurana P, Tyagi AK, Khurana JP (2008) Genome-wide analysis of intronless genes in rice and Arabidopsis. Funct Integr Genomics 8 69–78 [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP (2007) F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143 1467–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345 646–651 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7 106–111 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Ma L, Strickland E, Deng XW (2005) Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 17 3239–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F, Lam E, Chua NH (1989) Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature 340 727–730 [DOI] [PubMed] [Google Scholar]
- Landschulz WH, Johnson PF, McKnight SL (1988) The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240 1759–1764 [DOI] [PubMed] [Google Scholar]
- Lohmer S, Maddaloni M, Motto M, Di Fonzo N, Hartings H, Salamini F, Thompson RD (1991) The maize regulatory locus Opaque-2 encodes a DNA-binding protein which activates the transcription of the b-32 gene. EMBO J 10 617–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32 317–328 [DOI] [PubMed] [Google Scholar]
- Ma L, Chen C, Liu X, Jiao Y, Su N, Li L, Wang X, Cao M, Sun N, Zhang X, et al (2005) A microarray analysis of the rice transcriptome and its comparison to Arabidopsis. Genome Res 15 1274–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallappa C, Yadav V, Negi P, Chattopadhyay S (2006) A basic leucine zipper transcription factor, G-box-binding factor 1, regulates blue light-mediated photomorphogenic growth in Arabidopsis. J Biol Chem 281 22190–22199 [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Moyano E, Alcocer MJ, Martin C (1998) Two bZIP proteins from Antirrhinum flowers preferentially bind a hybrid C-box/G-box motif and help to define a new sub-family of bZIP transcription factors. Plant J 13 489–505 [DOI] [PubMed] [Google Scholar]
- Meier I, Gruissem W (1994) Novel conserved sequence motifs in plant G-box binding proteins and implications for interactive domains. Nucleic Acids Res 22 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami M, Huh GH, Yang P, Iwabuchi M (1993) Coordinate gene expression of five subclass histones and the putative transcription factors, HBP-1a and HBP-1b, of histone genes in wheat. Plant Mol Biol 23 429–434 [DOI] [PubMed] [Google Scholar]
- Muszynski MG, Dam T, Li B, Shirbroun DM, Hou Z, Bruggemann E, Archibald R, Ananiev EV, Danilevskaya ON (2006) delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol 142 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Ohmiya K, Hattori T (1996) A rice bZIP protein, designated OSBZ8, is rapidly induced by abscisic acid. Plant J 9 217–227 [DOI] [PubMed] [Google Scholar]
- Nakase M, Aoki N, Matsuda T, Adachi T (1997) Characterization of a novel rice bZIP protein which binds to the alpha-globulin promoter. Plant Mol Biol 33 513–522 [DOI] [PubMed] [Google Scholar]
- Nantel A, Quatrano RS (1996) Characterization of three rice basic/leucine zipper factors, including two inhibitors of EmBP-1 DNA binding activity. J Biol Chem 271 31296–31305 [DOI] [PubMed] [Google Scholar]
- Niggeweg R, Thurow C, Kegler C, Gatz C (2000) Tobacco transcription factor TGA2.2 is the main component of as-1-binding factor ASF-1 and is involved in salicylic acid- and auxin-inducible expression of as-1-containing target promoters. J Biol Chem 275 19897–19905 [DOI] [PubMed] [Google Scholar]
- Nishimura R, Ohmori M, Fujita H, Kawaguchi M (2002) A Lotus basic leucine zipper protein with a RING-finger motif negatively regulates the developmental program of nodulation. Proc Natl Acad Sci USA 99 15206–15210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X, Renshaw-Gegg L, Miller L, Guiltinan MJ (1999) Bipartite determinants of DNA-binding specificity of plant basic leucine zipper proteins. Plant Mol Biol 41 1–13 [DOI] [PubMed] [Google Scholar]
- Okanami M, Meshi T, Tamai H, Iwabuchi M (1996) HALF-1, a bZIP-type protein, interacting with the wheat transcription factor HBP-1a contains a novel transcriptional activation domain. Genes Cells 1 87–99 [DOI] [PubMed] [Google Scholar]
- Onate L, Vicente-Carbajosa J, Lara P, Diaz I, Carbonero P (1999) Barley BLZ2, a seed-specific bZIP protein that interacts with BLZ1 in vivo and activates transcription from the GCN4-like motif of B-hordein promoters in barley endosperm. J Biol Chem 274 9175–9182 [DOI] [PubMed] [Google Scholar]
- Ono A, Izawa T, Chua NH, Shimamoto K (1996) The rab16B promoter of rice contains two distinct abscisic acid-responsive elements. Plant Physiol 112 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Suzuki A, Wu CY, Washida H, Takaiwa F (2001) A rice functional transcriptional activator, RISBZ1, responsible for endosperm-specific expression of storage protein genes through GCN4 motif. J Biol Chem 276 14139–14152 [DOI] [PubMed] [Google Scholar]
- O'Shea EK, Rutkowski R, Kim PS (1992) Mechanism of specificity in the Fos-Jun oncoprotein heterodimer. Cell 68 699–708 [DOI] [PubMed] [Google Scholar]
- Patthy L (1987) Intron-dependent evolution: preferred types of exons and introns. FEBS Lett 214 1–7 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290 2105–2110 [DOI] [PubMed] [Google Scholar]
- Ringli C, Keller B (1998) Specific interaction of the tomato bZIP transcription factor VSF-1 with a non-palindromic DNA sequence that controls vascular gene expression. Plant Mol Biol 37 977–988 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Uribe L, O'Connell MA (2006) A root-specific bZIP transcription factor is responsive to water deficit stress in tepary bean (Phaseolus acutifolius) and common bean (P. vulgaris). J Exp Bot 57 1391–1398 [DOI] [PubMed] [Google Scholar]
- Schindler U, Menkens AE, Beckmann H, Ecker JR, Cashmore AR (1992. a) Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J 11 1261–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U, Terzaghi W, Beckmann H, Kadesch T, Cashmore AR (1992. b) DNA binding site preferences and transcriptional activation properties of the Arabidopsis transcription factor GBF1. EMBO J 11 1275–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RJ, Ketudat M, Aukerman MJ, Hoschek G (1992) Opaque-2 is a transcriptional activator that recognizes a specific target site in 22-kD zein genes. Plant Cell 4 689–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H, Sato K, Berberich T, Miyazaki A, Ozaki R, Imai R, Kusano T (2005) LIP19, a basic region leucine zipper protein, is a Fos-like molecular switch in the cold signaling of rice plants. Plant Cell Physiol 46 1623–1634 [DOI] [PubMed] [Google Scholar]
- Strathmann A, Kuhlmann M, Heinekamp T, Droge-Laser W (2001) BZI-1 specifically heterodimerises with the tobacco bZIP transcription factors BZI-2, BZI-3/TBZF and BZI-4, and is functionally involved in flower development. Plant J 28 397–408 [DOI] [PubMed] [Google Scholar]
- Suckow M, von Wilcken-Bergmann B, Muller-Hill B (1993. a) Identification of three residues in the basic regions of the bZIP proteins GCN4, C/EBP and TAF-1 that are involved in specific DNA binding. EMBO J 12 1193–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckow M, von Wilcken-Bergmann B, Muller-Hill B (1993. b) The DNA binding specificity of the basic region of the yeast transcriptional activator GCN4 can be changed by substitution of a single amino acid. Nucleic Acids Res 21 2081–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Jacquemin I, Surdin-Kerjan Y (1992) MET4, a leucine zipper protein, and centromere-binding factor 1 are both required for transcriptional activation of sulfur metabolism in Saccharomyces cerevisiae. Mol Cell Biol 12 1719–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurow C, Schiermeyer A, Krawczyk S, Butterbrodt T, Nickolov K, Gatz C (2005) Tobacco bZIP transcription factor TGA2.2 and related factor TGA2.1 have distinct roles in plant defense responses and plant development. Plant J 44 100–113 [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Onate L, Lara P, Diaz I, Carbonero P (1998) Barley BLZ1: a bZIP transcriptional activator that interacts with endosperm-specific gene promoters. Plant J 13 629–640 [DOI] [PubMed] [Google Scholar]
- Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M (2002) Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol 22 6321–6335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh J, Freeling M (1999) The liguleless2 gene of maize functions during the transition from the vegetative to the reproductive shoot apex. Plant J 19 489–495 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 1056–1059 [DOI] [PubMed] [Google Scholar]
- Yamamoto MP, Onodera Y, Touno SM, Takaiwa F (2006) Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiol 141 1694–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Wu L, Hwang YS, Chen L, Huang N (2001) Expression of the REB transcriptional activator in rice grains improves the yield of recombinant proteins whose genes are controlled by a Reb-responsive promoter. Proc Natl Acad Sci USA 98 11438–11443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Chen L, Beachy R (1997. a) Promoter elements required for phloem-specific gene expression from the RTBV promoter in rice. Plant J 12 1179–1188 [DOI] [PubMed] [Google Scholar]
- Yin Y, Zhu Q, Dai S, Lamb C, Beachy RN (1997. b) RF2a, a bZIP transcriptional activator of the phloem-specific rice tungro bacilliform virus promoter, functions in vascular development. EMBO J 16 5247–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Tessaro MJ, Lassner M, Li X (2003) Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15 2647–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.