Abstract
To understand the interaction between the virus and its host, we used three sources of cDNA microarrays to examine the expression of 12,309 unique genes at 6 h postinfection of HeLa cells with high multiplicities of adenovirus type 2. Seventy-six genes with significantly changed expression ratios were identified, suggesting that adenovirus only modulates expression of a limited set of cellular genes. Quantitative real-time PCR analyses on selected genes were performed to confirm the microarray results. Significantly, a pronounced transcriptional activation by the promiscuous E1A-289R transcriptional activator was not apparent. Instead, promoter sequences in 45% of the upregulated genes harbored a potential E2F binding site, suggesting that the ability of the amino-terminal domain of E1A to regulate E2F-dependent transcription may be a major pathway for regulation of cellular gene expression. CDC25A was the only upregulated gene directly involved in cell cycle control. In contrast, several genes implicated in cell growth arrest were repressed. The transforming growth factor beta superfamily was specifically affected in the expression of both the upstream ligand and an intracellular regulator. In agreement with previous reports, adenovirus also targeted the innate immune response by downregulating several cytokines, including CLL2, CXCL1, and interleukin-6. Finally, stress response genes encoding GADD45B, ATF3, and TP53AP1 were upregulated. Importantly, we also found a novel countermeasure—activation of the apoptosis inhibitor survivin.
During a virus infection, reprogramming of the host cell occurs for mainly two reasons. First, the virus needs to establish optimal conditions for replication to ensure efficient production of progeny virus. Second, the virus must interfere with the host cell antiviral defense mechanisms to maximize the likelihood of escape and spread of the progeny virus. Adenovirus expresses regulatory proteins from early region 1A (E1A), E1B, E3, and E4 to achieve these goals. The immediate-early E1A gene encodes two primary regulators of viral and cellular gene expression, the E1A-243R and E1A-289R proteins (73). Four conserved regions (CRs) have been identified in E1A. CR1 and CR2 are present in both E1A-243R and E1A-289R, CR3 is unique to E1A-289R, and CR4 is the recently defined carboxy-terminal domain that interacts with the transcriptional corepressor CtBP (18).
The E1A-289R protein is required for transcriptional activation of all viral genes but can also act as a promiscuous transcriptional activator of cellular genes (27). Several mechanisms by which the CR3 of E1A-289R modulates gene expression have been described (3), including targeting of the basal transcription machinery and specific transcription factors. Recently, it was also found that the Mediator complex is required for E1A-289R transactivation (86) and that E1A-289R associates with the Mediator complexes in adenovirus-infected cells (96).
CR1 and CR2, together with the extreme N-terminal region of E1A, are essential to force the host cell to enter the S phase of the cell cycle to provide an optimal environment for viral replication (8). The cell cycle-inducing capacity results partly from the ability of E1A to disrupt a series of inhibitory complexes between members of the retinoblastoma tumor suppressor (pRb) family and the transcription factor E2F family (20), leading to deregulated expression of E2F-dependent genes. Moreover, E1A-induced cell proliferation also involves interaction with chromatin-modifying and transcriptional coactivator complexes. Importantly, coactivators act as general transcriptional integrators, mediating communication between basal, specific, and modifying units of the transcription machinery (16).
Mechanistically, the interaction between E1A and the coactivators p300/CBP (5, 7) has been suggested to disrupt the histone acetyltransferase activity of p300/CBP and their associated factor PCAF (15, 78), leading to decreased transcription from a variety of different genes, including those involved in growth arrest (64), cell differentiation (11, 14), and immune evasion (10). Although the effect of p300/CBP on cell growth seems to be context-dependent (31), p300 was recently shown to cause a premature G1 exit (49). In addition, E1A interacts with TRRAP, which is a component of three distinct histone acetyltransferase complexes (25, 28, 70). Thus, E1A has the capacity to interact with multiple histone acetyltransferase complexes and recruit these to viral or selected cellular promoters. The capacity of E1A to suppress histone acetyltransferase activities is still controversial (2), and E1A-associated histone acetyltransferase activity was recently shown to require intact binding sites for the above-mentioned histone acetyltransferase complexes (51).
As a possible side effect of S-phase-stimulatory activities, pRb and p300/CBP binding to E1A promotes p53 accumulation and consequently p53-dependent apoptosis (23, 60). E1B-55K plays a major role in counteracting the proapoptotic program. First, E1B-55K can bind to p53 and actively repress p53-dependent transcription (98), possibly by recruiting transcriptional corepressor complexes (76). Second, E1B-55K binds and promotes degradation of p53 through an E4orf6-E3 ubiquitin ligase complex (77). Importantly, E1A can also counteract its own induction of p53 apoptosis by blocking p53 transcriptional activation through sequestering of p300/CBP (82).
Three proteins encoded by the E3 transcription unit, 14.7K, 10.4K, and 14.5K, also inhibit apoptosis, either by eliminating cell surface expression of the death domain-containing receptors of the tumor necrosis factor receptor superfamily or through activation of the NF-κB apoptosis protection response (13). Although E1A is responsible for the increased tumor necrosis factor alpha sensitivity (4), E1A also counteracts this induction by interfering with the transcriptional activity of NF-κB (21).
In relation to transcriptional regulation, E4 orf6/7 stabilize the interaction of E2F to the duplicated E2F binding sites in the E2 promoter (68, 71). E4 orf3 associates with E1B 55K in the nuclear promyelocytic leukemia protein oncogenic domains (POD) structures (57), and the observed reorganizing of PODs during infection implicates a possible involvement in the regulation of transcription factor availability and activity. The E4 orf4 protein interacts with protein phosphatase 2A, leading to inhibition of E1A-dependent transactivation of the junB promoter (47). Alone, E4 orf4 induces a p53-independent apoptosis pathway (52, 62, 80), although the relevance during a wild-type adenovirus infection remains to be clarified.
Most of the extensive knowledge about viral products and their potential activities stems from the analysis of individual genes. So far, less is known about their relevance for the interaction between virus and host cell during the infection. Here we present a systematic approach, using cDNA microarray analysis, to identify cellular genes targeted by adenovirus during the early phase of an infection. We identified 76 differentially expressed cellular genes. Their identity and potential promoter structures support a model in which adenovirus specifically affects a limited number of genes involved in cell growth control and antiviral defense and furthermore indicate that a significant proportion of regulatory events involve modulated activities of E2F and coactivators such as p300/CBP.
MATERIALS AND METHODS
Cell culture, virus infection, and RNA isolation.
HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% newborn calf serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Subconfluent cells grown on 10-cm dishes were mock infected or infected with adenovirus type 2 at a multiplicity of 100 fluorescence-forming units per cell (74) in serum-free DMEM. After adsorption (45 min at 37°C), the medium was replaced with DMEM containing 10% newborn calf serum. At 6 h after infection, total cellular RNA was extracted with TRIZOL Reagent (Gibco-BRL). Where indicated, mRNA was isolated with Oligotex (Qiagen), according to the manufacturer's protocol. The quantity and quality of RNA were determined with the RNA 6000 Nano LabChip kit and a Bioanalyzer 400 (Agilent Technologies).
cDNA microarray.
Three different sources of cDNA microarray were used in this study. Type I was a 6,000 human cDNA microarray from the cDNA Microarray Core Facility, Department of Human Genetics, University of California, Los Angeles. Type II was a 7,500 human cDNA microarray from the DNA Microarray Core Facility, Uppsala University, Sweden. Type III was a 21,000 human cDNA microarray from the Department of Biotechnology, Royal Institute of Technology, Stockholm, Sweden. In the type II array, duplicate sets of clones were printed. In addition, 10 adenovirus-specific PCR amplicons representing coding regions of E1A′ (nucleotides 981 to 1097), E1A" (nucleotides 506 to 623), E1B' (nucleotides 2632 to 2733), E1B" (3618 to 3752), E2A (nucleotides 22531 to 22627), E2B (nucleotides 4129 to 4235), E3 (nucleotides 27797 to 27901), E4 (nucleotides 32919 to 33012), L1 (nucleotides 11601 to 11698), and L3 (nucleotides 22138 to 22231) were also included.
Preparation of cDNA probe and microarray hybridization.
Two different protocols were used for preparation of cDNA. The CyScribe first-strand cDNA direct labeling method (Amersham Bioscience) was used for type I cDNA microarrays, and the Micromax (TSA Amplified) protocol (PerkinElmer Life Sciences, Inc.) was used for type II and III microarrays.
CyScribe first-strand cDNA direct labeling.
Briefly, 1.5 μg of polyadenylated RNA from mock-infected and infected cells was reverse transcribed with a mixture of random nonamers and oligo(dT) primers, to generate indocarbocyanine- or indodicarbocyanine-labeled cDNA. Dye Swap labeling was performed on every RNA batch. The mRNA was degraded in 50 mM NaOH for 10 min at 65°C. After purification on Microcon YM-30 columns (Millipore), CyDye-labeled cDNAs were suspended in hybridization buffer containing 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.2% sodium dodecyl sulfate, 1.3 mg of tRNA per ml, and 0.7 mg of cot1 DNA per ml and were denatured (100°C for 2 min), followed by 10 min of incubation at 37°C. The DNA microarray chips were prehybridized in 5× SSC, 5× Denhardt's solution, 0.2 μg of tRNA per ml, 0.5% sodium dodecyl sulfate, and 50% formamide for 1 h at 42°C, rinsed once with distilled H2O and once with 2-propanol, followed by spin drying. Hybridization was performed in a humid chamber for 12 to 16 h at 65°C, followed by stepwise washing with buffer 1 (1× SSC, 0.2% sodium dodecyl sulfate), buffer 2 (0.4× SSC), and buffer 3 (0.2× SSC), and the material then quickly rinsed in water. Drying was achieved by centrifugation at 500 rpm for 3 min.
Micromax labeling. Labeling was done according to the manufacturer's protocol. Five micrograms of total RNA from mock- or adenovirus-infected HeLa cells was used to produce biotin-labeled and fluorescein-labeled cDNA, respectively. Dye Swap labeling was performed as above.
Data collection, normalization, and analysis.
A GenePix 4000B microarray scanner (Axon Instruments, Inc.) and the GenePix Pro 4.0 acquisition software were used to scan the chips at 10-μm resolution. Each array generated two distinct images, one for each fluorescent dye, that were used for quantification of gene expression data. Arrays of types I and II were quantified with the GenePix Pro 4.0 software, in which composed color images were used to identify spot positions and to classify individual spots according to a flagging system (6). The intensities of both fluorescents in each spot were measured according to set procedures with the standard fixed circle segmentation method. Arrays of type III were quantified with UCSF Spot 2.0 (available at: http://jainlab.ucsf.edu/Downloads.html). Spot positions were obtained automatically, and no flagging occurred. The intensities were measured with the default settings, allowing noncircular spot segmentation (40). In both cases, spotting parameters were imported to correlate spot locations with gene identities, and the data were finally exported as tab-delimited text files.
The normalization was performed within the framework of the statistical software R (R is a language and environment for statistical computing and graphics: http://www.r-project.org/). The specific methods used were implemented as part of the add-on package com.braju.sma (9) that extends the earlier package Statistics for Microarray Analysis (SMA; contains functions for exploratory microarray analysis: http://www.stat.berkeley.edu/users/terry/zarray/Software/smacode.html). Prior to normalization, spots with negative flag values were excluded from the type I and II arrays. Similarly, spots with signal intensities below the 98% quantile of the empty spot distribution were considered nonexistent and hence excluded from the type III arrays. Background-subtracted data from each experiment were then individually normalized in an intensity-dependent manner (97). The concept is to fit a smoothing curve to the log ratio M = log2(R/G) over the mean log intensity A = log2√(R · G). R and G represent fluorescence intensity in the red (Cy5) and green (Cy3) labeled cDNA that was hybridized to DNA microarray. The robust scatter plot smoother function Lowess (19) was used for this purpose. Across-slide normalization was subsequently performed to obtain equal spread (as measured by absolute median deviation) between arrays of identical type by scaling the log ratios (M).
Genes with significantly changed expression ratios were identified with the significance analysis of microarrays (SAM; software for gene expression data mining: http://www-stat.stanford.edu/≈tibs/SAM/) method (90). Each type of array was processed separately with the one-class response setting, greatest possible number of permutations and a false discovery rate of less than 5%. However, before running SAM, all three normalized datasets were filtered to exclude genes with more than one missing value. The missing values that remained were replaced by SAM's k-nearest neighbors imputer. In the case of the type II arrays, an average value was calculated for each duplicated pair of spots within an array before running SAM.
Quantitative real-time PCR.
The quantitative real-time PCR assays were performed on the same sets of RNA that were used for the cDNA microarray experiments. Unlabeled PCR primers and 5(6)-carboxy-fluorescein dye-labeled TaqMan MGB probes (Applied Biosystems Primers) were selected from Assays-on-Demand. For cDNA synthesis, 300 ng of mRNA was reverse transcribed in a total volume of 20 μl containing 10 μM dithiothreitol, 250 μM deoxynucleoside triphosphate mix, 0.5 ng of oligo(dT)/μl, and 200 U of Superscript II (Invitrogen AB) at 42°C for 1 h. cDNAs were diluted 1:40 and 1:80 with sterile H2O.
Quantitative real-time PCR was performed in a 25-μl volume containing 4 μl of diluted cDNA, 19.75 μl of TaqMan universal PCR master mix (Applied Biosystems), and 1.25 μl of probe and primer mix. β-Actin was used as an internal control. A negative template control that contained all TaqMan reagents except DNA was performed in parallel. A cDNA pool containing equal amounts of cDNA from mock- and adenovirus-infected cells was used for generating a standard curve. The amplification profile in the ABI Prism 7700 sequence detector (Perkin-Elmer Life Sciences, Inc.) was performed for 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 65°C. The data were analyzed and converted into values by the sequence detector v1.7 software system. The threshold cycle values were then translated into relative copy numbers of cDNA by using the standard curve.
RESULTS
Analysis of RNA from 12,309 unique genes during the early phase of an adenovirus type 2 infection.
The microarray technology offers the possibility to analyze, at any given time point, accumulated changes in the RNA content of a virus-infected cell. Alterations observed may reflect modifications in transcription, processing, and/or stability. This study analyzed the RNA profile in cultured monolayers of HeLa cells at 6 h postinfection with high multiplicities (100 focus-forming units/cell) of adenovirus type 2. At this time point, all early viral regulatory proteins are expressed, but the infection has not yet proceeded into the late phase.
Microarray chips produced from cDNA libraries have intrinsic problems due to the risk of contamination in the bacterial clone library, failure in PCR amplification of cDNAs, and simply misnamed cDNAs. To decrease some of these risk factors, we chose to use three different sources of cDNA microarrays: a 6,000 array from the University of California-Los Angeles (type I), a 7,5000 in-house array (type II), and a 21,000 array from the Royal Institute of Technology, Stockholm (type III). In total, 22,566 cDNAs, which represented 12,309 unique genes were tested.
Among the unique entries, 6,893 cDNAs were found to be present on at least two types of arrays. The type III array included all cDNAs printed on the type I array and 2,523 of the cDNAs printed on the type II array. In addition, 2,613 clones overlapped between the type II array and the type I array. Altogether, the use of multiple arrays allowed validation of data reproducibility and also excluded some of the false results that might be caused by the array manufacture process. Moreover, three independent preparations of RNA from adenovirus-infected and uninfected cells were analyzed twice on each type of array. For these duplicates, reciprocal labeling of the RNA was performed. Two different labeling methods, direct labeling for the type I array and the TSA amplifier protocol for the type II and type III arrays (see Materials and Methods), were used, but with very similar results. The major difference was the higher sensitivity of the TSA protocol, allowing analysis of as little as 2 to 5 μg of total RNA. In the final analysis, the data presented were based on 17 independent hybridization experiments (6 on type I arrays, 6 on type II arrays, and 5 on type III arrays), allowing solid control of variations in the labeling and hybridization procedure.
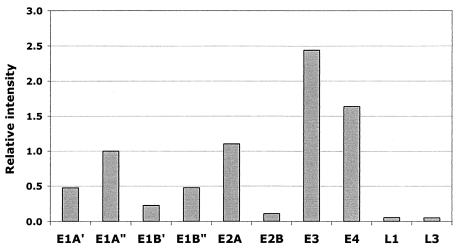
To monitor the adenovirus infection, the type II arrays were designed to contain PCR amplicons representing all adenovirus early genes (E1A, E1B, E2A, E3, and E4), as well as the late genes L1 and L3. At 6 h postinfection, all early viral genes were expressed to significant levels, whereas very low levels of L1 and L3 transcripts were detected (Fig. 1). These results are in agreement with a virus infection that had proceeded well into the early phase but not passed the early-to-late transition.
FIG. 1.
Early viral gene expression at 6 h postinfection. Expression levels are shown as relative signal intensities from type II arrays with RNA from adenovirus-infected cells. The signal obtained from E1A" (see Materials and Methods) was arbitrarily set to 1.
Identification of 76 differentially expressed cellular genes.
After quantification of intensities, genes with signals below the background level (estimated with empty spots for type III arrays) or flagged as bad or nonexistent during spot identification (performed with GenePix Pro 4.0 for type I and II arrays) were filtered out. Data were then normalized with scale intensity-dependent normalization (97), and differentially expressed genes were identified by SAM with the one-class response setting, greatest possible number of permutations, and a false discovery rate of less than 5% (90).
Following the statistical analyses, RNA from virus-infected cells demonstrated differential expression (more than 1.5-fold change compared to the uninfected control RNA) for 78 cellular clones (Tables 1 and 2). None of the clones demonstrated contradictory results between the arrays, demonstrating the reproducibility and reliability of the experimental approach and statistical analysis. Moreover, for 15 of the clones, statistically verified values were obtained from at least two of the three types of arrays. Gene information was available for 51 clones (Table 1), whereas the genes for 25 clones were not yet identified (Table 2). With information extracted mainly with the Source tool, known or suggested functions were used to classify 45 of the identified genes into five defined categories: cell cycle and proliferation, transcription, RNA metabolism, protein metabolism, and stress and immune response (Table 1).
TABLE 1.
Named genes differentially expressed during an adenovirus infection
| Category | Gene | Clone no. | Changea on array type:
|
Name | ||
|---|---|---|---|---|---|---|
| I | II | III | ||||
| Cell cycle and proliferation | FZD8 | 810459 | * | — | 1.7 | frizzled homolog 8 (Drosophila) |
| CDC25A | 204301 | — | 1.6 | 1.7 | Cell division cycle 25A | |
| P2RX5 | 486678 | 1.5 | * | * | Purinergic receptor P2X | |
| CARF | 302997 | −2.2 | — | * | Collaborates/cooperates with ARF | |
| GAS1 | 365826 | −1.7 | * | −1.9 | Growth arrest-specific 1 | |
| BMP4 | 797048 | * | −1.7 | −1.7 | Bone morphogenetic protein 4 | |
| SGK | 232946 | — | — | −1.7 | Serum/glucocorticoid-regulated kinase | |
| CCND1 | 199371 | — | −1.6 | — | Cyclin D1 | |
| CKTSF1B1 | 324951 | −1.5 | — | * | Cysteine knot superfamily 1 | |
| Transcription | NR4A1 | 309893 | * | 1.8 | 2.3 | Nuclear receptor subfamily 4, group A, member 1 |
| JUNB | 122428 | 1.5 | — | 2.1 | junB proto-oncogene | |
| JUNB | 309864 | — | 1.8 | 2.3 | junB proto-oncogene | |
| ATF3 | 51448 | — | * | 1.9 | Activating transcription factor 3 | |
| SNAI1 | 810119 | * | 1.9 | * | Snail homolog 1 | |
| TLE3 | 321574 | 1.8 | — | * | Transducin-like enhancer of split 3 | |
| TLE3 | 490971 | 1.6 | — | 1.6 | Transducin-like enhancer of split 3 | |
| ELK4 | 236155 | * | * | 1.8 | ETS-domain protein | |
| SZF1 | 124922 | — | — | 1.6 | KRAB-zinc finger protein | |
| POLR2A | 740130 | 1.5 | * | * | Polymerase (RNA) II, polypeptide A | |
| ID3 | 756405 | −1.5 | * | * | Inhibitor of DNA binding 3 | |
| RNA metabolism | NUFIP1 | 240223 | — | — | 1.9 | Nuclear fragile X mental retardation protein interacting protein 1 |
| RNPC1 | 814526 | — | * | 1.8 | RNA-binding region (RNP1, RRM) containing 1 | |
| HNRPK | 415700 | * | 1.4 | 1.7 | Heterogeneous nuclear ribonucleoprotein K | |
| GEMIN4 | 897597 | — | * | 1.6 | Gem (nuclear organelle)-associated protein 4 | |
| SSB | 49970 | −2.3 | * | * | Sjogren syndrome antigen B (autoantigen La) | |
| Protein metabolism | SMURF1 | 725407 | — | * | 2.6 | E3 ubiquitin ligase SMURF1 |
| HSPA1L | 50615 | 2.4 | * | 1.6 | Heat shock 70-kDa protein 1-like | |
| ADAMTS1 | 62263 | * | 1.8 | * | A disintegrin-like and metalloprotease with thrombospondin type 1 motif, 1 | |
| ALPI | 626967 | — | * | 1.8 | Alkaline phosphatase, intestinal | |
| C18B11 | 131988 | 1.8 | * | * | C18B11 homolog | |
| PGA5 | 155768 | * | * | 1.6 | Pepsinogen 5, group I (pepsinogen A) | |
| CTSD | 264117 | — | * | 1.6 | Cathepsin D | |
| CTNS | 358752 | 1.6 | * | * | Cystinosis, nephropathic | |
| MRPS25 | 1565455 | — | 1.5 | — | Mitochondrial ribosomal protein S25 | |
| FUT4 | 133213 | 1.5 | * | * | Fucosyltransferase 4 | |
| FBXO32 | 487371 | −1.8 | — | −2.2 | F-box only protein 32 | |
| RNF19 | 742074 | — | — | −1.6 | Ring finger protein 19 | |
| Stress and immune response | GADD45B | 725109 | — | 3 | 1.6 | Growth arrest and DNA-damage-inducible, beta |
| BIRC5 | 2513075 | — | 1.6 | — | Baculoviral IAP repeat-containing 5 (survivin) | |
| TP53TG1 | 506646 | — | 1.5 | — | p53 target gene 1 | |
| F3 | 1928791 | — | −2.4 | — | Coagulation factor III | |
| CCL2 | 768561 | −2.2 | * | * | Chemokine (C-C motif) ligand 2 | |
| TNFAIP3 | 770670 | −1.8 | * | * | Tumor necrosis factor alpha-induced protein 3 | |
| CXCL1 | 324437 | −1.8 | −1.5 | * | Chemokine (C-X-C motif) ligand 1 | |
| SDPR | 128058 | — | * | −1.8 | Serum deprivation response | |
| IL6 | 310406 | * | * | −1.7 | Interleukin-6 | |
| SNK | 795877 | −1.5 | −1.4 | −1.5 | Serum-inducible kinase | |
| Other | MGC2555 | 810457 | 2.3 | * | * | Hypothetical protein MGC2555 |
| TNKS1BP1 | 143227 | — | — | 2.1 | Tankyrase 1 binding protein 1 | |
| COL6A1 | 487429 | 1.6 | * | * | Collagen, type VI, alpha 1 | |
| BTBD3 | 811918 | — | — | −1.6 | BTB (POZ) domain containing 3 | |
| VPS28 | 810038 | 1.6 | — | * | Vacuolar protein sorting 28 (yeast) | |
| POMZP3 | 2566917 | — | 1.5 | — | POM and ZP3 fusion | |
Relative change (fold) in expression between RNA from infected cells compared to RNA from uninfected cells. *, data excluded due to low intensity or high variation; —, gene absent on the array.
TABLE 2.
Unnamed genes differentially expressed during an adenovirus infection
| Gene | Clone no. | Changea on array type:
|
||
|---|---|---|---|---|
| I | II | III | ||
| FLJ14299 | 788087 | — | — | 2.5 |
| 366156 | — | — | 2.1 | |
| FLJ11767 | 360065 | 2.0 | — | * |
| FLJ13937 | 40038 | 2.0 | — | * |
| FLJ23507 | 450653 | — | — | 1.9 |
| ESTsb | 587268 | — | — | 1.9 |
| 50182 | * | * | 1.7 | |
| FLJ38824 | 416155 | 1.7 | — | * |
| 112559 | — | — | 1.7 | |
| 277305 | — | — | 1.7 | |
| ESTs | 701275 | — | — | 1.7 |
| ESTs | 244300 | — | — | 1.6 |
| 141854 | — | — | 1.6 | |
| KIAA0247 | 292894 | 1.6 | — | * |
| FLJ10307 | 809879 | 1.6 | — | , |
| ESTs | 825467 | — | — | 1.5 |
| ESTs | 811028 | −4.0 | — | −2.0 |
| MAGE:4243 767 | 270826 | — | — | −2.0 |
| ESTs | 285908 | — | — | −2.0 |
| ESTs | 897177 | * | * | −1.9 |
| KIAA1376 | 135900 | — | — | −1.8 |
| FLJ11441 | 357278 | −1.8 | — | −1.7 |
| 273048 | −1.7 | — | * | |
| ESTs | 291394 | * | — | −1.6 |
| FLJ39046 | 73933 | — | — | −1.6 |
See Table 1 for definitions of asterisks and dashes.
ESTs, expressed sequence tags.
Cell cycle and proliferation signals.
Three genes with stimulatory effects on cell cycle progression and proliferation were found to be upregulated: CDC25A, required for activation of G1 cyclin/cdk complexes; FZD8, a receptor in the Wnt signaling pathway; and the purinergic receptor P2RX5. In contrast, five genes related to growth arrest were clearly downregulated: growth arrest-specific 1 (GAS1), a putative tumor suppressor protein that blocks S-phase entry; CKTSF1B1 and BMP4, both involved in transforming growth factor beta signaling; the M-phase-specific kinase SGK; and CARF, a protein that cooperates with ARF. Finally, in agreement with earlier results (84), the early G1-phase cyclin D1 (CCND1) was also downregulated.
Transcription.
A number of genes encoding cellular transcription factors were found to be upregulated in the infected samples. Surprisingly, most of these have been described as transcriptional repressor proteins or inducers of cell growth inhibition. ATF3 is a member of the basic region-leucine zipper family (bZip), TLE3 belongs to the Groucho family, and SZF1-1 is a KRAB-zinc finger protein. A second bZip protein, JunB, is known to act antagonistically to the proto-oncogene c-jun, thereby blocking cellular proliferation. The nuclear receptor NR4A1 can induce apoptosis, and the ETS domain protein ELK4 is a component of the ternary complex factor complex. Of the up-regulated genes, ATF3 and JunB have previously been shown to be targets of E1A-mediated transactivation (34, 47). Finally, the largest subunit of RNA polymerase II, PolR2A, was also upregulated. The only downregulated gene linked to transcription was Id3, a transcriptional helix-loop-helix repressor protein inhibiting DNA binding of the transcription factor E2A (59).
RNA metabolism.
Four out of five genes encoding proteins with proven or proposed ability to bind RNA were found to be upregulated. HNRPK and GEMIN4 are nuclear proteins that have been implicated in RNA maturation and spliceosome assembly, respectively. NUFIP1, which interacts with the nuclear fragile X protein, has RNA binding capacity, and RNPC1 has an RNA recognition motif. In contrast, the La autoantigen (SSB), which binds and stabilizes histone mRNA, was downregulated.
Protein structure, stability, and modification.
Three genes encoding proteins involved in ubiquitination were identified. A member of the SCF ubiquitin ligase complex, the F-box-only protein 32 (FBXO32), and the ubiquitination ring finger protein 19 (RNF19) were both downregulated. SCF represent a Skp1, Cullin, and F-box protein-containing E3 ubiquitin ligase. SMURF1, on the other hand, an E3 ligase which triggers degradation of TGF-β-induced SMAD1 and SMAD5, was upregulated. Three proteases, the metalloprotease ADAMTS1, cathepsin D, and pepsinogen A (PGA5), were also upregulated, as was the alkaline phosphatase ALPI. Finally, two upregulated genes related to ribosomal functions were also assigned to this category. MRPS25 is a mitochondrial ribosomal protein, and C18B11 is homologous to a bacterial pseudouridine synthase acting on the ribosomal 23S RNA.
Immune and stress response.
Most of the genes assigned to this category were found to be downregulated. These included the cytokines CXCL1 and CCL2, which display chemotactic activity to attract neutrophils or monocytes and basophils, respectively, and interleukin-6, which is a key mediator in acute-phase reactions, tissue damage, and infections. As a possible consequence of cytokine downregulation, the TNF-α-induced protein, a gene highly similar to a cytokine-inducible serine/threonine kinase, and the cytokine-induced coagulation factor 3 (F3) were also found to be downregulated. A gene normally induced by serum deprivation, SDPR, was also downregulated. However, four genes assigned to this category were upregulated. These included the stress-induced genes for GADD45B and the hsp70-like HSPA1L, the p53 target gene TP53TG1, and the inhibitor of apoptosis survivin (BIRC5).
Analyses of consensus transcription factor binding site in the promoter regions of differentially expressed genes.
A large number of genes have been identified as potential targets for regulation by adenoviral proteins. By far most reports concern the ability of adenovirus E1A to modulate transcription. Increased transcription from E2F-responsive genes following the dissociation of an inhibitory pRb-E2F complex by E1A is likely to contribute to a substantial part of the observed transcriptional activation induced by E1A. Similarly, since p300/CBP has been shown to act as a transcriptional cofactor for several different transcription factors, the sequestering and/or inactivation of the p300/CBP proteins by E1A probably contributes to a significant part of the observed E1A-mediated repression of transcription. With this in mind, an obvious task was to investigate whether the differential gene expression observed in our array experiments was supported by the presence of specific transcription factor binding sites.
By using the EZ-Retrieve tool (93), the 51 named genes were subjected to analysis for the presence of consensus transcription factor binding sites in their upstream promoter sequences (−500 to −1). In agreement with the fact that E2F is a target for E1A-mediated activation, 45% of the upregulated genes contained potential E2F binding sites in their promoters (Table 3). Similarly, E1A has also been shown to cooperate with the cyclic AMP-responsive element (CRE) binding factor CREB (12), and CREs were also present in 45% of the upregulated genes. Significantly, E2F and CREB binding sites were much less abundant in the downregulated genes (22 and 28%, respectively). In contrast, binding sites for STAT and NF-κB, both characterized targets for E1A-mediated repression (21, 58), were found in only 12 and 18%, respectively, of the upregulated genes, whereas their presence in the downregulated genes was 44 and 33%, respectively. In addition, we also found a significant overrepresentation in the downregulated promoter sequences of binding sites for C/EBPβ (33% compared to 18% in the upregulated genes). As a comparison, consensus Sp1 binding sites were present in multiple copies for a majority of the genes, and as many as 64% of the upregulated genes and 56% of the downregulated genes contained one or more potential Sp1 binding sites (data not shown).
TABLE 3.
Presence of consensus transcription factor binding sites in the −500 to −1 promoter sequence
| Binding site | Upregulated genes
|
Downregulated genes
|
||||
|---|---|---|---|---|---|---|
| Matcha | % | Genes | Match | % | Gene symbol | |
| E2F | 15/33 | 45 | ADAMTS1, CDC25A, CTSD, FZD8, GEMIN4, HNRPK, HSPA1L, MGC2555, NR4A1, NUP1P1, POLR2A, POMCP3, SNAI1, SZF1, TNKS1BP1 | 4/18 | 22 | BTBDR, CARF, CCND1, SSB |
| CREB | 15/33 | 45 | C18B11, CTSD, FUT1, ELK4, HSPA1L, MRPS25, NR4A1, NUF1P1, P2RX5, POMCP3, SZF1, TLE3, TNKS1BP1, TP53TG1, VPS28 | 5/18 | 28 | ID3, IL6, RNF19, SDPR, TNFAIP3 |
| STAT | 4/33 | 12 | COL6A1, GADD45B, SZF1, TP53TG1 | 8/18 | 44 | BMP4, CCL2, CCND1, CKTSF1B, CXCL1, F3, FBXO32, GAS1 |
| C/EBPβ | 6/33 | 18 | ADAMTS1, ATF3, C18B11, MRPS25, PGA5, POLR2A | 6/18 | 33 | BMP4, FBXO32, IL6, RNF19, SSB, SDPR |
| NF-κB | 6/33 | 18 | GADD45B, PGA5, POLR2A, SMURF1, SNAI1, VSP28 | 6/18 | 33 | CCL2, CKTSF1B, CXCL1, F3, IL6, TNFAIP3 |
The number of genes out of the total number of genes analyzed belonging to the category of up- or downregulated genes where one or more consensus transcription factor binding site was identified.
Confirmation of expression changes by quantitative real-time PCR analysis.
To confirm the cDNA microarray results, quantitative real-time PCR analyses were performed on RNA prepared 2, 4, and 6 h postinfection. Four genes, JunB, GAS1, CCL2, and IL-6, were identified by this cDNA microarray analysis to be differentially expressed, whereas the expression of β-actin was not influenced by adenovirus infection. For these five genes, the quantitative real-time PCR results corroborated the microarray data and also showed that a pronounced change in RNA levels was not observed until 6 h postinfection, except for the JunB mRNA, whose expression was already slightly enhanced at 4 h postinfection (Table 4).
TABLE 4.
Results from quantitative real-time PCR analysis compared to microarray data
| Gene | Changea (fold) at time postinfection:
|
|||
|---|---|---|---|---|
| PCR
|
Microarray | |||
| 2 h | 4 h | 6 h | 6 h | |
| β-Actin | 1 | 1 | 1 | 1 |
| JunB | 1 | 1.4 | 2.5 | 2.2 |
| GAS1 | 1 | 1 | −2.5 | −1.8 |
| CCL2 | 1 | 1 | −2.6 | −2.3 |
| IL-6 | 1 | 1 | −3.4 | −1.6 |
| c-Myc | 1 | 1 | −1.5 | — |
| c-Jun | 1 | 1 | 1 | — |
| Cyclin E1 | 1 | 1 | −3.0 | * |
See Table 1 for definitions of asterisk and dashes.
Finally, c-Jun, c-Myc, and cyclin E have previously been shown to be targets for E1A transcriptional regulation (37, 42, 91, 92). Since c-Jun, and c-Myc were absent on the arrays and cyclin E, although present, gave inconclusive results, the expression of these three genes was also analyzed by quantitative real-time PCR. Surprisingly, c-Jun expression remained unchanged, whereas expression of both c-Myc and cyclin E was downregulated at 6 h postinfection. Thus, the effects of E1A observed during transient transfection assays and in E1A-transformed cells may not be relevant during a virus infection.
DISCUSSION
In this study, we analyzed changes in the host cell gene expression profile at 6 h after infection with adenovirus type 2. High multiplicities of virus were used in order to obtain a synchronous infection. At 6 h postinfection, virus infection had proceeded far into the early phase and the virus has expressed all of its early viral gene products (Fig. 1). Thus, the major viral transcriptional activator, E1A, has redirected the cellular transcription machinery towards the viral chromosome and possibly also reprogrammed essential host cell genes. The virus infection has furthermore elicited host cell responses which are aimed at limiting the productive infection. At this time, these signals may or may not have been counteracted by viral proteins designed to evade the antiviral defense system of the host. Therefore, the observed alterations in host cell gene expression are the net result of direct regulatory activities as well as indirect consequences of these activities. To definitely assign the observed effects on specific genes to viral or host cell activities is, at this stage, impossible. However, based on our knowledge about cellular reprogramming during a virus infection, the observed alterations in host cell gene expression fit well with a strategic targeting of genes involved in cell growth and antiviral defense. In addition, a number of genes related to RNA and protein metabolism were upregulated, suggesting that an optimization of cellular metabolism occurs to ensure efficient expression of viral genes.
The promoters of most adenovirus early genes harbor cyclic AMP-inducible elements. In agreement, the CR3 domain of E1A-289R can activate transcription by cooperating with members of the ATF/CREB family of transcription factors (35). However, a large number of reports have also demonstrated a wide-range capacity of E1A-289R to activate transcription. Here we report that only a limited number of genes were upregulated during a virus infection, suggesting that the previously observed promiscuity of the E1A CR3 transcriptional activator might not be relevant for the lytic cycle. It is however noteworthy that the promoter analysis identified potential CREB binding sites in 45% of all upregulated genes but in only 28% of the downregulated genes.
Adenovirus-induced cell cycle deregulation is mainly achieved by the direct targeting of key regulators of the cell cycle. The interaction between E1A and pRb allows released transcription factor E2F to activate transcription of its target genes (20). It is therefore reasonable to assume that during an adenovirus infection, expression of E2F-dependent host cell genes is subjected to a selective regulation. In support of this assumption, we found that 45% of the upregulated genes contained potential E2F binding sites within 500 bp upstream of the transcriptional start site, whereas only 22% of the downregulated genes harbored potential E2F binding sites (Table 3). This is in agreement with recent reports showing that expression of ADAMTS1, CDC25A, CCND1, FZD8, and TNKSBP1 was regulated by E2F (39, 61, 67, 79, 85). However, the same reports also demonstrated that ATF3, BMP4, CXCL1, ID3, JunB, SGK, SNK, and TLE were E2F responsive, although we were unable to identify consensus E2F sites in the proximal 500-bp promoter sequence. A potential E2F binding site was, however, found in ATF3 when the search was extended to include up to 1000 bp upstream of the transcriptional start site (data not shown). In summary, the adenovirus infection had similar effects on these previously described E2F-responsive genes with the exception of ADAMTS1 and TNKS1BP1, which were downregulated by E2F (61, 67), and CCND1 and ID3, which were induced by E2F (85) or in G1 (79), respectively.
Transcriptional repression by E1A has been demonstrated for a variety of genes induced by transcription factors such as AP1, STAT, C/EBPβ, and NF-κB (21, 58). This correlates with the ability of E1A to bind and sequester the transcriptional coactivators p300/CBP and components of the recently described transformation-transactivation domain-associated protein (TRRAP) complexes (51). Significantly, potential promoter binding sites for STAT, C/EBPβ, and NF-κB were two to three times more abundant in the downregulated genes compared to the upregulated genes (Table 3). Moreover, E2F can recruit the p300/CBP proteins (66) and possibly also TRRAP (50). The presence of potential E2F binding sites in four of the downregulated genes thus indicates that E1A can repress transcription of E2F-dependent genes by interfering with E2F cofactor recruitment. Finally, in this context it should be noted that, depending on the target promoter, E1A can activate transcription through the p300/CBP interaction (53, 54), possibly by enhancing the acetyltransferase activity of p300/CBP (2).
A primary task for a virus is to override the fundamental control of the host cell cycle and force progression into S phase, where viral DNA replication can occur. In agreement with earlier reports (83, 84), we found that expression of two key regulators of cell cycle progression, cyclin D1 and CDC25A, were regulated by the virus infection. However, in growing HeLa cells, the virus seems to put more effort into counteracting the activity of inhibitors of cell growth. E1A has been shown to block growth inhibition by TGF-β1 (24, 65), and here we show that TGF-β superfamily signaling was inhibited both through downregulated expression of an upstream ligand (BMP4) and upregulated expression of a signal terminator, SMURF1, which triggers degradation of BMP4 intracellular transducers SMAD1 and SMAD5 (75). Expression of the BMP4 antagonist CKTSF1B1 was downregulated, which at first sight seemed counterintuitive to inhibition of TGF-β-induced growth suppression. However, since CKTSF1B1 can activate p21Cip and thereby induce growth arrest, reduced expression of CKTSF1B1 would in fact favor cellular proliferation (17). Notably, CKTSF1B1 is generally expressed at lower levels in tumors compared to normal cells, supporting its regulatory role in cell proliferation (88, 89).
As a possible consequence of the effect of inhibited TGF-β superfamily signaling, expression of the TGF-β-induced serum glucocorticoid-induced kinase (SGK1) (50, 95) and ID3 (46) genes was downregulated. Expression of two additional cell cycle-inhibitory genes, GAS1 and CARF, was repressed. Downregulation of the cell cycle inhibitor GAS1 plays an important role during v-Src-triggered S-phase entry (32). Although the exact function of the recently identified ARF-interacting protein CARF is yet to be defined, current results indicate that it cooperates with p19ARF in p53-dependent and -independent tumor-suppressive functions (36, 94). Importantly, since ARF mediates the induction of p53 by E1A (26), the downregulation of CARF might be part of the viral defense mechanism against apoptosis. In summary, a minimum of 20% of the identified genes that were up- or downregulated during an adenovirus infection showed a clear functional relation to gene products involved in the control of cell growth.
As an immediate response to virus infection, the host cell activates a cascade of genes with the aim of inhibiting cell proliferation or inducing apoptosis. Although the initial contact between virus and cell will start an immediate innate immune response, usually through activation of type I interferons (30), the subsequent expression of E1A triggers proapoptotic host response programs, for example, by stabilizing and hence increasing the activity of p53 (60). As a possible result of p53 activation, we detected upregulation of three p53-inducible genes, TP53TG1 (87) and the stress response genes GADD45B and ATF3 (45). TP53TG1 has been suggested to play a role in p53 signaling (87). GADD45B and ATF3 have been shown to regulate activities of Cdc2 and p21WAF1, leading to G1 arrest, inhibited cell cycle progression, and apoptosis (43, 81).
Finally, HSPAIL, a member of the heat shock protein 70 (Hsp70) family of molecular chaperones, which are known to act on aberrant proteins under stress conditions, was also upregulated. Although it is possible that HSPAIL is induced as part of a stress response against the virus infection, adenovirus might also benefit from its expression and may therefore have developed means to specifically induce its expression. This is supported by the result that heat shock response is essential for adenovirus replication (29). Thus, it is possible that, similar to Hsp70 (69), HSPAIL may also be induced by adenovirus E1A. In agreement with a viral attempt to evade the apoptotic response of the host cell, we found that a target gene for p53-mediated repression (38, 63), the survival factor BIRC5 (48, 72), was upregulated. This might reflect the ability of E1B 55K to interfere with p53-mediated repression (76a).
It is well established that adenovirus has the capacity to interfere with the host immune response, mainly through proteins encoded by the E3 region. Our study demonstrates that during an adenovirus infection, expression of several genes involved in the innate immune response was inhibited. Two of the three cytokines, CXCL1 and CCL2, that were downregulated during infection were found to harbor potential binding sites for STAT in their promoter sequences (Table 3). STAT is activated by the interferon signaling (22) pathway, and several studies have demonstrated that E1A can interfere with STAT activity at multiple levels, such as reducing protein levels (55) and blocking formation of the STAT transcriptional complex (1, 44) by inhibiting DNA binding (33) or the interaction between STAT and p300/CBP (10, 56). In contrast, the third downregulated cytokine, interleukin-6, is repressed by E1A through an NF-κB site (41). In agreement, potential NF-κB binding sites were detected in the interleukin-6 promoter sequence, but no consensus STAT binding sites were found, even when the search was extended up to 1,000 bp upstream of the transcriptional start site (data not shown).
In summary, our results have shown that expression of a limited number of genes (0.6% of detected expressions) was modulated during infection of HeLa cells. Significantly, adenovirus consistently targeted genes involved in regulation of cell growth and antiviral defense. However, half of the regulated genes were found to encode proteins related to metabolic pathways or cell structures. The relevance of modulating these genes has only been addressed briefly, and additional experiments are required to determine whether these events are initiated directly by viral factors or constitute host cell responses or indirect effects of yet unknown importance.
Acknowledgments
We thank Hanna Berglind and Anja Castensson for excellent technical assistance.
This work was supported by the Beijer Foundation and the Swedish Cancer Society. C.S. holds a position supported by the Strategic Research Council.
REFERENCES
- 1.Ackrill, A. M., G. R. Foster, C. D. Laxton, D. M. Flavell, G. R. Stark, and I. M. Kerr. 1991. Inhibition of the cellular response to interferons by products of the adenovirus type 5 E1A oncogene. Nucleic Acids Res. 19:4387-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ait-Si-Ali, S., S. Ramirez, F. X. Barre, F. Dkhissi, L. Magnaghi-Jaulin, J. A. Girault, P. Robin, M. Knibiehler, L. L. Pritchard, B. Ducommun, D. Trouche, and A. Harel-Bellan. 1998. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 396:184-186. [DOI] [PubMed] [Google Scholar]
- 3.Akusjarvi, G. 1993. Proteins with transcription regulatory properties encoded by human adenoviruses. Trends Microbiol. 1:163-170. [DOI] [PubMed] [Google Scholar]
- 4.Ames, R. S., B. Holskin, M. Mitcho, D. Shalloway, and M. J. Chen. 1990. Induction of sensitivity to the cytotoxic action of tumor necrosis factor alpha by adenovirus E1A is independent of transformation and transcriptional activation. J. Virol. 64:4115-4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arany, Z., D. Newsome, E. Oldread, D. M. Livingstone, and R. Eckner. 1995. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature 374:81-84. [DOI] [PubMed] [Google Scholar]
- 6.Axon Instruments 2001. GenePix Pro 4.0. Axon Instruments, Denver, Colo.
- 7.Bannister, A. J., and T. Kouzarides. 1995. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 14:4758-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayley, S. T., and J. S. Mymryk. 1994. Adenovirus E1A proteins and transformation. Int. J. Oncol. 5:425-444. [DOI] [PubMed]
- 9.Bengtsson, H. 2002. http://www.maths.lth.se/help/R/com.brajv.sma/.
- 10.Bhattacharya, S., R. Eckner, S. Grossman, E. Oldread, Z. Arany, A. D'Andrea, and D. M. Livingston. 1996. Cooperation of Stat2 and p300/CBP in signalling-induced by interferon-alpha. Nature 383:344-347. [DOI] [PubMed] [Google Scholar]
- 11.Braun, T., E. Bober, and H. H. Arnold. 1992. Inhibition of muscle differentiation by the adenovirus E1a protein: repression of the transcriptional activating function of the HLH protein Myf-5. Genes Dev. 6:888-902. [DOI] [PubMed] [Google Scholar]
- 12.Brockmann, D., and H. Esche. 2003. The multifunctional role of E1A in the transcriptional regulation of CREB/CBP-dependent target genes. Curr. Top. Microbiol. Immunol. 272:97-129. [DOI] [PubMed] [Google Scholar]
- 13.Burgert, H. G., Z. Ruzsics, S. Obermeier, A. Hilgendorf, M. Windheim, and A. Elsing. 2002. Subversion of host defense mechanisms by adenoviruses. Curr. Top. Microbiol. Immunol. 269:273-318. [DOI] [PubMed] [Google Scholar]
- 14.Caruso, M., F. Martelli, A. Giordano, and A. Felsani. 1993. Regulation of MyoD gene transcription and protein function by the transforming domains of the adenovirus E1A oncoprotein. Oncogene 8:267-278. [PubMed] [Google Scholar]
- 15.Chakravarti, D., V. Ogryzko, H. Y. Kao, A. Nash, H. Chen, Y. Nakatani, and R. M. Evans. 1999. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell 96:393-403. [DOI] [PubMed] [Google Scholar]
- 16.Chan, H. M., and N. B. La Thangue. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114:2363-2373. [DOI] [PubMed] [Google Scholar]
- 17.Chen, B., M. Athanasiou, Q. Gu, and D. G. Blair. 2002. Drm/Gremlin transcriptionally activates p21(Cip1) via a novel mechanism and inhibits neoplastic transformation. Biochem. Biophys. Res. Commun. 295:1135-1141. [DOI] [PubMed] [Google Scholar]
- 18.Chinnadurai, G. 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell 9:213-224. [DOI] [PubMed] [Google Scholar]
- 19.Cleveland, W. S. 1981. LOWESS: A program for smoothing scatterplots by robust locally weighted regression. Statistician 35.
- 20.Cobrinik, D. 1996. Regulatory interactions among E2Fs and cell cycle control proteins. Curr. Top. Microbiol. Immunol. 208:31-61. [DOI] [PubMed] [Google Scholar]
- 21.Cook, J. L., T. A. Walker, G. S. Worthen, and J. R. Radke. 2002. Role of the E1A Rb-binding domain in repression of the NF-kappa B-dependent defense against tumor necrosis factor-alpha. Proc. Natl. Acad. Sci. USA 99:9966-9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 23.Debbas, M., and E. White. 1993. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 7:546-554. [DOI] [PubMed] [Google Scholar]
- 24.de Groot, R. P., O. Kranenburg, L. de Wit, J. van den Eijnden-van Raaij, C. Mummery, A. J. van der Eb, and A. Zantema. 1995. Adenovirus E1A antagonizes both negative and positive growth signals elicited by transforming growth factor beta 1. Cell Growth Differ. 6:531-540. [PubMed] [Google Scholar]
- 25.Deleu, L., S. Shellard, K. Alevizopoulos, B. Amati, and H. Land. 2001. Recruitment of TRRAP required for oncogenic transformation by E1A. Oncogene 20:8270-8275. [DOI] [PubMed] [Google Scholar]
- 26.de Stanchina, E., M. E. McCurrach, F. Zindy, S. Y. Shieh, G. Ferbeyre, A. V. Samuelson, C. Prives, M. F. Roussel, C. J. Sherr, and S. W. Lowe. 1998. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 12:2434-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flint, J., and T. Shenk. 1997. Viral transactivating proteins. Annu. Rev. Genet. 31:177-212. [DOI] [PubMed] [Google Scholar]
- 28.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297-307. [DOI] [PubMed] [Google Scholar]
- 29.Glotzer, J. B., M. Saltik, S. Chiocca, A. I. Michou, P. Moseley, and M. Cotten. 2000. Activation of heat-shock response by an adenovirus is essential for virus replication. Nature 407:207-211. [DOI] [PubMed] [Google Scholar]
- 30.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 31.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 32.Grossi, M., S. A. La Rocca, G. Pierluigi, S. Vannucchi, E. M. Ruaro, C. Schneider, and F. Tato. 1998. Role of Gas1 downregulation in mitogenic stimulation of quiescent NIH3T3 cells by v-Src. Oncogene 17:1629-1638. [DOI] [PubMed] [Google Scholar]
- 33.Gutch, M. J., and N. C. Reich. 1991. Repression of the interferon signal transduction pathway by the adenovirus E1A oncogene. Proc. Natl. Acad. Sci. USA 88:7913-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hai, T., C. D. Wolfgang, D. K. Marsee, A. E. Allen, and U. Sivaprasad. 1999. ATF3 and stress responses. Gene Expr. 7:321-335. [PMC free article] [PubMed] [Google Scholar]
- 35.Hardy, S., and T. Shenk. 1988. Adenoviral control regions activated by E1A and the cyclic AMP response element bind to the same factor. Proc. Natl. Acad. Sci. USA 85:4171-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasan, M. K., T. Yaguchi, T. Sugihara, P. K. Kumar, K. Taira, R. R. Reddel, S. C. Kaul, and R. Wadhwa. 2002. CARF is a novel protein that cooperates with mouse p19ARF (human p14ARF) in activating p53. J. Biol. Chem. 277:37765-37770. [DOI] [PubMed] [Google Scholar]
- 37.Hiebert, S. W., M. Lipp, and J. R. Nevins. 1989. E1A-dependent trans-activation of the human MYC promoter is mediated by the E2F factor. Proc. Natl. Acad. Sci. USA 86:3594-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffman, W. H., S. Biade, J. T. Zilfou, J. Chen, and M. Murphy. 2002. Transcriptional repression of the antiapoptotic survivin gene by wild type p53. J. Biol. Chem. 277:3247-3257. [DOI] [PubMed] [Google Scholar]
- 39.Ishida, S., E. Huang, H. Zuzan, R. Spang, G. Leone, M. West, and J. R. Nevins. 2001. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21:4684-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jain, A. N., T. A. Tokuyasu, A. M. Snijders, R. Segraves, D. G. Albertson, and D. Pinkel. 2002. Fully automatic quantification of microarray image data. Genome Res. 12:325-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janaswami, P. M., D. V. Kalvakolanu, Y. Zhang, and G. C. Sen. 1992. Transcriptional repression of interleukin-6 gene by adenoviral E1A proteins. J. Biol. Chem. 267:24886-24891. [PubMed] [Google Scholar]
- 42.Jayachandra, S., K. G. Low, A. E. Thlick, J. Yu, P. D. Ling, Y. Chang, and P. S. Moore. 1999. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by with different transcription coadaptors. Proc. Natl. Acad. Sci. USA 96:11566-11571. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Jin, S., M. J. Antinore, F. D. Lung, X. Dong, H. Zhao, F. Fan, A. B. Colchagie, P. Blanck, P. P. Roller, A. J. Fornace, Jr., and Q. Zhan. 2000. The GADD45 inhibition of Cdc2 kinase correlates with GADD45-mediated growth suppression. J. Biol. Chem. 275:16602-16608. [DOI] [PubMed] [Google Scholar]
- 44.Kalvakolanu, D. V., S. K. Bandyopadhyay, M. L. Harter, and G. C. Sen. 1991. Inhibition of interferon-inducible gene expression by adenovirus E1A proteins: block in transcriptional complex formation. Proc. Natl. Acad. Sci. USA 88:7459-7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kannan, K., N. Amariglio, G. Rechavi, J. Jakob-Hirsch, I. Kela, N. Kaminski, G. Getz, E. Domany, and D. Givol. 2001. DNA microarrays identification of primary and secondary target genes regulated by p53. Oncogene 20:2225-2234. [DOI] [PubMed] [Google Scholar]
- 46.Kee, B. L., R. R. Rivera, and C. Murre. 2001. Id3 inhibits B lymphocyte progenitor growth and survival in response to TGF-beta. Nat. Immunol. 2:242-247. [DOI] [PubMed] [Google Scholar]
- 47.Kleinberger, T., and T. Shenk. 1993. Adenovirus E4orf4 protein binds to protein phosphatase 2A, and the complex down regulates E1A-enhanced junB transcription. J. Virol. 67:7556-7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kobayashi, K., M. Hatano, M. Otaki, T. Ogasawara, and T. Tokuhisa. 1999. Expression of a murine homologue of the inhibitor of apoptosis protein is related to cell proliferation. Proc. Natl. Acad. Sci. USA 96:1457-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolli, S., A. M. Buchmann, J. Williams, S. Weitzman, and B. Thimmapaya. 2001. Antisense-mediated depletion of p300 in human cells leads to premature G1 exit and upregulation of c-Myc. Proc. Natl. Acad. Sci. USA 98:4646-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lang, F., K. Klingel, C. A. Wagner, C. Stegen, S. Warntges, B. Friedrich, M. Lanzendorfer, J. Melzig, I. Moschen, S. Steuer, S. Waldegger, M. Sauter, M. Paulmichl, V. Gerke, T. Risler, G. Gamba, G. Capasso, R. Kandolf, S. C. Hebert, S. G. Massry, and S. Broer. 2000. Deranged transcriptional regulation of cell-volume-sensitive kinase hSGK in diabetic nephropathy. Proc. Natl. Acad. Sci. USA 97:8157-8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lang, S. E., and P. Hearing. 2003. The adenovirus E1A oncoprotein recruits the cellular TRRAP/GCN5 histone acetyltransferase complex. Oncogene 22:2836-2841. [DOI] [PubMed] [Google Scholar]
- 52.Lavoie, J. N., M. Nguyen, R. C. Marcellus, P. E. Branton, and G. C. Shore. 1998. E4orf4, a novel adenovirus death factor that induces p53-independent apoptosis by a pathway that is not inhibited by zVAD-fmk. J. Cell Biol. 140:637-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee, B. H., and M. B. Mathews. 1997. Transcriptional coactivator cyclic AMP response element binding protein mediates induction of the human proliferating cell nuclear antigen promoter by the adenovirus E1A oncoprotein. Proc. Natl. Acad. Sci. USA 94:4481-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee, J. S., R. H. See, K. M. Galvin, J. Wang, and Y. Shi. 1995. Functional interactions between YY1 and adenovirus E1A. Nucleic Acids Res. 23:925-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leonard, G. T., and G. C. Sen. 1996. Effects of adenovirus E1A protein on interferon-signaling. Virology 224:25-33. [DOI] [PubMed] [Google Scholar]
- 56.Leonard, G. T., and G. C. Sen. 1997. Restoration of interferon responses of adenovirus E1A-expressing HT1080 cell lines by overexpression of p48 protein. J. Virol. 71:5095-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leppard, K. N., and R. D. Everett. 1999. The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 components. J. Gen. Virol. 80:997-1008. [DOI] [PubMed] [Google Scholar]
- 58.Look, D. C., W. T. Roswit, A. G. Frick, Y. Gris-Alevy, D. M. Dickhaus, M. J. Walter, and M. J. Holtzman. 1998. Direct suppression of Stat1 function during adenoviral infection. Immunity 9:871-880. [DOI] [PubMed] [Google Scholar]
- 59.Loveys, D. A., M. B. Streiff, and G. J. Kato. 1996. E2A basic-helix-loop-helix transcription factors are negatively regulated by serum growth factors and by the Id3 protein. Nucleic Acids Res. 24:2813-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lowe, S. W., and H. E. Ruley. 1993. Stabilization of the p53 tumor suppressor is-induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 7:535-545. [DOI] [PubMed] [Google Scholar]
- 61.Ma, Y., R. Croxton, R. L. Moorer, Jr., and W. D. Cress. 2002. Identification of novel E2F1-regulated genes by microarray. Arch. Biochem. Biophys. 399:212-224. [DOI] [PubMed] [Google Scholar]
- 62.Marcellus, R. C., J. N. Lavoie, D. Boivin, G. C. Shore, G. Ketner, and P. E. Branton. 1998. The early region 4 orf4 protein of human adenovirus type 5 induces p53-independent cell death by apoptosis. J. Virol. 72:7144-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mirza, A., M. McGuirk, T. N. Hockenberry, Q. Wu, H. Ashar, S. Black, S. F. Wen, L. Wang, P. Kirschmeier, W. R. Bishop, L. L. Nielsen, C. B. Pickett, and S. Liu. 2002. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene 21:2613-2622. [DOI] [PubMed] [Google Scholar]
- 64.Missero, C., E. Calautti, R. Eckner, J. Chin, L. H. Tsai, D. M. Livingston, and G. P. Dotto. 1995. Involvement of the cell-cycle inhibitor Cip1/WAF1 and the E1A-associated p300 protein in terminal differentiation. Proc. Natl. Acad. Sci. USA 92:5451-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Missero, C., E. Filvaroff, and G. P. Dotto. 1991. Induction of transforming growth factor beta 1 resistance by the E1A oncogene requires binding to a specific set of cellular proteins. Proc. Natl. Acad. Sci. USA 88:3489-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morris, L., K. E. Allen, and N. B. La Thangue. 2000. Regulation of E2F transcription by cyclin E-Cdk2 kinase mediated through p300/CBP co-activators. Nat. Cell Biol. 2:232-239. [DOI] [PubMed] [Google Scholar]
- 67.Muller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neill, S. D., C. Hemstrom, A. Virtanen, and J. R. Nevins. 1990. An adenovirus E4 gene product trans-activates E2 transcription and stimulates stable E2F binding through a direct association with E2F. Proc. Natl. Acad. Sci. USA 87:2008-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nevins, J. R. 1982. Induction of the synthesis of a 70, 000 dalton mammalian heat shock protein by the adenovirus E1A gene product. Cell 29:913-919. [DOI] [PubMed] [Google Scholar]
- 70.Nikiforov, M. A., S. Chandriani, J. Park, I. Kotenko, D. Matheos, A. Johnsson, S. B. McMahon, and M. D. Cole. 2002. TRRAP-dependent and TRRAP-independent transcriptional activation by Myc family oncoproteins. Mol. Cell. Biol. 22:5054-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Obert, S., R. J. Oconnor, S. Schmid, and P. Hearing. 1994. The adenovirus E4-6/7 protein transactivates the E2 promoter by inducing dimerization of a heteromeric E2F complex. Mol. Cell. Biol. 14:1333-1346. [DOI] [PMC free article] [PubMed]
- 72.Ogasawara, T., M. Hatano, M. Otaki, N. Sekita, K. Kobayashi, M. Miyazaki, N. Nakajima, and T. Tokuhisa. 2001. A novel homologue of the TIAP/m-survivin gene. Biochem. Biophys. Res. Commun. 282:207-211. [DOI] [PubMed] [Google Scholar]
- 73.Perricaudet, M., G. Akusjärvi, A. Virtanen, and U. Pettersson. 1979. Structure of two spliced mRNAs from the transforming region of hmna subgroup C adenoviruses. Nature 281:694-696. [DOI] [PubMed] [Google Scholar]
- 74.Philipson, L. 1961. Adenovirus assay by the fluorescent cellcounting procedure. Virology 15:263-268. [DOI] [PubMed] [Google Scholar]
- 75.Podos, S. D., K. K. Hanson, Y. C. Wang, and E. L. Ferguson. 2001. The DSmurf ubiquitin-protein ligase restricts BMP signaling spatially and temporally during Drosophila embryogenesis. Dev. Cell 1:567-578. [DOI] [PubMed] [Google Scholar]
- 76.Punga, T., and G. Akusjarvi. 2000. The adenovirus-2 E1B-55K protein interacts with a mSin3A/histone deacetylase 1 complex. FEBS Lett. 476:248-252. [DOI] [PubMed] [Google Scholar]
- 76a.Punga, T., and G. Akusjärvi. FEBS Lett., in press. [DOI] [PubMed]
- 77.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reid, J. L., A. J. Bannister, P. Zegerman, M. A. Martinez-Balbas, and T. Kouzarides. 1998. E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J. 17:4469-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ren, B., H. Cam, Y. Takahashi, T. Volkert, J. Terragni, R. A. Young, and B. D. Dynlacht. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shtrichman, R., and T. Kleinberger. 1998. Adenovirus type 5 E4 open reading frame 4 protein induces apoptosis in transformed cells. J. Virol. 72:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smith, M. L., I. T. Chen, Q. Zhan, I. Bae, C. Y. Chen, T. M. Gilmer, M. B. Kastan, P. M. O'Connor, and A. J. Fornace, Jr. 1994. Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science 266:1376-1380. [DOI] [PubMed] [Google Scholar]
- 82.Somasundaram, K., and W. S. El-Deiry. 1997. Inhibition of p53-mediated transactivation and cell cycle arrest by E1A through its p300/CBP-interacting region. Oncogene 14:1047-1057. [DOI] [PubMed] [Google Scholar]
- 83.Spitkovsky, D., P. Jansen-Durr, E. Karsenti, and I. Hoffman. 1996. S-phase induction by adenovirus E1A requires activation of cdc25a tyrosine phosphatase. Oncogene 12:2549-2554. [PubMed] [Google Scholar]
- 84.Spitkovsky, D., P. Steiner, J. Lukas, E. Lees, M. Pagano, A. Schulze, S. Joswig, D. Picard, M. Tommasino, M. Eilers, et al. 1994. Modulation of cyclin gene expression by adenovirus E1A in a cell line with E1A-dependent conditional proliferation. J. Virol. 68:2206-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stanelle, J., T. Stiewe, C. C. Theseling, M. Peter, and B. M. Putzer. 2002. Gene expression changes in response to E2F1 activation. Nucleic Acids Res. 30:1859-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stevens, J. L., G. T. Cantin, G. Wang, A. Shevchenko, and A. J. Berk. 2002. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296:755-758. [DOI] [PubMed] [Google Scholar]
- 87.Takei, Y., S. Ishikawa, T. Tokino, T. Muto, and Y. Nakamura. 1998. Isolation of a novel TP53 target gene from a colon cancer cell line carrying a highly regulated wild-type TP53 expression system. Genes Chromosomes Cancer 23:1-9. [PubMed] [Google Scholar]
- 88.Topol, L. Z., M. Marx, D. Laugier, N. N. Bogdanova, N. V. Boubnov, P. A. Clausen, G. Calothy, and D. G. Blair. 1997. Identification of drm, a novel gene whose expression is suppressed in transformed cells and which can inhibit growth of normal but not transformed cells in culture. Mol. Cell. Biol. 17:4801-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Topol, L. Z., W. S. Modi, S. Koochekpour, and D. G. Blair. 2000. DRM/GREMLIN (CKTSF1B1) maps to human chromosome 15 and is highly expressed in adult and fetal brain. Cytogenet. Cell Genet. 89:79-84. [DOI] [PubMed] [Google Scholar]
- 90.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van Dam, H., M. Duyndam, R. Rottier, A. Bosch, L. de Vries-Smits, P. Herrlich, A. Zantema, P. Angel, and A. J. van der Eb. 1993. Heterodimer formation of cJun and ATF-2 is responsible for induction of c-jun by the 243 amino acid adenovirus E1A protein. EMBO J. 12:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van Dam, H., R. Offringa, I. Meijer, B. Stein, A. M. Smits, P. Herrlich, J. L. Bos, and A. J. van der Eb. 1990. Differential effects of the adenovirus E1A oncogene on members of the AP-1 transcription factor family. Mol. Cell. Biol. 10:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van der Meijden, C. M., D. S. Lapointe, M. X. Luong, D. Peric-Hupkes, B. Cho, J. L. Stein, A. J. van Wijnen, and G. S. Stein. 2002. Gene profiling of cell cycle progression through S-phase reveals sequential expression of genes required for DNA replication and nucleosome assembly. Cancer Res. 62:3233-3243. [PubMed] [Google Scholar]
- 94.Wadhwa, R., T. Sugihara, M. K. Hasan, E. L. Duncan, K. Taira, and S. C. Kaul. 2003. A novel putative collaborator of p19(ARF). Exp. Gerontol. 38:245-252. [DOI] [PubMed] [Google Scholar]
- 95.Waldegger, S., K. Klingel, P. Barth, M. Sauter, M. L. Rfer, R. Kandolf, and F. Lang. 1999. h-sgk serine-threonine protein kinase gene as transcriptional target of transforming growth factor beta in human intestine. Gastroenterology 116:1081-1088. [DOI] [PubMed] [Google Scholar]
- 96.Wang, G., and A. J. Berk. 2002. In vivo association of adenovirus large E1A protein with the human mediator complex in adenovirus-infected and-transformed cells. J. Virol. 76:9186-9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yew, P. R., X. Liu, and A. J. Berk. 1994. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 8:190-202. [DOI] [PubMed] [Google Scholar]